Abstract
How mechanochemical signals induced by the amount of weight borne by the skeletal musculature are translated into modifications to muscle sarcomeres is poorly understood. Our laboratory recently demonstrated that, in response to experimentally induced increases in the weight load borne by a rat, alternative splicing of the fast skeletal muscle troponin T (Tnnt3) pre-mRNA in gastrocnemius was adjusted in a correlated fashion with the amount of added weight. (Schilder RJ, Kimball SR, Marden JH, Jefferson LS. J Exp Biol 214: 1523–1532, 2011). Thus muscle load is perceived quantitatively by the body, and mechanisms that sense it appear to control processes that generate muscle sarcomere composition plasticity, such as alternative pre-mRNA splicing. Here we demonstrate how mechanical stretch (see earlier comment) of C2C12 muscle cells in culture results in changes to Tnnt3 pre-mRNA alternative splicing that are qualitatively similar to those observed in response to added weight in rats. Moreover, inhibition of Akt signaling, but not that of ERK1/2, prevents the stretch-induced effect on Tnnt3 pre-mRNA alternative splicing. These findings suggest that effects of muscle load on Tnnt3 pre-mRNA alternative splicing are controlled by a cell-autonomous mechanism, rather than systemically. They also indicate that, in addition to its regulatory role in protein synthesis and muscle mass plasticity, Akt signaling may regulate muscle sarcomere composition by modulating alternative splicing events in response to load. Manipulation of Tnnt3 pre-mRNA alternative splicing by mechanical stretch of cells in culture provides a model to investigate the biology of weight sensing by skeletal muscles and facilitates identification of mechanisms through which skeletal muscles match their performance and experienced load.
Keywords: skeletal muscle, mechanotransduction, cyclic stretch, C2C12 myotube
a key prerequisite for the efficient movement and overall function of animals is the ability of skeletal muscles to support body weight. Skeletal muscle is a highly plastic tissue, but how it perceives quantitative information regarding a variable load and responds appropriately to maintain a balance between performance and load is a poorly understood and complex issue (6, 19, 20, 24, 33). The role of mechanical signals (or lack thereof) in muscle homeostasis has been examined mainly with regard to their effects on muscle mass and protein synthesis (9, 18, 42). However, muscle mass-specific force output is predicted to decrease with increasing muscle size (2, 41), and increasing muscle mass alone, therefore, would not suffice as body weight support requirements increase. The ability of skeletal muscle to vary performance by altering the protein composition, in concert with overall muscle size changes, has been studied to a lesser extent in this regard (but see, e.g., Refs. 5, 11, 35), but is a potent candidate mechanism to circumvent this geometric constraint.
Our laboratory recently reported that alternative splicing of the pre-mRNA for skeletal muscle fast troponin T (Tnnt3) was rapidly altered in response to a 5-day period of an externally imposed increase in muscle loading by means of a weighted vest (41). Before that study, similar results were obtained in insects (28). These findings provide a novel molecular marker for how muscles sense and make quantitative adjustments that correlate with an applied load, as the Tnnt3 pre-mRNA splicing response correlated quantitatively with weight load per se. Moreover, obese rats showed an impairment of this response, suggesting that such individuals fail to make appropriate adjustments to skeletal muscle composition as their body weight increases (41). Resolving whether the quantitative adjustments to Tnnt3 pre-mRNA alternative splicing in response to muscle loading observed in vivo are regulated by a systemic or cell-autonomous mechanism would provide important insights into the general biology of body weight sensing and impaired muscle function in obesity (e.g., Refs. 20, 23, 25).
Alternative splicing of muscle sarcomere genes is a mechanism that allows muscle to be highly plastic in terms of the amino acid composition of sarcomere proteins, while maintaining overall stoichiometry. Most genes (e.g., 90% of human genes; Ref. 44) are alternatively spliced, but the functional consequences of quantitative variation in mRNA splice form expression have been studied for only a few genes (21, 30). Mammalian skeletal muscle troponin T is encoded by a slow (Tnnt1) and fast (Tnnt3) gene (38), the latter of which undergoes extensive alternative splicing. Tnnt3 pre-mRNA comprises 18 exons, including a cassette of 6 alternatively spliced exons near the 5′ end and a mutually exclusive pair of exons near the 3′ end, allowing a possible 128 unique splice forms. Changes in the relative abundance of different Tnnt3 alternative splice forms is likely to be an important component of muscle plasticity, as Tnnt3 affects the calcium sensitivity of muscle contraction (5, 10, 35, 36). Our laboratory's previous work demonstrated that Tnnt3 splicing adjustments associated with body weight gain consisted of an overall decrease in the relative abundance of Tnnt3 splice forms that include Tnnt3 exon 4 (41). Reduced inclusion of exon 4 is known to correlate with increased muscle fiber calcium sensitivity (4, 40), suggesting that the overall effect of natural and experimental increases in body weight is an increased calcium sensitivity and force output in gastrocnemius and perhaps other weight-bearing muscles (41).
To examine whether a cell-autonomous mechanism regulates this quantitative response in Tnnt3 alternative splicing, we tested the hypothesis that Tnnt3 pre-mRNA alternative splicing is affected by mechanical stretch in C2C12 muscle cell cultures. The experiments were designed to employ a substrate deformation paradigm (i.e., a FlexCell culture system) that is expected to approximate what happens to muscle cells in vivo during contractions associated with increased loading (6). A cell culture model is also more conducive to identifying signaling pathways that may translate mechanical signals into alternative splicing events. Interestingly, a growing body of literature suggests that many alternative splicing events require the activity of signaling pathways that are mainly known for their ability to regulate skeletal muscle protein synthesis and growth in response to variable mechanical loading (27). The protein kinases Akt (also known as PKB) and ERK1/2 have been repeatedly shown to be activated in similar in vitro model systems (1, 16, 17) and to modulate the phosphorylation of mRNA binding splice factors that influence decisions regarding exon inclusion/exclusion (3, 32, 43). Therefore, we also tested the hypothesis that Akt and ERK1/2 kinases regulate Tnnt3 pre-mRNA alternative splicing in response to mechanical stretch.
MATERIALS AND METHODS
Materials and reagents.
High- (4.5 g/l) and low-glucose (1 g/l) Dulbecco's modified Eagle's medium (DMEM) and Pen Strep (P/S; 5,000 U/ml penicillin, 5 mg/ml streptomycin) were purchased from Invitrogen (Carlsbad, CA). Fetal bovine serum (FBS) was purchased from Atlas Biologicals (Fort Collins, CO), and horse serum was purchased from Sigma-Aldrich (St. Louis, MO). Akt1/2-kinase inhibitor (KI) and MEK inhibitor U0126 were purchased from Sigma-Aldrich and Promega (Madison, WI), respectively. The following antibodies were purchased from Cell Signaling Technology (Danvers, MA): Akt, phospho-Akt (Ser473) XP, p44/42 MAPK (ERK1/2), phospho-p44/42 MAPK (Thr202/Tyr204) (pErk1/2). Polyvinylidene difluoride membrane used in Western blotting was purchased from Pall (Ann Harbor, MI), and enhanced chemiluminescence (i.e., Pierce ECL and Amersham ECL Plus) reagents were purchased from Thermo Fisher Scientific (Rockford, IL) and GE Healthcare (Piscataway, NJ), respectively.
Cell culture and mechanical stretch conditions.
