Summary
The floor plate (FP) is a midline signaling center, known to direct ventral cell fates and axon guidance in the neural tube. The recent identification of midbrain FP as a source of dopaminergic neurons has renewed interest in its specification and organization, which remain poorly understood. In this study, we have examined the chick midbrain and spinal FP and show that both can be partitioned into medial (MFP) and lateral (LFP) subdivisions. Although Hedgehog (HH) signaling is necessary and sufficient for LFP specification, it is not sufficient for MFP induction. By contrast, the transcription factor FOXA2 can execute the full midbrain and spinal cord FP program via HH-independent and dependent mechanisms. Interestingly, although HH-independent FOXA2 activity is necessary and sufficient for inducing MFP-specific gene expression (e.g., LMX1B, BMP7), it cannot confer ventral identity to midline cells without also turning on Sonic hedgehog (SHH).
We also note that the signaling centers of the midbrain, the FP, roof plate (RP) and the midbrain-hindbrain boundary (MHB) are physically contiguous, with each expressing LMX1B and BMP7. Possibly as a result, SHH or FOXA2 misexpression can transform the MHB into FP and also suppress RP induction. Conversely, HH or FOXA2 knockdown expands the endogenous RP and transforms the MFP into a RP and/or MHB fate. Finally, combined HH blockade and FOXA2 misexpression in ventral midbrain induces LMX1B expression, which triggers the specification of the RP, rather than the MFP. Thus we identify HH-independent and dependent roles for FOXA2 in specifying the FP. In addition, we elucidate for the first time, a novel role for SHH in determining whether a midbrain signaling center will become the FP, MHB or RP.
Keywords: floor plate, midbrain-hindbrain boundary, roof plate, LMX1B, BMP7, signaling centers, spinal cord
Introduction
During development, the ventral midline of the vertebrate neural tube is occupied by the FP, which secretes SHH and plays a critical role in cell fate specification and axon guidance (His, 1888; Kingsbury, 1920; Placzek and Briscoe, 2005; Strahle et al., 2004). Despite 100 years of study, questions remain unanswered with regard to the embryonic origin, specification and function of the FP across species and axial levels of the neural tube (Placzek and Briscoe, 2005).
The simplest functional organization of the FP to have emerged is its partition into medial (MFP) and lateral (LFP) subdivisions (Odenthal et al., 2000; Schauerte et al., 1998). These divisions are well established in the anamniote and avian spinal FP where the MFP and LFP differ from each other in their embryonic origin, and more controversially, in their dependence upon HH signaling (Charrier et al., 2002; Odenthal et al., 2000; Peyrot et al., 2011; Placzek and Briscoe, 2005; Strahle et al., 2004). HH signaling is required in the fish for specification of the neurectoderm-derived LFP, while the node-derived MFP depends upon Nodal signaling for its induction (Hatta et al., 1991; Odenthal et al., 2000; Schauerte et al., 1998; Strahle et al., 2004). However, these results are complicated by a requirement for HH signaling in the maintenance of MFP at later stages and by the presence of residual FP cells in the nodal (cyclops) mutant (Odenthal et al., 2000; Ribes et al., 2010).
Gene expression patterns also support a partition of the mouse FP into medial and lateral subdivisions (Odenthal et al., 2000). However, the entire FP disappears following the loss of HH signaling, leaving unanswered the question of whether these subregions differentially require HH signaling for maintenance and/or induction as they do in the fish (Chiang et al., 1996; Fogel et al., 2008; Odenthal et al., 2000).
The chick spinal FP also displays node and neurectoderm-derived MFP and LFP subdivisions (Catala et al., 2000; Charrier et al., 2002). Here, SHH misexpression cannot induce an ectopic MFP, while notochord transplants readily do so via unidentified mechanisms (Catala et al., 2000; Charrier et al., 2002; Gray and Dale, 2010). More recent studies have suggested that early and transient exposure to SHH is capable of inducing a FP in all species, including the bird, fish and mouse (Ribes et al., 2010). But whether SHH is necessary and sufficient for the full FP program has not been addressed.
