Abstract
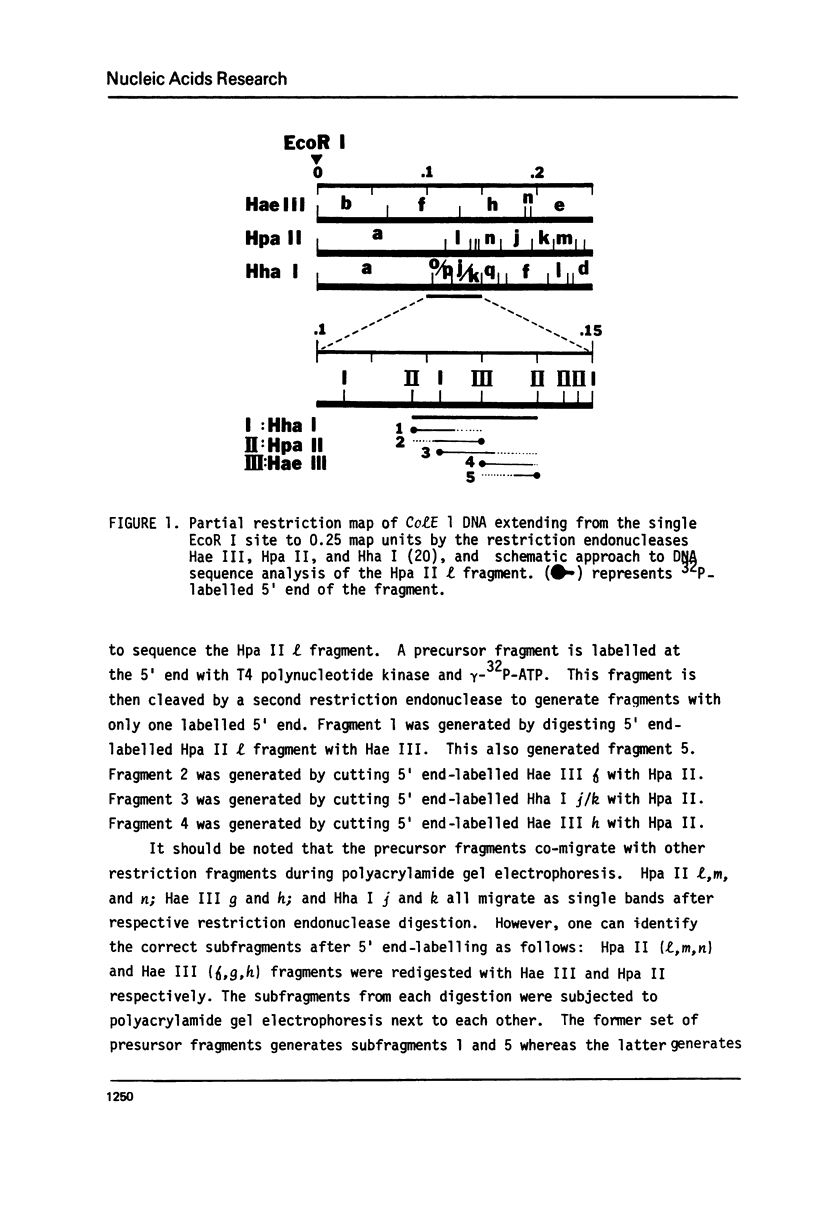
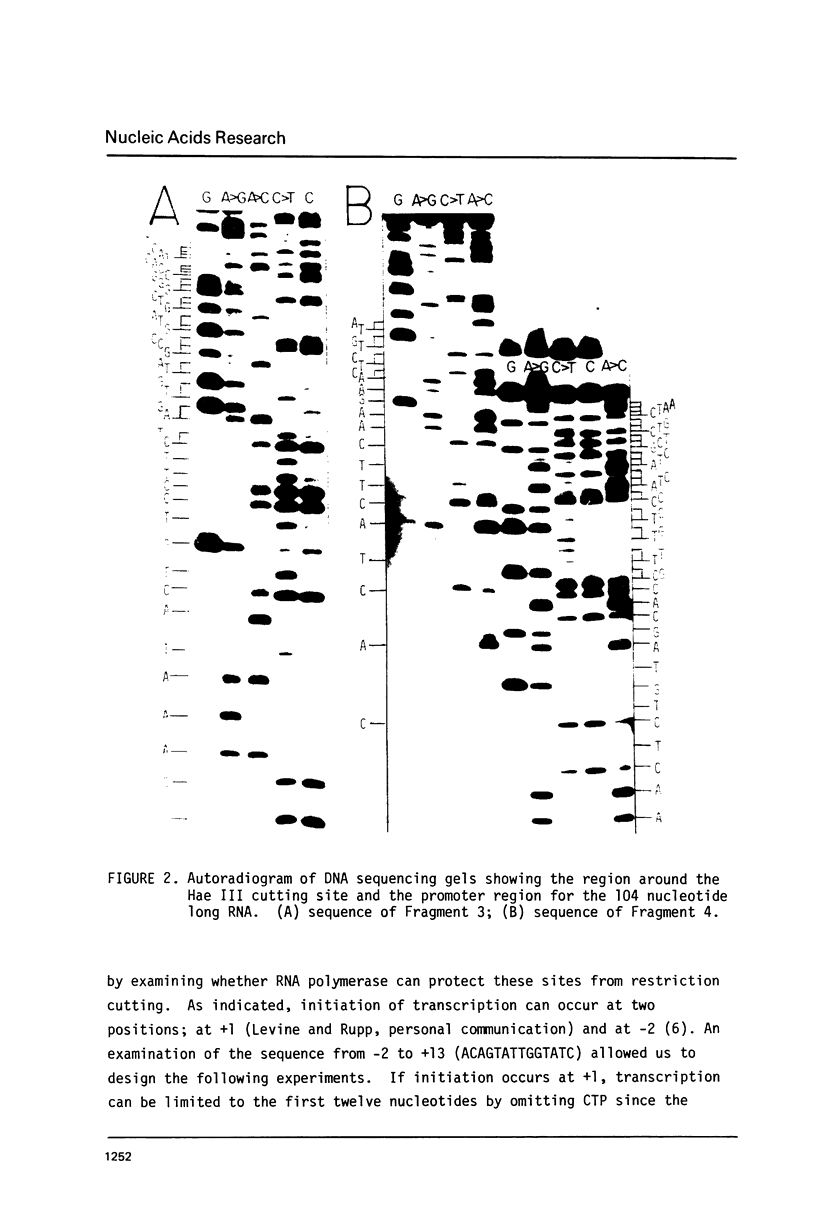
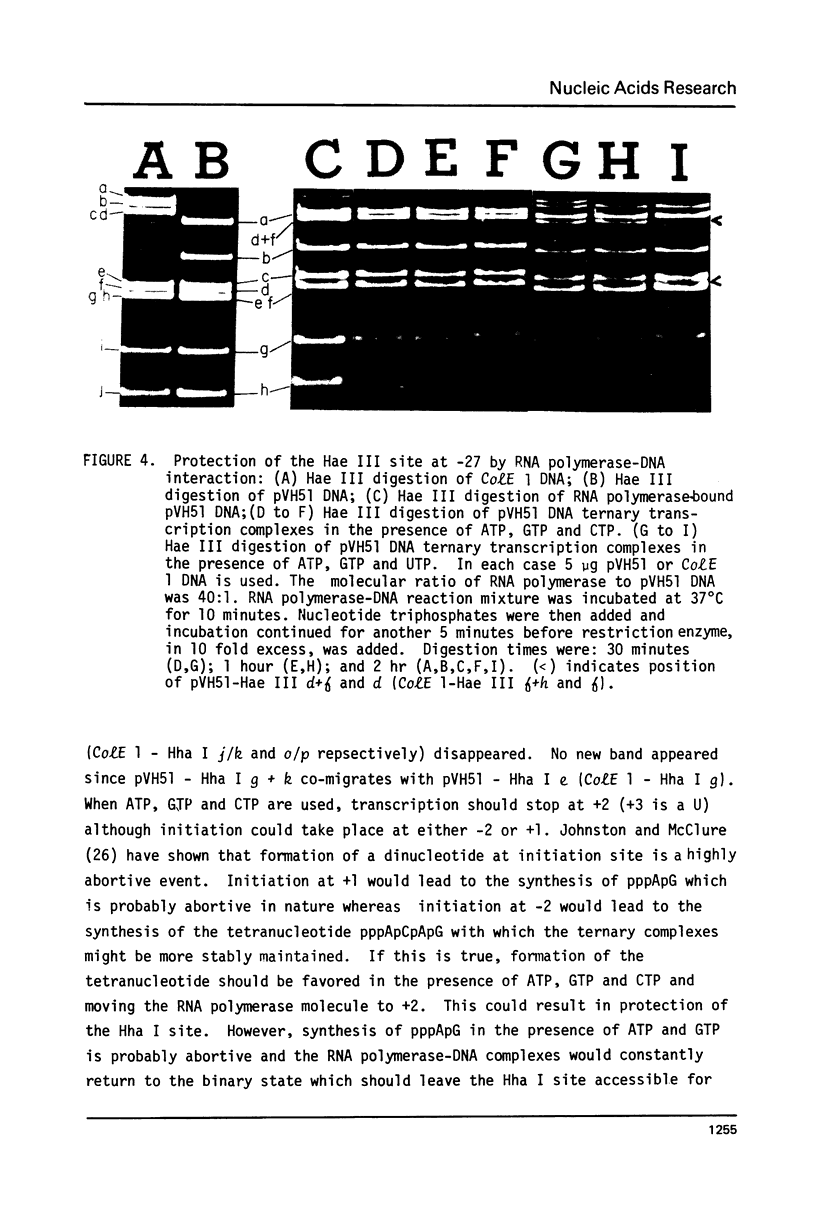
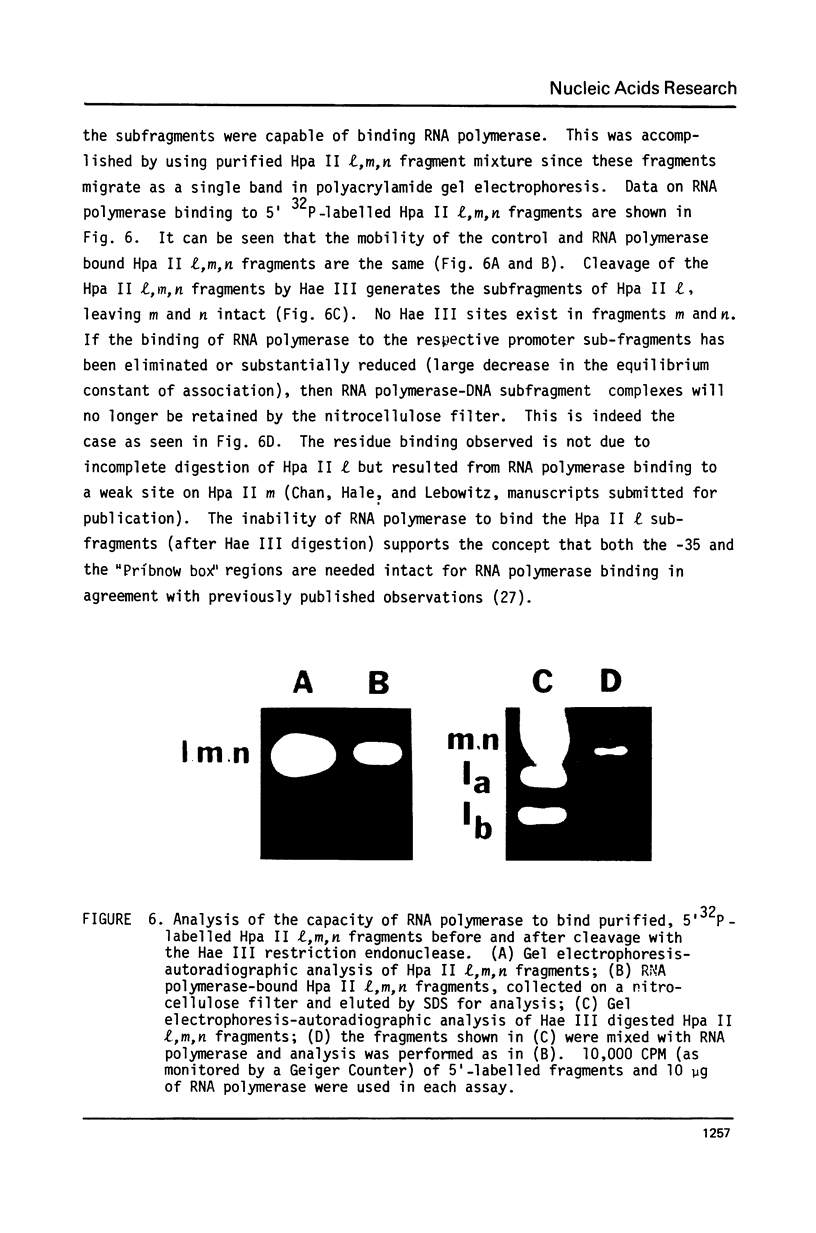
This paper presents the location and nucleotide sequence of a strong promoter of ColE 1. This promoter is of interest because of its greatly enhanced activity in the supercoiled state of the plasmid DNA (3) and its possible role in the maintenance of the plasmid replicon (4). This strong promoter is located at the restriction endonuclease Hae III f-h site 0.13 map units from the single EcoR 1 site proximal to the origin of DNA replication. The nucleotide sequence of the Hpa II l fragment of ColE 1 which contains this promoter has been determined. Initiation of transcription at this promoter occurred at two positions. Limited transcription by omitting one of the four nucleotide triphosphates allowed transcription to proceed to the fourth (-UTP) and to the twelfth (-CTP) nucleotides respectively. This was used to probe the interaction between RNA polymerase and the ColE 10.13 promoter by means of restriction cutting at the Hae III site at =27 and the Hha I site at +17. RNA polymerase binding alone blocks restriction cutting at the HAE III site but not at the Hha I site. Limited transcrption to the fourth nucleotide resulted in blocking at both sites. Transcription to the twelfth nucleotide resulted in partial cutting at the Hae III site and blocking at the Hha I site.
Full text
PDF










Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bastia D. Determination of restriction sites and the nucleotide sequence surrounding the relaxation site of ColE1. J Mol Biol. 1978 Oct 5;124(4):601–639. doi: 10.1016/0022-2836(78)90174-2. [DOI] [PubMed] [Google Scholar]
- Benham C. J. Torsional stress and local denaturation in supercoiled DNA. Proc Natl Acad Sci U S A. 1979 Aug;76(8):3870–3874. doi: 10.1073/pnas.76.8.3870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bird R. E., Tomizawa J. Ribonucleotide-deoxyribonucleotide linkages at the origin of DNA replication of colicin E1 plasmid. J Mol Biol. 1978 Mar 25;120(1):137–143. doi: 10.1016/0022-2836(78)90300-5. [DOI] [PubMed] [Google Scholar]
- Burgess R. R., Jendrisak J. J. A procedure for the rapid, large-scall purification of Escherichia coli DNA-dependent RNA polymerase involving Polymin P precipitation and DNA-cellulose chromatography. Biochemistry. 1975 Oct 21;14(21):4634–4638. doi: 10.1021/bi00692a011. [DOI] [PubMed] [Google Scholar]
- CLARK A. J., MARGULIES A. D. ISOLATION AND CHARACTERIZATION OF RECOMBINATION-DEFICIENT MUTANTS OF ESCHERICHIA COLI K12. Proc Natl Acad Sci U S A. 1965 Feb;53:451–459. doi: 10.1073/pnas.53.2.451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clewell D. B. Nature of Col E 1 plasmid replication in Escherichia coli in the presence of the chloramphenicol. J Bacteriol. 1972 May;110(2):667–676. doi: 10.1128/jb.110.2.667-676.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez N., Wiggs J., Chamberlin M. J. A simple procedure for resolution of Escherichia coli RNA polymerase holoenzyme from core polymerase. Arch Biochem Biophys. 1977 Aug;182(2):404–408. doi: 10.1016/0003-9861(77)90521-5. [DOI] [PubMed] [Google Scholar]