C2C12 myoblasts were purchased from ATCC (Manassas, VA). The cells were maintained in high-glucose (i.e., 4.5 g/l) DMEM with 10% FBS and 1% P/S under standard cell culture conditions (i.e., 5% CO2, 95% O2, temperature = 37°C). They were seeded at 100,000/well in type I collagen-coated, BioFlex flexible-bottomed, six-well culture dishes (Flexcell International, Hillsborough, NC) in “medium-glucose” (i.e., 2.75 g/l) DMEM with 10% FBS and 1% P/S. The cells were then allowed to proliferate to ∼80% confluency (∼48 h postseeding), and the medium was replaced with low-glucose DMEM with 10% FBS and 1% P/S at 24 h postseeding. Myoblasts were induced to differentiate to myotubes in low-glucose DMEM, with 2% horse serum and 1% P/S for 6 days. The medium was refreshed every 48 h and was changed to low-glucose (i.e., 1 g/l), serum-free DMEM with 1% P/S 12 h before the onset of cyclic stretch. For the Akt and ERK1/2 inhibition studies, a final medium replacement with low-glucose, serum-free DMEM with 1% P/S containing 0.4 μM Akt-KI or 10 μM U0126, respectively, was performed 2 h before the onset of mechanical stretch. Cells were kept in medium containing the inhibitors throughout the 24-h experiment.
The BioFlex culture plates were placed on the Flexcell FX-5000 Tension system (Flexcell International, Hillsborough, NC) 2 h before the onset of the actual experiments. The Flexcell FX-5000 Tension system provides equibiaxial, cyclic strain by means of vacuum-controlled stretching of the flexible rubber bottom of BioFlex culture plates around cylindrical loading posts. The loading posts are kept in an incubator under standard cell culture conditions (37°C, 5% CO2, 95% O2). Mechanical stretch was applied by means of a sinusoidal (0.5 Hz), 15% strain wave for 24 h. Cells were harvested at 0.5, 1, 2, 4, 8, and 24 h, except for the Akt and ERK1/2 inhibition experiments, in which case cells were harvested at 8 and 24 h only. Control cells were maintained in similar fashion on type I collagen-coated BioFlex culture plates, but did not receive mechanical stretch.
Immunoblotting.
Myotubes were harvested in lysis buffer [20 mM HEPES, 2 mM EGTA, 50 mM sodium fluoride, 100 mM KCl, 0.2 mM EDTA (disodium salt), 50 mM β-glycerophosphate, pH 7.4] containing protease inhibitors [1 mM DTT, 1 mM benzamidine, 0.5 mM sodium vanadate, and 10 μl/ml protease inhibitor cocktail (Sigma no. P8340)]. The cells were lysed for 20 min at 4°C on a rocking platform. The lysate was then first split into two fractions, one of which was used directly for extraction of total RNA (see below). The remaining fraction was centrifuged for 3 min, at 4°C, and 1,000 g. The supernatant was diluted with 2× sample buffer (0.125 M Tris base, pH 6.8, 25% glycerol, 2.5% SDS, 0.25% β-mercaptoethanol, 5% bromophenol blue) and boiled for 5 min at 95°C. Subsequently, 20 μl of the samples were separated by means of SDS-polyacrylamide gel electrophoresis on 4–15% precast polyacrylamide gels (BioRad, Hercules, CA). After electrophoresis, proteins were transferred onto a polyvinylidene difluoride membrane, and the membrane was blocked in Tris-buffered saline containing 0.1% Tween 20 (TBST) containing 5% nonfat dry milk for 1 h at room temperature. After two 10-min washes in TBST, incubation with primary antibody (in TBST) was performed overnight at 4°C, followed by incubation with secondary antibody (in TBST containing 5% nonfat dry milk) for 1 h at room temperature. After four 5-min washes with TBST, Western blots were developed using either ECL or ECL Plus reagents. Chemiluminescent imaging was performed using a GeneGnome HR (Syngene, Frederick, MD) system, and integrated optical densities of bands were obtained using GeneTools (Syngene, Frederick, MD) software.
Characterization and quantification of Tnnt3 pre-mRNA splice form relative abundance.
Total RNA was extracted from cell lysates using Trizol reagent (Invitrogen) and precipitated in isopropanol, according to the manufacturer's instructions. The RNA and nontemplate (i.e., H2O instead of RNA) samples were reverse transcribed using a High Capacity cDNA Reverse Transcription kit (Applied Biosystems). Tnnt3 amplicons were amplified by PCR using fluorescein (FAM)-labeled forward primer fTnt_F1 (5′-FAM-CCCCCAACCTTCTCAGACT-3′), and two unlabeled reverse primers, fTnt_R2 (5′-CCTTCTTGCTGTGCTTCTGG-3′) and fTnt_R4 (5′-CGGACAGTCATGATATCGTATTT-3′), as previously described (35). PCR was performed using HotStart GoTaq polymerase (Promega) under the following cycling conditions: 2 min at 95°C, followed by 4 cycles of 30 s at 94°C, 30 s at 65°C (−1.0°C/cycle), followed by 1 min and 15 s at 72°C. This was followed by 33 cycles of 30 s at 94°C, 30 s at 60°C, 1 min and 15 s at 72°C, and ending with a final 15 min at 72°C. Negative control samples were included in this analysis for each experiment.
To quantify effects of mechanical stretch on Tnnt3 alternative splicing, irrespective of overall gene expression patterns (see Fig. 3A), we determined quantitative changes within the Tnnt3 splice form mixture for each sample. Thus, FAM-labeled PCR products were diluted 1:10 and 1 μl was analyzed by capillary electrophoresis (ABI Hitachi 3730XL DNA Analyzer; Applied Biosystems), which allows precise determination of PCR fragment size and quantity in a PCR amplicon pool. Samples with a fragment peak height exceeding the linear detection range of the instrument were further diluted and run again. The relative abundance of each Tnnt3 amplicon in the PCR reaction was determined by dividing its peak height by the total of all Tnnt3 amplicon peak heights (see also Fig. 1). The use of this relative abundance measure is a method of normalizing for different starting amounts of template. For our purposes, therefore, no further normalizing method is necessary. Amplicon fragment size was determined using the GS1200 LIZ internal size standard and Genemapper (Applied Biosystems, Carlsbad, CA) fragment analysis software.
Fig. 3.
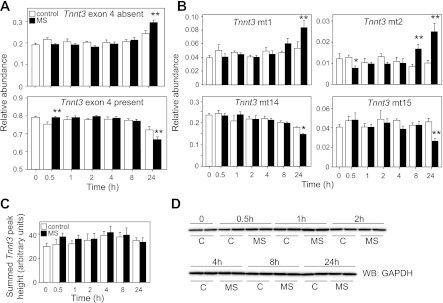
MS induces changes in alternative splicing of Tnnt3 pre-mRNA. A: mean summed relative abundances of Tnnt3 splice forms missing (top) or including (bottom) exon 4, as a function of the duration of MS. B: mean relative abundance of individual Tnnt3 splice forms (Tnnt3 mt1-2 and mt14-15) affected by MS. Tnnt3 mt4 was also increased by MS (Table 1), but not displayed here. Values are means ± SE [N = 8 per treatment for each time point, obtained from four (N = 2 per treatment and time point) independent experiments]. Statistically significant effects of MS are shown at *α = 0.05 or **α = 0.01, as determined by two-way ANOVA, followed by least squares means contrasts Student's t-tests at a given time point. MS did not affect overall Tnnt3 mRNA levels (C) or protein levels, as indicated by GAPDH expression (D) across the experimental time course.
Fig. 1.