Even less is known about the specification of anterior (midbrain, rostral hindbrain) FP, which is distinct from the spinal FP in some respects (Patten et al., 2003; Placzek and Briscoe, 2005). For example, in addition to contributions from the node and neurectoderm, the chick anterior FP is also colonized by a unique population of cells emerging from “area a”, a region anterior to Hensen’s node (Patten et al., 2003; Schoenwolf and Sheard, 1990). Interestingly, “area a” explants can be induced to a FP fate in the absence of the notochord by a brief exposure to prechordal plate-derived SHH and Nodal activity (Patten et al., 2003). But whether the anterior FP cell-types are organized into medial and lateral subdivisions and whether they differentially utilize HH and Nodal signaling is not known.
This question has recently acquired significance with the identification of an MFP-like ventral midline region as the predominant source of midbrain dopaminergic (mDA) neurons, which help regulate voluntary movements and the reward-reinforcement circuitry of the brain (Bayly et al., 2007; Blaess et al., 2011; Joksimovic et al., 2009; Ono et al., 2007). Recent work, including ours, suggests that HH signaling may not be necessary for patterning this MFP-like region (Hynes et al., 2000; Kittappa et al., 2007; Lin et al., 2009; Ye et al., 1998). Instead, some studies have reported that the winged helix transcription factor FOXA2 may be a more potent inducer of mDA progenitors than SHH (Kittappa et al., 2007; Lin et al., 2009; Norton et al., 2005; Ribes et al., 2010; Sasaki and Hogan, 1993).
Since SHH and FOXA2 transcriptionally upregulate each other, the differences between their inductive abilities are not well understood (Kittappa et al., 2007; Sasaki et al., 1997). FP studies in the fish suggest that FOXA2 activity can be regulated by, and interacts with, both the SHH and TGFβ/Nodal signaling cascades (Strahle et al., 2004). However, the role of FOXA2 in fish FP specification is complicated by several observations. Although FOXA2 can rescue the MFP in the fish nodal (cyclops) mutant, nodal-dependent induction of FP occurs in the absence of FOXA2 (Norton et al., 2005; Rastegar et al., 2002). Despite its ability to induce the fish MFP, foxa2 mutants display relatively mild phenotypes consisting mainly of a failure of the FP to fully differentiate (Norton et al., 2005). Although these observations suggest a role for FOXA2 in elaborating, rather than inducing the fish FP, they may also reflect functional redundancy among Foxa2 homologs. This idea is supported by the concurrent downregulation of Foxa2 and Foxa3 in the fish, which results in a complete loss of node-derived structures, including the notochord and MFP (Dal-Pra et al., 2011). An identical phenotype is seen in Foxa2−/− mutant mice where the absence of the node and notochord also precludes FP specification (Ang and Rossant, 1994; Norton et al., 2005).
Although conditional Foxa2 or Foxa1/2 knockouts targeting the ventral midbrain have been created in mice, they are not informative because they disrupt FOXA1/2 activity around E9, when ventral midbrain specification is well underway (Bayly et al., 2007; Blaess et al., 2006; Ferri et al., 2007; Fogel et al., 2008; Lin et al., 2009; Perez-Balaguer et al., 2009). Thus, the precise requirement for SHH and FOXA2 in FP specification remains poorly understood.
In this study, we have approached this problem by comparing the role of SHH and FOXA2 in midbrain and spinal FP specification in the chick, a species in which FOXA1 or FOXA3 genes have not been identified. We show that the midbrain FP can be clearly distinguished into MFP and LFP according to multiple criteria. Early manipulations of SHH and FOXA2 exclusively targeted to the neural plate suggest that SHH is not sufficient to induce the MFP in either the midbrain or spinal cord. By contrast, FOXA2 is necessary and sufficient for full MFP and LFP specification and does so via HH-dependent and independent mechanisms.
Notably, we show for the first time that the midbrain signaling centers express a common set of genes, and can take on each others’ identities in a SHH-dependent manner. Thus, by regulating the identity of orthogonal signaling centers, MFP signals direct 3-dimensional patterning in the midbrain and hindbrain.
Materials and Methods
Chick embryos
Fertilized Leghorn eggs (Ideal Poultry, Texas) were incubated at 38°C. Embryos were staged according to Hamburger and Hamilton (Hamburger, 1951).