- Hardies S. C., Patient R. K., Klein R. D., Ho F., Reznikoff W. S., Wells R. D. Construction and mapping of recombinant plasmids used for the preparation of DNA fragments containing the Escherichia coli lactose operator and promoter. J Biol Chem. 1979 Jun 25;254(12):5527–5534. [PubMed] [Google Scholar]
- Hashimoto-Gotoh T., Inselburg J. ColE1 plasmid incompatibility: localization and analysis of mutations affecting incompatibility. J Bacteriol. 1979 Aug;139(2):608–619. doi: 10.1128/jb.139.2.608-619.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hershfield V., Boyer H. W., Chow L., Helinski D. R. Characterization of a mini-ColC1 plasmid. J Bacteriol. 1976 Apr;126(1):447–453. doi: 10.1128/jb.126.1.447-453.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnsrud L. Contacts between Escherichia coli RNA polymerase and a lac operon promoter. Proc Natl Acad Sci U S A. 1978 Nov;75(11):5314–5318. doi: 10.1073/pnas.75.11.5314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morita M., Oka A. The structure of a transcriptional unit on colicin E1 plasmid. Eur J Biochem. 1979 Jul;97(2):435–443. doi: 10.1111/j.1432-1033.1979.tb13131.x. [DOI] [PubMed] [Google Scholar]
- Müller W., Weber H., Meyer F., Weissmann C. Site-directed mutagenesis in DNA: generation of point mutations in cloned beta globin complementary dna at the positions corresponding to amino acids 121 to 123. J Mol Biol. 1978 Sep 15;124(2):343–358. doi: 10.1016/0022-2836(78)90303-0. [DOI] [PubMed] [Google Scholar]