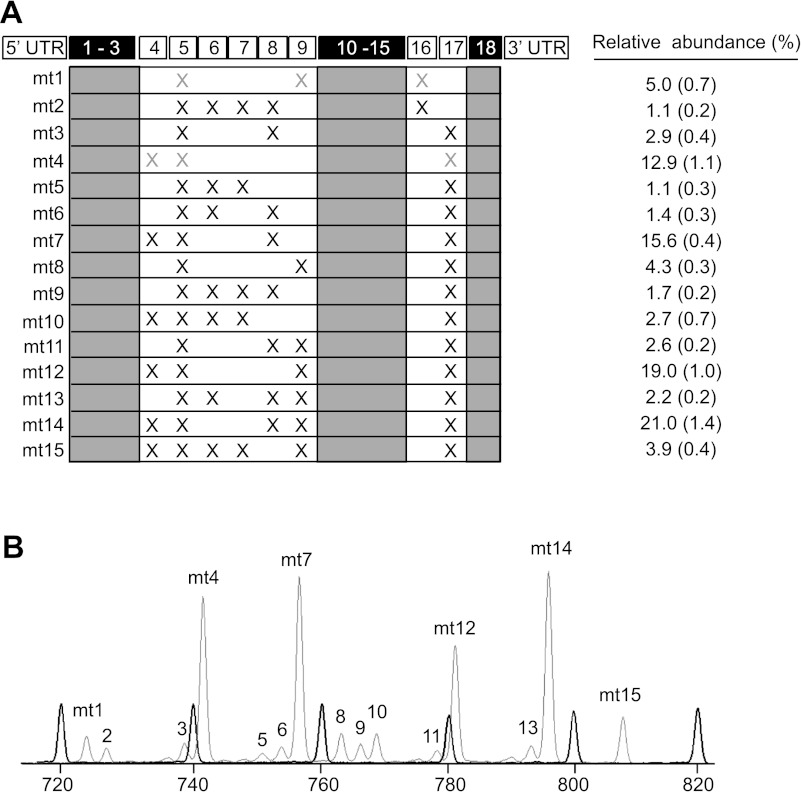
Characterization of C2C12 myotube skeletal muscle fast troponin T (Tnnt3) splice forms. A: mouse Tnnt3 pre-mRNA contains 18 exons. While the 5′-end alternative exon cassette (i.e., exons 4–9), combined with mutually exclusive 3′-end exons 16 and 17, can give rise to potentially 128 different splice forms, only 15 Tnnt3 splice forms (labeled mt1-mt15) were detected. Tnnt3 splice forms for which exon composition is represented by gray X's have not yet been verified by cloning and sequencing. The relative abundance of each Tnnt3 splice form at day 7 of differentiation is indicated on the right [values are means (SE)]. B: fluorescently labeled Tnnt3 PCR amplicons from C2C12 myotubes separated by means of capillary electrophoresis (gray trace). Intensity (height) of peaks is proportional to the mRNA splice form abundance in the original RNA sample. Internal size standards are represented by the black trace. UTR, untranslated region.
To examine Tnnt3 splice form exon composition, amplicons were extracted from an agarose gel using a QiaQuick gel extraction kit (Qiagen), cloned using a TOPO-TA cloning kit (Invitrogen), and sequenced (ABI Hitachi 3730XL DNA Analyzer).
Data analysis.
All Tnnt3 splice form relative abundance data were arcsine transformed to meet normality assumptions of the statistical tests applied. Untransformed data means are presented in Figs. 1–6 and Tables 1 and 2. Specific statistical tests applied to the data are indicated in the text, or figure and table legends. All statistical analyses were performed using JMP software (The SAS Institute).
Fig. 2.
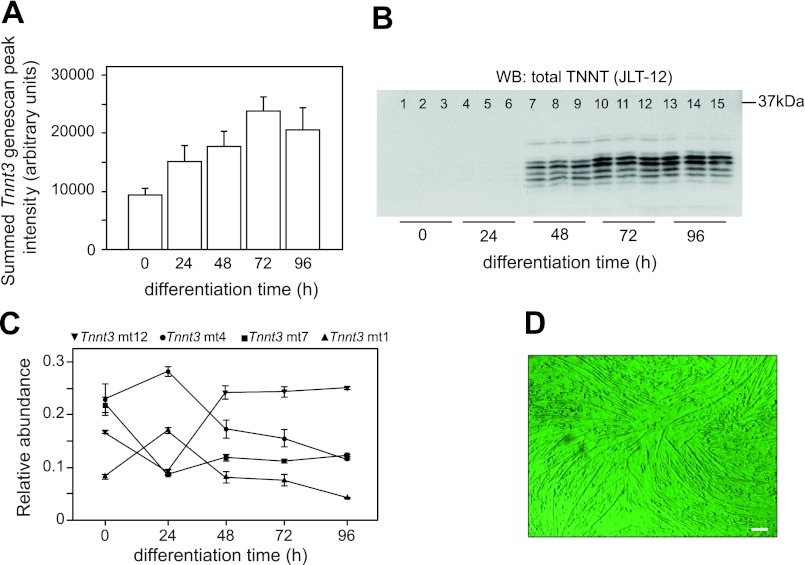
C2C12 myotube Tnnt3 expression during early differentiation. A: while overall Tnnt3 mRNA levels increase steadily over a 96-h period, significant troponin T protein expression did not occur until 48 h of differentiation. Note that total Tnnt3 expression was obtained by summing the peak heights (determined by fragment analysis, see materials and methods) of all 15 Tnnt3 splice forms and is thus not normalized using some housekeeping gene's expression levels. B: Western blotting (WB) using a total troponin T antibody (JLT12; Sigma) was performed on samples from one of these experiments only, with equal amounts of total protein loaded per lane. Note that JLT12 also recognizes slow and cardiac troponin T. C: importantly, large shifts in the mean relative abundance of Tnnt3 splice forms were observed during the first 48 h of myotube differentiation. For clarity, the relative abundance of only 4 out of 15 Tnnt3 splice forms is displayed as a function of differentiation time. This stimulated us to use day 7 myotubes in the analyses of effects of mechanical stretch (MS) instead of younger myotubes. Values in A and C are means ± SE (N = 6 obtained from two independent experiments using 3 replicates per treatment). D: micrograph of day 7 myotubes showing mature, multinucleated C2C12 myotubes just before the onset of MS experiments (scale bar = 100 μm).
Fig. 4.
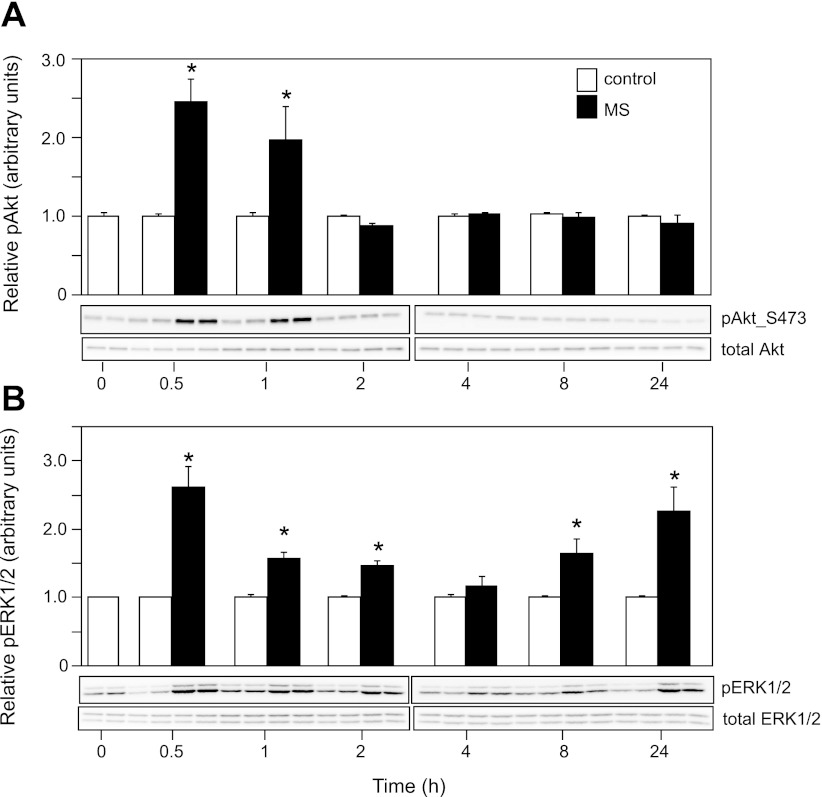
MS activates Akt and ERK1/2 protein kinases. Western blot analysis showing increased mean phosphorylation (p) levels of Akt (A) and ERK1/2 (B) kinases in C2C12 myotubes in response to MS. Representative blots (duplicates for each treatment) are presented below the quantitative results. In contrast to ERK1/2, for which phosphorylation was increased throughout the experiment, Akt phosphorylation was acute and preceded the effects on Tnnt3 pre-mRNA alternative splicing. Means of samples from MS cells are presented relative to means in C cells, with the latter set to 1.0. Values are means ± SE (N = 4 per treatment for each time point, obtained from two independent experiments). *Statistically significant differences at α = 0.05, as determined by Student's t-tests at each time point.