Expression vectors
Embryos were electroporated with EGFP (EFX-EGFP), Ptc1Δloop2 (pCIG-Ptc1Δloop2), Foxa2 (pMes-Foxa2-IRES-EGFP/EFX-Foxa2), SHH (XEX-SHH; pMes-SHH-ires-RFP), dominant negative Foxa2 (pCAGGS-Fkha2-IRES-EGFP, pCAGGS-Fkha2-EnR–IRES-EGFP) or FOXA2 RNAi (pSilencer-FOXA2 shRNA) expression vectors (Agarwala and Ragsdale, 2002; Agarwala et al., 2001; Bayly et al., 2007; Briscoe et al., 2001; Jacob et al., 2007). EFX-Foxa2 was constructed by ligating mouse Foxa2 cDNA (pBSSK-mHnf3b) into EFX3C (Agarwala et al., 2001; Sasaki and Hogan, 1993). pMes-Foxa2-IRES-EGFP and pMes-SHH-ires-RFP were created by subcloning Foxa2 or SHH cDNAs into the pMes-IRES-EGFP and pMes-IRES-RFP vectors.
FOXA2 shRNA constructs
Four short-hairpin RNA constructs were created by cloning into the pSilencer vector (Ambion). Of these, only one construct (FOXA2-1663) produced effective knockdown of FOXA2 mRNA (Supplementary Fig. S1A, B) and was generated using the following forward: 5′-CTCCTCCTAAAGGCAAAGGTTCAAGAGACCTTTGCCTTTAGGAGGAGTTTTTT-3′ and reverse primers: 5′-AATTAAAAAACTCCTCCTAAAGGCAAAGGTCTCTTGAACCTTTGCCTTTAGGAGGAGGGCC-3′. RNAi electroporations (4 μg/μl) typically produced small effects, but replicated the larger effects of the dnFOXA2 constructs described above (Jacob et al., 2007). These data were therefore pooled.
In ovo electroporation
1–5 μg/μl DNA was electroporated into H&H stages 4–11 midbrains and H&H 8–11 spinal cords according to previous protocols (Agarwala and Ragsdale, 2002; Agarwala et al., 2001; Briscoe et al., 2001; Eom et al., 2011; Momose et al., 1999).
Midbrain explants
Whole embryo (H&H 3–6) or midbrain-hindbrain (H&H 7–9) explants were cultured for 24 hours in 100 μM cyclopamine (Sigma) with 2-hydroxypropyl-β-cyclodextrin (HBC, Sigma) or HBC alone, using established procedures (Agarwala and Ragsdale, 2002; Bayly et al., 2007; Incardona et al., 1998; Norton et al., 2005).
In situ hybridization
Embryos were harvested between E1–E6 and subjected to one and two-color in situ hybridizations according to published protocols (Agarwala and Ragsdale, 2002; Agarwala et al., 2001). The LMX1A and MN-CAD cDNAs were generated by PCR amplification from E5 midbrains using the forward: 5′-ATGGACAGCGACGATACCTCAC-3′ and reverse: 5′-CCAGACCTACCTCCTGAAACAAGC-3′ primers for LMX1A and the forward 5′-CAGATGCTGATGACCCAACCTATG-3′ and reverse 5′-TGATGTGAACGGTGGTGGTGTC3′ primers for MN-CAD.
Immunohistochemistry
Embryos were fixed in 4% paraformaldehyde for 15 min-2 hours. 14 μm cross-sections were stained with antibodies against SHH and LMX1B (DHSB; 1:200 each), Alexa-conjugated secondary antibodies were used for fluorescent detection (Afonso and Henrique, 2006). DAPI was used for staining nuclei.
Wholemount p-Smad 1/5/8 immunohistochemistry
Embryos were fixed in 4% PFA for 3 hours and subjected to wholemount p-Smad1/5/8 (1: 2500–5000; Cell Signaling) immunolabeling according to previously established protocols (Eom et al., 2011).