- Oka A. Fine cleavage map of a small colicin E1 plasmid carrying genes responsible for replication and colicin E1 immunity. J Bacteriol. 1978 Feb;133(2):916–924. doi: 10.1128/jb.133.2.916-924.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oka A., Nomura N., Morita M., Sugisaki H., Sugimoto K., Takanami M. Nucleotide sequence of small ColE1 derivatives: structure of the regions essential for autonomous replication and colicin E1 immunity. Mol Gen Genet. 1979 May 4;172(2):151–159. doi: 10.1007/BF00268276. [DOI] [PubMed] [Google Scholar]
- Pribnow D. Bacteriophage T7 early promoters: nucleotide sequences of two RNA polymerase binding sites. J Mol Biol. 1975 Dec 15;99(3):419–443. doi: 10.1016/s0022-2836(75)80136-7. [DOI] [PubMed] [Google Scholar]
- Roberts R. J., Breitmeyer J. B., Tabachnik N. F., Myers P. A. A second specific endonuclease from Haemophilus aegyptius. J Mol Biol. 1975 Jan 5;91(1):121–123. doi: 10.1016/0022-2836(75)90375-7. [DOI] [PubMed] [Google Scholar]
- Roberts R. J., Myers P. A., Morrison A., Murray K. A specific endonuclease from Haemophilus haemolyticus. J Mol Biol. 1976 May 5;103(1):199–208. doi: 10.1016/0022-2836(76)90060-7. [DOI] [PubMed] [Google Scholar]