Fig. 5.
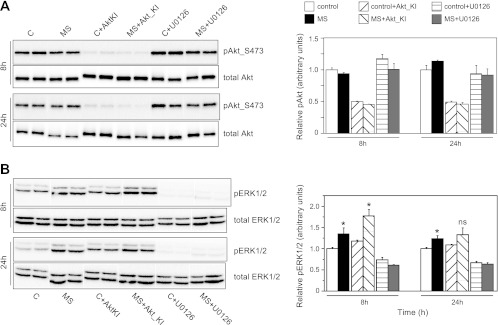
Effects of MS in the presence of pharmacological inhibitors of Akt (A) or ERK1/2 (B) phosphorylation. Western blot analyses are shown of phosphorylation levels of Akt and ERK1/2 in MS and C cells, in the presence or absence of inhibitors Akt-kinase inhibitor (KI) or U0126 (see materials and methods), at 8 and 24 h of treatment. Representative blots (duplicates per treatment) are displayed on the left, while quantification of relative phosphorylation (with respect to total Akt and ERK1/2 levels, respectively) are displayed on the right. In both cases, the effects of MS on Akt and ERK1/2 phosphorylation displayed in Fig. 4 were completely blocked by the respective inhibitors, while no adverse effects of Akt-KI on ERK1/2 phosphorylation or U0126 on Akt phosphorylation were observed. Values are means ± SE (N = 6 per treatment for each time point; two independent experiments were performed with N = 3 per treatment and time point each). *Significant differences between C and MS cells for each treatment at α = 0.05, as determined by Student's t-tests. ns, Nonsignificant.
Fig. 6.
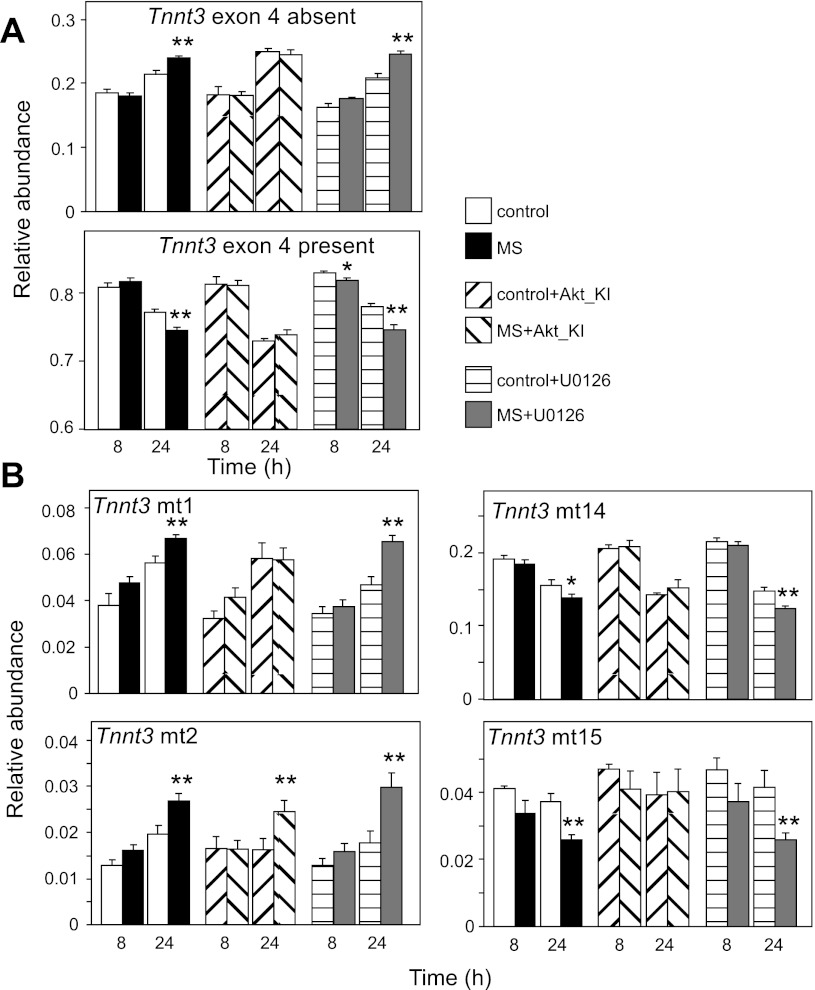
Akt-KI inhibits the effect of MS on alternative splicing of Tnnt3 pre-mRNA. A: mean summed relative abundances of Tnnt3 splice forms missing (top) or including (bottom) exon 4, at 8 and 24 h of MS, in the presence or absence of pharmacological inhibitors Akt-KI and U0126. MS caused the same effects on Tnnt3 exon 4 inclusion for C and U0126-treated cells, as displayed in Fig. 3. This effect was inhibited by Akt-KI treatment. B: Akt-KI inhibited the response to MS for Tnnt3 mt1 and mt14-15, but not for Tnnt3 mt2 and mt4 (the latter is not displayed here). Values are means ± SE (N = 6 per treatment for each time point; two independent experiments were performed with N = 3 per treatment and time point each). Statistically significant effects of MS are shown at *α = 0.05 or **α = 0.01, as determined by two-way ANOVA, followed by least squares means contrasts Student's t-tests at a given time point.
Table 1.
Two-way ANOVA table for effects of time, mechanical stretch, and their interaction on Tnnt3 splice form abundances
| Model Effects |
||||
|---|---|---|---|---|
| Tnnt3 Splice Form | Exon 4 | Time | MS | Time × MS |
| mt1 | N | 6.0 (0 < 0.0001) | 0.08 (0.77) | 3.3 (0.0093) |
| mt2 | N | 2.5 (0.036) | 5.76 (0.019) | 7.6 (<0.0001) |
| mt3 | N | 11.8 (<0.0001) | 0.18 (0.67) | 2.6 (0.03)* |
| mt4 | Y | 6.3 (<0.0001) | 8.4 (0.0048) | 3.7 (0.0049) |
| mt5 | N | 6.7 (<0.0001) | 0.09 (0.77) | 0.75 (0.59) |
| mt6 | N | 9.1 (<0.0001) | 0.36 (0.55) | 0.74 (0.60) |
| mt7 | Y | 4.8 (0.0006) | 11.3 (0.0012) | 2.68 (0.027)* |
| mt8 | N | 2.4 (0.043) | 5.3 (0.023) | 3.7 (0.0047)* |
| mt9 | N | 1.8 (0.12) | 2.3 (0.13) | 1.9 (0.10) |
| mt10 | Y | 8.0 (<0.0001) | 0.02 (0.90) | 1.5 (0.19) |
| mt11 | N | 2.6 (0.029) | 0.24 (0.62) | 0.99 (0.42) |
| mt12 | Y | 10.4 (<0.0001) | 2.0 (0.16) | 0.42 (0.83) |
| mt13 | N | 0.20 (0.96) | 1.9 (0.17) | 1.48 (0.21) |
| mt14 | Y | 76.5 (<0.0001) | 0.47 (0.49) | 3.99 (0.048) |
| mt15 | Y | 0.49 (0.79) | 0.02 (0.89) | 2.9 (0.019) |
| Exon 4 absent | N | 23.9 (<0.0001) | 3.53 (0.064) | 3.2 (0.017) |
| Exon 4 present | Y | 16.2 (<0.0001) | 7.54 (0.0074) | 7.4 (<0.0001) |
Significant interactions are indicative of time point-specific effects. The presence (Y) or absence (N) of exon 4 in specific splice forms is indicated in column 2. Model effect values are F-statistics and P values at α = 0.05 [N = 8 per treatment for each time point, obtained from four (N = 2 per treatment and time point) independent experiments].