Results
Differential gene expression in medial and lateral FP
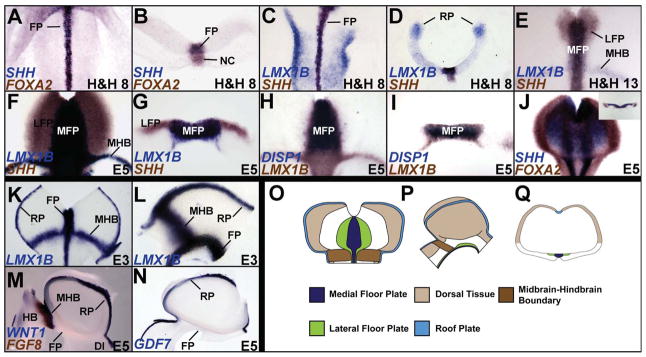
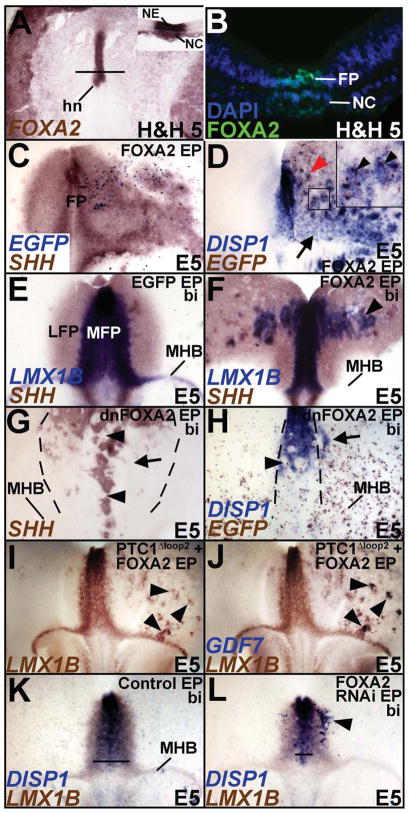
We first determined whether the midbrain FP could be divided into medial and lateral divisions by differential gene expression (Odenthal et al., 2000; Placzek and Briscoe, 2005; Strahle et al., 2004). SHH, FOXA2, LMX1B and Bone morphogenetic protein 7 (BMP7) mRNAs were expressed in the presumptive midbrain FP by H&H 4 and continued to be expressed there until at least E6 (Fig. 1A–I; Supplementary Fig. S2A; Aglyamova and Agarwala, 2007; Echelard et al., 1993; Hamburger, 1951; Lawson et al., 2001; Ruiz i Altaba et al., 1993; Yuan and Schoenwolf, 1999). By H&H 13, a SHH+/FOXA2+/LMX1B-negative/BMP7-negative territory emerged rostrally to surround the SHH+/FOXA2+/LMX1B+/BMP7+ FP (Fig. 1E; Aglyamova and Agarwala, 2007). This lateral region expanded caudally toward the MHB, so that it fully encompassed the medial region between late E3–E6 (Fig. 1F, G; Supplementary Fig. S2B, C). Based on gene expression and additional criteria described below, these FP territories were identified as MFP and LFP (Odenthal et al., 2000).
Figure 1.
A–J. Gene expression patterns distinguish the midbrain MFP and LFP. SHH, FOXA2, LMX1B and DISP1 expression in the FP. A: Whole embryo, top-down view; B, D, G, I, inset J: cross-sections; C, E, F, H, J: flattened wholemounts (open book preparation, rostral to the top and ventricular surface facing the viewer). K–N. Overlapping and specific gene expression patterns in midbrain signaling centers. Flattened wholemount (K) and sagittal (L–N) views displaying physically contiguous LMX1B expression in the MFP, RP and MHB (K, L), the exclusive expression of FGF8 in the MHB (M) and of GDF7 in RP (N). Note that WNT1 is expressed in the RP and MHB, but not FP. (O–Q) Cartoons displaying the 3-dimensional spatial relationships among the midbrain signaling centers. O: flattened wholemount; P: sagittal view, rostral to the right; Q: cross-section.
Several other markers, e.g., LMX1A, DISP1, MN-CAD and Chordin, were also expressed in the MFP and distinguished it from the LFP (Fig. 1H, I; Supplementary Fig. S2D; data not shown). Their expression patterns were dynamic and some occupied narrower domains than the LMX1B territory, especially at later ages (Fig. 1H, I; Supplementary Fig. S2D; data not shown). A closer examination also showed that the expression of SHH, FOXA2, DISP1 and LMX1B in MFP cells was mosaic, suggesting that the subarchitecture of the MFP may be more complex than outlined here (Fig. 1H; Supplementary Fig. S2E, E′). However, since our perturbations did not differentially affect the MFP markers identified above, we treated the MFP as one entity in this study.