- Schaller H., Gray C., Herrmann K. Nucleotide sequence of an RNA polymerase binding site from the DNA of bacteriophage fd. Proc Natl Acad Sci U S A. 1975 Feb;72(2):737–741. doi: 10.1073/pnas.72.2.737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scherer G. E., Walkinshaw M. D., Arnott S. A computer aided oligonucleotide analysis provides a model sequence for RNA polymerase-promoter recognition in E.coli. Nucleic Acids Res. 1978 Oct;5(10):3759–3773. doi: 10.1093/nar/5.10.3759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmitz A., Galas D. J. The interaction of RNA polymerase and lac repressor with the lac control region. Nucleic Acids Res. 1979 Jan;6(1):111–137. doi: 10.1093/nar/6.1.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharp P. A., Sugden B., Sambrook J. Detection of two restriction endonuclease activities in Haemophilus parainfluenzae using analytical agarose--ethidium bromide electrophoresis. Biochemistry. 1973 Jul 31;12(16):3055–3063. doi: 10.1021/bi00740a018. [DOI] [PubMed] [Google Scholar]
- Siebenlist U. Nucleotide sequence of the three major early promoters of bacteriophage T7. Nucleic Acids Res. 1979;6(5):1895–1907. doi: 10.1093/nar/6.5.1895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simpson R. B. Contacts between Escherichia coli RNA polymerase and thymines in the lac UV5 promoter. Proc Natl Acad Sci U S A. 1979 Jul;76(7):3233–3237. doi: 10.1073/pnas.76.7.3233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang H. L., Heller K., Gellert M., Zubay G. Differential sensitivity of gene expression in vitro to inhibitors of DNA gyrase. Proc Natl Acad Sci U S A. 1979 Jul;76(7):3304–3308. doi: 10.1073/pnas.76.7.3304. [DOI] [PMC free article] [PubMed] [Google Scholar]