Although significant overall interaction effects were observed for skeletal muscle fast troponin T (Tnnt3) splice forms mt3, mt7 and mt8, post hoc least squares means Student's t-test revealed that mechanical stretch (MS) caused significant effects at 30 min, but these were not sustained subsequently.
Table 2.
Two-way ANOVA table for effects of time and mechanical stretchon the relative abundance of Tnnt3 splice forms, including/excluding exon 4, and Tnnt3 splice forms affected by mechanical stretch
| Model Effects |
|||
|---|---|---|---|
| Tnnt3 Splice Form | Time | MS | Treatment |
| Exon 4 absent | 112.6 (<0.0001) | 9.2 (0.0057) | Control |
| Exon 4 present | 40.1 (<0.0001) | 23.4 (<0.0001) | |
| mt1 | 37.7 (<0.0001) | 28.5 (<0.0001) | |
| mt2 | 15.4 (0.0006) | 18.9 (0.0002) | |
| mt14 | 43.2 (<0.0001) | 6.5 (0.018) | |
| mt15 | 0.22 (0.65) | 12.7 (0.002) | |
| Exon 4 absent | 68.2 (<0.0001) | 8.7 (0.008) | U0126 |
| Exon 4 present | 41.8 (<0.0001) | 25.2 (<0.0001) | |
| mt1 | 32.7 (<0.0001) | 8.5 (0.008) | |
| mt2 | 13.9 (0.0012) | 9.1 (0.0067) | |
| mt14 | 204.4 (<0.0001) | 8.0 (0.0099) | |
| mt15 | 4.4 (0.049) | 9.8 (0.005) | |
| Exon 4 absent | 42.5 (<0.0001) | 0.14 (0.71) | Akt-KI |
| Exon 4 present | 89.5 (<0.0001) | 2.4 (0.14) | |
| mt1 | 19.4 (0.0002) | 1.22 (0.28) | |
| mt2 | 1.6 (0.21) | 3.4 (0.08)* | |
| mt14 | 64.4 (<0.0001) | 0.2 (0.66) | |
| mt15 | 0.16 (0.69) | 1.0 (0.32) | |
Separate two-way ANOVA analyses were performed for control cells and those exposed to pharmacological inhibitor Akt-kinase inhibitor (KI) or U0126. No significant interaction terms were detected for the period between 8 and 24 h, and interactions were excluded from this analysis. Model effect values are F-statistics and P values at α = 0.05 (N = 6 per treatment for each time point; two independent experiments were performed with N = 3 per treatment and time point each).
While the two-way ANOVA model did not reveal effects of MS across the two time points for Tnnt3 mt2 (in the presence of Akt-KI), the time point specific least squares means Student's t-tests did reveal significant effects of MS on Tnnt3 mt2 (see also Fig. 6B).
RESULTS
Characterization of Tnnt3 splice forms in C2C12 myotubes.
Previous work demonstrated expression of seven Tnnt3 mRNA splice forms in neonatal, and seven in skeletal, muscle of adult mice (45). However, to our knowledge, the identity and quantitative expression patterns of alternative Tnnt3 splice forms in C2C12 myoblasts and myotubes has not been previously determined. Therefore, we first set out to characterize C2C12 Tnnt3 splice form expression by means of PCR using FAM-labeled primers, followed by DNA fragment analysis. For this purpose, C2C12 myotubes were differentiated for 4 days, as described in the materials and methods section, and harvested at the start of differentiation (i.e., time point 0; at this point myoblasts were at 80% confluency), and 24, 48, 72, and 96 h postdifferentiation.
Total RNA was extracted from cell lysates and reverse transcribed, and Tnnt3 splice forms were amplified by PCR. DNA fragment analysis of the fluorescently labeled PCR amplicon pool resulted in the identification of 15 constitutively present (i.e., present in each individual sample, but not in negative control samples) DNA fragment peaks (Fig. 1). Cloning and sequencing of PCR amplicon pools resulted in the identification of 13 of the expected 15 alternative Tnnt3 mRNA splice forms. The predicted exon composition of the remaining two amplicons (i.e., Tnnt3 mt1 and mt4) is indicated in Fig. 1. Two out of the 15 Tnnt3 splice forms contained 3′ end exon 16 (i.e., α-type), while the remaining 13 contained 3′ end exon 17 (i.e., β-type). The discovery of the presence of α-type Tnnt3 splice forms in C2C12 myotubes is noteworthy, since thus far it was found that this type of Tnnt3 splice form only occurred in adult fast skeletal muscle (39). From here onward, these Tnnt3 splice forms are referred to as Tnnt3 mt1-15 (see Fig. 1).
Time course experiments demonstrated that, independently of total Tnnt3 mRNA levels (Fig. 2A) and total troponin T protein expression (Fig. 2B), the relative abundance of Tnnt3 splice forms varied significantly during the first 2 days of C2C12 myotube differentiation (Fig. 2C). After 48 h, a more stable expression pattern was apparent. To avoid obscuring any cyclic stretch-related changes by these large, early differentiation shifts in Tnnt3 splice form abundances, C2C12 myoblasts were allowed to differentiate for 6 days before exposing them to mechanical stretch on day 7 of differentiation. To take into account any remaining baseline (i.e., treatment independent) variation in Tnnt3 splice form relative abundances, we sampled nonstretched control myotubes at all time points analyzed.
Effects of mechanical stretch on alternative splicing of Tnnt3 pre-mRNA.
There is a lack of information regarding which specific Tnnt3 mRNA splice forms are translated into protein isoforms and may subsequently act to modulate skeletal muscle performance. This disconnect makes it difficult to assess the significance of any treatment-associated increase or decrease in the relative abundance of specific Tnnt3 splice forms. In addition, the inherent correlative nature of relative abundances prohibits us from identifying which specific Tnnt3 splice forms are directly up- or downregulated by treatments. However, previous work has demonstrated that inclusion of Tnnt3 exon 4 in general modulates the muscle calcium sensitivity (4) and, therefore, the force output of muscle. Rather than attempt to interpret the results for all 15 Tnnt3 splice forms individually, we have simplified the analyses by focusing on effects of mechanical stretch on overall Tnnt3 exon 4 inclusion.
As shown in Fig. 3A, mechanical stretch significantly decreased overall inclusion of Tnnt3 exon 4 in C2C12 myotubes at 24 h of treatment. This effect was driven by increased levels of Tnnt3 mt1 and mt2, which lack exon 4, and decreased levels of Tnnt3 mt14 and mt15, which contain exon 4 (Fig. 3B, Table 1). Notably, overall exon 4 inclusion decreased, despite a significant increase in the relative abundance of Tnnt3 mt4, which does contain exon 4. Mechanical stretch had no effect on the relative abundance of 7 out of 15 splice forms (Tnnt3 mt5-6, mt9-13; Table 1), including one of the most abundant splice forms, Tnnt3 mt12. Considerable baseline (i.e., in the absence of mechanical stretch) variation in the relative abundance of 12 out of 15 Tnnt3 splice forms was also detected (i.e., note the significant effects of variable “Time” in Table 1). Finally, acute (i.e., after 30 min) changes in the relative abundance of Tnnt3 mt3-4 and mt7-8 were detected (indicated by an asterisk in Table 1), but these were not sustained at subsequent time points. These acute changes are responsible for the increased levels of Tnnt3 splice forms containing exon 4 observed at the 30-min time point in Fig. 3A. Effects of mechanical stretch on Tnnt3 alternative splicing were not associated with changes in total Tnnt3 mRNA levels (Fig. 3C) or apparent cell numbers or protein content, as determined by Western blotting of GAPDH levels (Fig. 3D).
Effect of inhibition of Akt and ERK signaling on the Tnnt3 alternative splicing response to mechanical stretch.
To examine the possible roles of Akt and ERK1/2 signaling in the regulation of Tnnt3 splicing, we first examined how the mechanical stretch paradigm affected Akt and ERK1/2 phosphorylation. Phosphorylation of both kinases increase rapidly (i.e., within 30 min; Fig. 4) in response to mechanical stretch. While Akt phosphorylation decreased to control levels after 2 h, ERK1/2 phosphorylation followed a biphasic pattern, with a second significant activation event occurring at 8 h and maintained at 24 h. Similar rapid onset activation patterns for Akt and ERK1/2 signaling were previously reported for these kinds of mechanical stretch paradigms (1, 16), and our findings provide further evidence for the role of these protein kinases (and associated signaling pathways) in translating mechanical stimuli to intracellular functional events.
We next performed experiments in which C2C12 myotubes were mechanically stretched in the presence of Akt-KI, a specific inhibitor of Akt phosphorylation, or the MEK signaling inhibitor U0126. In these experiments, myotubes were harvested only at 8 and 24 h of stretch, as these were the time points at which most significant mechanical stretch-induced effects on Tnnt3 splicing were observed. In confirmation of the results depicted in Fig. 4, no effect of mechanical stretch on Akt phosphorylation was observed at either 8 or 24 h (Fig. 5A). In contrast, addition of Akt-KI to the medium significantly blunted Akt phosphorylation at 8 and 24 h in both control and mechanically stretched cells, whereas addition of U0126 did not affect basal levels of Akt phosphorylation. Addition of U0126 significantly blunted ERK1/2 phosphorylation at 8 and 24 h in both control and mechanically stretched cells (Fig. 5B), whereas Akt-KI did not affect basal levels of ERK1/2 phosphorylation. As observed in Fig. 4, mechanical stretch caused increased ERK phosphorylation in both untreated and Akt-KI-treated, mechanically stretched cells (although no statistically significant increase was observed for the Akt-KI treated, mechanically stretched cells; Fig. 5B).
In the study depicted in Fig. 6, mechanical stretch caused alterations in Tnnt3 pre-mRNA alternative splicing that qualitatively recapitulated those presented in Fig. 3. In other words, in both untreated and U0126-treated C2C12 myotubes, mechanical stretch caused decreased overall Tnnt3 exon 4 inclusion at 24 h of treatment (Fig. 6A; see also Table 2). Importantly, Akt-KI treatment prevented this stretch-induced effect. More specifically, Akt-KI treatment prevented the stretch-induced increase in Tnnt3 mt1 relative abundance and decreases in Tnnt3 mt14-15 relative abundance (Fig. 6B) demonstrated earlier (Fig. 3B, Table 2), while the stretch-induced increases in Tnnt3 mt2 and mt4 relative abundance were unaffected by Akt-KI treatment.
DISCUSSION
The results of the study described herein extend recent findings in rat skeletal muscle, where quantitative Tnnt3 pre-mRNA alternative splicing was altered in response to a 5-day period of weight loading (∼30% of body weight; Ref. 41). A key question raised by that study, and previous work in insects (28), is whether the Tnnt3 splicing response to muscle loading is controlled by a systemically circulating factor(s), or whether muscle cell-autonomous mechanisms are involved.
The present study examined the latter hypothesis and demonstrated that mechanical stretch of C2C12 myotubes caused rapid (i.e., within 24 h) changes to the expressed pool of Tnnt3 mRNAs generated by means of alternative pre-mRNA splicing. Given the known effect of variation in Tnnt3 splice form composition on muscle force output and calcium sensitivity (5, 35), it is tempting to speculate on functional consequences of the shifts in Tnnt3 splice form composition caused by mechanical stretch. The effect of mechanical stretch was limited to a subset of the 15 Tnnt3 splice forms identified in C2C12 myotubes: stretch consistently increased the amount of splice forms Tnnt3 mt1, mt2, and mt4, whereas it decreased the relative abundance of splice forms Tnnt3 mt14 and mt15. Thus mechanical stretch was associated with an overall decrease in exon 4 inclusion. In addition, the relative abundance of Tnnt3 exon 16 containing splice forms (Tnnt3 mt1, mt2) was increased in response to mechanical stretch.
While the exact nucleotide sequence of expressed Tnnt3 splice forms differs between C2C12 myotubes and adult rat muscle, the overall pattern in myotubes in culture is strikingly similar to what we observed in muscle, where body weight gain was associated with decreased exon 4 inclusion and increased abundance of Tnnt3 splice forms containing exon 16 (41). Inclusion of Tnnt3 exon 4 reduces muscle calcium sensitivity (4), whereas exon 16 inclusion increases it (8). We, therefore, predict that the mechanical stretch paradigm employed in the present studies produces a C2C12 myotube phenotype with enhanced calcium sensitivity and ability to produce force. Overall, these findings indicate that C2C12 myotubes subject to cyclic stretching are an appropriate model to study the effect of mechanical loading on Tnnt3 splicing observed in adult rat skeletal muscle.
Cyclically increasing the stretch experienced by muscle cells by means of deformation of their growth substrate is obviously quite different from loading live animals with a weighted harness (e.g., Ref. 6). However, common to both paradigms may be a change in the strain muscle cells experience, assuming that the skeletal muscles of loaded rats (at least initially) counteract the additional weight by active force generation (i.e., muscle shortening). Forces generated during muscle contraction in vivo cause cellular deformations, which are transmitted to the extracellular matrix (ECM) from where biochemical signals (e.g., integrin, ion channel based) can activate intracellular signaling cascades (e.g., Ref. 22). Interestingly, the C2C12 myotubes used in the present studies were grown on membranes coated with ECM-like material (i.e., collagen I) and likely deposited additional ECM components during their development (12). Therefore, our working hypothesis is that ECM-driven mechanotransduction regulates Tnnt3 pre-mRNA alternative splicing.
In addition to the ability to vary the types of splice variants, their precise quantitative regulation across time and tissues is key to gene and organismal function (21). A prime example of its importance comes from work on Libellula pulchella dragonflies (e.g., Ref. 29), which has demonstrated how regulation of quantitative variation in Tnt (insect muscle troponin T) splice form abundance allows organisms to shift from an energetically expensive or inexpensive lifestyle, thereby trading off reproductive success. Similarly, we know that rat skeletal muscles can precisely adjust their Tnnt3 splice form profile in a quantitative, load-dependent fashion, thereby affecting energy expenditure (41). Even though mechanical loading produces qualitatively similar (i.e., effects on exon inclusion mentioned earlier) results in rat skeletal muscles and C2C12 myotubes, it remains to be determined whether the latter response is as quantitatively appropriate to the amount of load they received as it was in rats. Exposing muscle cells to variable strain intensities in vitro has been shown to correlate quantitatively with the level of muscle gene expression and intracellular signaling (7, 31), but this has not been examined with regard to quantitative regulation of alternative splicing of skeletal muscle genes.
Mechanisms that regulate muscle contractility likely intersect with those that perceive energy availability and track expenditure (6). Indeed, findings in insects (i.e., fall armyworm, Spodoptera frugiperda) demonstrated that diet quality modulates the relationship between skeletal muscle troponin T splicing and body weight (28). The PI3K/Akt and Ras/Erk signaling pathways are important in regulating the balance between muscle protein synthesis and growth and nutrient availability (e.g., Refs. 13, 15). In addition, Akt and ERK1/2 have previously been shown to be activated in response to mechanical stretch in vitro (e.g., Refs. 1, 18). Importantly, these kinases are involved in the regulation of alternative splicing through their action on splice factor activity (e.g., Refs. 3, 27, 32, 43). We, therefore, considered Akt and ERK1/2 prime candidates to probe the intracellular mechanism of loading-induced alternative splicing. Indeed, inhibition of Akt phosphorylation was recently shown to prevent insulin and thyroid hormone-induced alternative splicing of the cardiac muscle titin (Ttn) gene (26). In the present study, we demonstrated that Akt and ERK1/2 were both phosphorylated in response to mechanical stretch, but that only inhibition of Akt phosphorylation, and not that of ERK1/2, resulted in an inhibition of mechanical stretch-induced Tnnt3 pre-mRNA alternative splicing. Thus our results suggest a general role for Akt in modulating alternative splicing of muscle sarcomere genes, adding important functionality to the PI3K/Akt signaling pathway and those with which it intersects. A caveat to this conclusion is that, although inhibition of Akt blocked stretch-induced Tnnt3 pre-mRNA alternative splicing, the time courses for activation of Akt and alternative splicing of the Tnnt3 mRNA were distinct, with activation of Akt occurring transiently during the first 2 h of stretch, while alternative splicing was not observed until 8 h. Thus the role of Akt in regulating alternative splicing may have been indirect, as opposed to a direct effect on phosphorylation of splicing factors.
Mechanical loading of C2C12 myotubes may be a potent model system to study the regulation of alternative splicing in general. More than 90% of (human) genes are alternatively spliced (e.g., Refs. 34, 37, 46), and mechanisms of alternative splicing have been characterized in considerable depth. Because the regulation of alternative splicing of many genes depends on the activity of a relatively small group of splice factors (mRNA binding proteins), it is likely that alternative splicing of sets of genes is coregulated. In support of this prediction, a recent global and longitudinal (i.e., across 32 days) exon array analysis (39) on bone tissue reported that up to 992 (!) genes (including 8 muscle-specific ones) were alternatively spliced in response to mechanical loading. An earlier observation that alternative splicing of cardiac troponin T is coregulated with that of the insulin receptor (14) provides further support for this idea. Therefore, Tnnt3 is likely but one of many genes for which pre-mRNA alternative splicing is modulated in response to mechanical load.
How exactly skeletal muscles perceive the amount of load imposed on them and orchestrate gene expression responses of individual muscle cells to achieve the homeostatic balance between muscle force production and body weight support required remains a seriously complex question with few answers at this point. The ability to manipulate alternative splicing of a sarcomere gene like Tnnt3 in an in vitro setting extends the experimental power of in vivo rat models and will facilitate the identification of such mechanisms.
GRANTS
The studies described herein were supported by funds from the National Institute of Diabetes and Digestive and Kidney Diseases DK15658 (to L. S. Jefferson), the American Physiological Society (Postdoctoral Fellowship in Physiological Genomics to R. J. Schilder), and a grant from The Pennsylvania Department of Health using Tobacco Settlement Funds (to S. R. Kimball).
DISCLAIMER
The Pennsylvania Department of Health specifically disclaims responsibility for any analyses, interpretations, or conclusions.
DISCLOSURES
No conflicts of interest, financial or otherwise, are declared by the author(s).
AUTHOR CONTRIBUTIONS
R.J.S., S.R.K., and L.S.J. conception and design of research; R.J.S. performed experiments; R.J.S. analyzed data; R.J.S., S.R.K., and L.S.J. interpreted results of experiments; R.J.S. and S.R.K. prepared figures; R.J.S. drafted manuscript; R.J.S., S.R.K., and L.S.J. edited and revised manuscript; R.J.S., S.R.K., and L.S.J. approved final version of manuscript.
ACKNOWLEDGMENTS
We acknowledge the Penn State Genomics Core Facility at University Park, PA, for services provided in Tnnt3 splice form sequencing and quantification.
REFERENCES
- 1. Atherton PJ, Szewczyk NJ, Selby A, Rankin D, Hillier K, Smith K, Rennie MJ, Loughna PT. Cyclic stretch reduces myofibrillar protein synthesis despite increases in FAK and anabolic signalling in L6 cells. J Physiol 587: 3719–3727, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Biewener AA. Scaling Body Support in Mammals: Limb Posture and Muscle Mechanics. Science 245: 45–48, 1989 [DOI] [PubMed] [Google Scholar]
- 3. Blaustein M, Pelisch F, Tanos T, Muñoz MJ, Wengier D, Quadrana L, Sanford JR, Muschietti JP, Kornblihtt AR, Cáceres JF, Coso OA, Srebrow A. Concerted regulation of nuclear and cytoplasmic activities of SR proteins by AKT. Nat Struct Mol Biol 12: 1037–1044, 2005 [DOI] [PubMed] [Google Scholar]
- 4. Briggs MM, Schachat F. Physiologically regulated alternative splicing patterns of fast troponin T RNA are conserved in mammals. Am J Physiol Cell Physiol 270: C298–C305, 1996 [DOI] [PubMed] [Google Scholar]
- 5. Brotto MA. Coupled expression of troponin T and troponin I isoforms in single skeletal muscle fibers correlates with contractility. Am J Physiol Cell Physiol 290: C567–C576, 2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Burkholder TJ. Mechanotransduction in skeletal muscle. Front Biosci 12: 174–191, 2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Chandran R, Knobloch TJ, Anghelina M, Agarwal S. Biomechanical signals upregulate myogenic gene induction in the presence or absence of inflammation. Am J Physiol Cell Physiol 293: C267–C276, 2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Gallon C, Tschirgi M, Chandra M. Differences in myofilament calcium sensitivity in rat psoas fibers reconstituted with troponin T isoforms containing the α- and β-exons. Arch Biochem Biophys 456: 127–134, 2006 [DOI] [PubMed] [Google Scholar]
- 9. Goldspink G. Changes in muscle mass and phenotype and the expression of autocrine and systemic growth factors by muscle in response to stretch and overload. J Anat 194: 323–334, 1999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Gomes AV, Barnes JA, Harada K, Potter JD. Role of troponin T in disease. Mol Cell Biochem 263: 115–129, 2004 [DOI] [PubMed] [Google Scholar]
- 11. Greaser ML, Moss RL, Reiser PJ. Variations in contractile properties of rabbit single muscle fibres in relation to troponin T isoforms and myosin light chains. J Physiol 406: 85–98, 1988 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Gullberg D, Fessler LI, Fessler JH. Differentiation, extracellular matrix synthesis, and integrin assembly by Drosophila embryo cells cultured on vitronectin and laminin substrates. Dev Dyn 199: 116–128, 1994 [DOI] [PubMed] [Google Scholar]
- 13. Haddad F, Adams GR. Inhibition of MAP/ERK kinase prevents IGF-I-induced hypertrophy in rat muscles. J Appl Physiol 96: 203–210, 2004 [DOI] [PubMed] [Google Scholar]
- 14. Ho TH, Charlet -BN, Poulos MG, Singh G, Swanson MS, Cooper TA. Muscleblind proteins regulate alternative splicing. EMBO J 23: 3103–3112, 2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Hoffman EP, Nader GA. Balancing muscle hypertrophy and atrophy. Nat Med 10: 584–585, 2004 [DOI] [PubMed] [Google Scholar]
- 16. Hornberger TA, Armstrong DD, Koh TJ, Burkholder TJ, Esser KA. Intracellular signaling specificity in response to uniaxial vs. multiaxial stretch: implications for mechanotransduction. Am J Physiol Cell Physiol 288: C185–C194, 2005 [DOI] [PubMed] [Google Scholar]
- 17. Hornberger TA, Stuppard R, Conley KE, Fedele MJ, Fiorotto ML, Chin ER, Esser KA. Mechanical stimuli regulate rapamycin-sensitive signalling by a phosphoinositide 3-kinase-, protein kinase B- and growth factor-independent mechanism. Biochem J 380: 795–804, 2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Hornberger TA. Mechanotransduction and the regulation of mTORC1 signaling in skeletal muscle. Int J Biochem Cell Biol 43: 1267–1276, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Huang H, Kamm RD, Lee RT. Cell mechanics and mechanotransduction: pathways, probes, and physiology. Am J Physiol Cell Physiol 287: C1–C11, 2004 [DOI] [PubMed] [Google Scholar]
- 20. Hulens M, Vansant G, Lysens R, Claessens AL, Muls E, Brumagne S. Study of differences in peripheral muscle strength of lean vs. obese women: an allometric approach. Int J Obes Relat Metab Disord 25: 676–681, 2001 [DOI] [PubMed] [Google Scholar]
- 21. Irimia M, Rukov JL, Roy SW, Vinther J, Garcia-Fernandez J. Quantitative regulation of alternative splicing in evolution and development. Bioessays 31: 40–50, 2009 [DOI] [PubMed] [Google Scholar]
- 22. Jaalouk D, Lammerding J. Mechanotransduction gone awry. Nat Rev Mol Cell Biol 10: 63–73, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Jackson AW, Lee DC, Sui X, Morrow JR, Church TS, Maslow AL, Blair SN. Muscular strength is inversely related to prevalence and incidence of obesity in adult men. Obesity (Silver Spring) 18: 1988–1995, 2010 [DOI] [PubMed] [Google Scholar]
- 24. Janmey PA, McCulloch CA. Cell mechanics: integrating cell responses to mechanical stimuli. Annu Rev Biomed Eng 9: 1–34, 2007 [DOI] [PubMed] [Google Scholar]
- 25. Lafortuna CL, Maffiuletti NA, Agosti F, Sartorio A. Gender variations of body composition, muscle strength and power output in morbid obesity. Int J Obes (Lond) 29: 833–841, 2005 [DOI] [PubMed] [Google Scholar]
- 26. Linke WA, Krüger M. The giant protein titin as an integrator of myocyte signaling pathways. Physiology 25: 186–198, 2010 [DOI] [PubMed] [Google Scholar]
- 27. Lynch KW. Regulation of alternative splicing by signal transduction pathways. Adv Exp Med Biol 623: 161–174, 2007 [DOI] [PubMed] [Google Scholar]
- 28. Marden JH, Fescemyer HW, Saastamoinen M, Macfarland SP, Vera JC, Frilander MJ, Hanski I. Weight and nutrition affect pre-mRNA splicing of a muscle gene associated with performance, energetics and life history. J Exp Biol 211: 3653–3660, 2008 [DOI] [PubMed] [Google Scholar]
- 29. Marden JH, Fitzhugh GH, Girgenrath M, Wolf MR, Girgenrath S. Alternative splicing, muscle contraction and intraspecific variation: associations between troponin T transcripts, Ca(2+) sensitivity and the force and power output of dragonfly flight muscles during oscillatory contraction. J Exp Biol 204: 3457–3470, 2001 [DOI] [PubMed] [Google Scholar]
- 30. Marden JH. Quantitative and evolutionary biology of alternative splicing: how changing the mix of alternative transcripts affects phenotypic plasticity and reaction norms. Heredity 100: 111–120, 2008 [DOI] [PubMed] [Google Scholar]
- 31. Martineau LC, Gardiner PF. Insight into skeletal muscle mechanotransduction: MAPK activation is quantitatively related to tension. J Appl Physiol 91: 693–702, 2001 [DOI] [PubMed] [Google Scholar]
- 32. Matter N, Herrlich P, König H. Signal-dependent regulation of splicing via phosphorylation of Sam68. Nature 420: 691–695, 2002 [DOI] [PubMed] [Google Scholar]
- 33. McCullen SD, Haslauer CM, Loboa EG. Musculoskeletal mechanobiology: interpretation by external force and engineered substratum. J Biomech 43: 119–127, 2010 [DOI] [PubMed] [Google Scholar]
- 34. Nilsen TW, Graveley BR. Expansion of the eukaryotic proteome by alternative splicing. Nature 463: 457–463, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Ogut O, Granzier H, Jin JP. Acidic and basic troponin T isoforms in mature fast-twitch skeletal muscle and effect on contractility. Am J Physiol Cell Physiol 276: C1162–C1170, 1999 [DOI] [PubMed] [Google Scholar]
- 36. Pan BS, Potter JD. Two genetically expressed troponin T fragments representing alpha and beta isoforms exhibit functional differences. J Biol Chem 267: 23052–23056, 1992 [PubMed] [Google Scholar]
- 37. Pan Q, Shai O, Lee LJ, Frey BJ, Blencowe BJ. Deep surveying of alternative splicing complexity in the human transcriptome by high-throughput sequencing. Nat Genet 40: 1413–1415, 2008 [DOI] [PubMed] [Google Scholar]
- 38. Perry S. Troponin T: genetics, properties and function. J Muscle Res Cell Motil 19: 575–602, 1998 [DOI] [PubMed] [Google Scholar]
- 39. Roosa SMM, Liu Y, Turner CH. Alternative splicing in bone following mechanical loading. Bone 48: 543–551, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Schachat FH, Diamond MS, Brandt PW. Effect of different troponin T-tropomyosin combinations on thin filament activation. J Mol Biol 198: 551–554, 1987 [DOI] [PubMed] [Google Scholar]
- 41. Schilder RJ, Kimball SR, Marden JH, Jefferson LS. Body weight-dependent troponin T alternative splicing is evolutionarily conserved from insects to mammals and is partially impaired in skeletal muscle of obese rats. J Exp Biol 214: 1523–1532, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Spangenburg EE. Changes in muscle mass with mechanical load: possible cellular mechanisms. Appl Physiol Nutr Metab 34: 328–335, 2009 [DOI] [PubMed] [Google Scholar]
- 43. Stamm S. Regulation of alternative splicing by reversible protein phosphorylation. J Biol Chem 283: 1223–1227, 2008 [DOI] [PubMed] [Google Scholar]
- 44. Wang ET, Sandberg R, Luo S, Khrebtukova I, Zhang L, Mayr C, Kingsmore SF, Schroth GP, Burge CB. Alternative isoform regulation in human tissue transcriptomes. Nature 456: 470–476, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Wang J, Jin JP. Primary structure and developmental acidic to basic transition of 13 alternatively spliced mouse fast skeletal muscle troponin T isoforms. Gene 193: 105–114, 1997 [DOI] [PubMed] [Google Scholar]
- 46. Zhou HL, Hinman MN, Barron VA, Geng C, Zhou G, Luo G, Siegel RE, Lou H. Hu proteins regulate alternative splicing by inducing localized histone hyperacetylation in an RNA-dependent manner. Proc Natl Acad Sci U S A 108: E627–E635, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]