We were unable to find markers that uniquely and stably identify the LFP for the duration of our experiments. This included NKX2.2 expression, which labeled regions outside the FP (Supplementary Fig. S2F). The midbrain LFP was therefore identified by the combined presence of SHH and FOXA2 and the absence of LMX1A, LMX1B, DISP1, MN-CAD and BMP7 (Fig. 1E–J, Supplementary Fig. S2A–D; Table 1; data not shown).
Table 1.
Summary of gene expression patterns identifying the midbrain signaling centers. Unique identifiers for each signaling center are highlighted in red.
| Lateral Floor Plate | SHH | FOXA2 | ||||||||
| Medial Floor Plate | SHH | FOXA2 | DISP1 | MN-CAD | LMX1 A | LMX1B | BMP7 | |||
| Midbrain-Hindbrain Boundary | LMX1B | BMP7 | WNT1 | FGF8 | ||||||
| Roof Plate | LMX1B | BMP7 | WNT1 | GDF7 |
Interestingly, the MFP, RP and MHB all expressed BMP7 and LMX1B and were physically contiguous (Fig. 1K, L; Supplementary Fig. S2B, C). However, LMX1A, DISP1 and MN-CAD were expressed exclusively in the MFP, while FGF8 and GDF7 were expressed exclusively in the MHB and the RP respectively (Fig. 1H, I, M, N; Supplementary Fig. S2D; data not shown). These markers permitted us to unequivocally distinguish the midbrain signaling centers from each other in later experiments (Table 1). These data are summarized in Fig. 1O–Q.
SHH is sufficient for LFP, but not MFP induction in the midbrain
SHH misexpression can induce SHH and FOXA2, both generic markers of the FP, but whether it can induce both midbrain MFP and LFP is not known (Agarwala and Ragsdale, 2002; Agarwala et al., 2001). Unlike controls, SHH misexpression (1–5 μg/μl) non-autonomously converted large swaths of midbrain into SHH+/FOXA2+/LMX1B-negative/DISP1-negative LFP (Fig. 2A; Agarwala and Ragsdale, 2002; Agarwala et al., 2001). However, SHH misexpression could only induce MFP markers (DISP1, LMX1B, BMP7, MN-CAD) along the MHB and nowhere else in the midbrain (Fig. 2A–C; Supplementary Fig. S3A, B, E).
Figure 2. SHH misexpression is sufficient for LFP, but not MFP induction.
(A–C, flattened wholemounts, oriented as in Fig. 1C; D–K′, cross-sections). (A–C) Unilateral SHH misexpression is sufficient to convert the entire right half of the midbrain into SHH+ LFP (A, B), but can only specify an LMX1B+/DISP1+/SHH+ MFP along the MHB (A–C). (D–F) SHH mRNA (D) but no SHH protein (E) or PTC1 transcripts (F) at H&H 4. Sections in D–F are drawn from presumptive neural plate anterior to Hensen’s node. (G–H′) SHH protein (red) at H&H 8− (G–G″) and H&H 10 (H–H′). G′ and G″ show the micrograph in G with DAPI staining. The boxed area in G′ is magnified in G″ to show a SHH+ cell (arrowhead) and a SHH-negative cell (arrow). H′ shows the image in H with DAPI staining. (I) SHH and PTC1 mRNA at H&H 8. (J, K′) H&H 4–6 SHH-ires-RFP electroporation (red) is sufficient for ectopic, non-autonomous SHH (J, green) induction and dorsal LMX1A/B suppression (arrow, K), but it is not sufficient for LMX1A/B (green) induction ventrally (K, K′). Boxed area in K is magnified in K′ to show the absence of LMX1A/B induction in or around SHH -ires-RFP+ cells. Please note that the sections shown in G–K′ are drawn from the rostrocaudal midpoint of the midbrain.
Spinal cord studies have suggested that FP induction is accomplished by early and transient HH signaling prior to somitogenesis (Patten et al., 2003; Ribes et al., 2010). To determine the precise times during which early HH activity might occur in the midbrain, we examined early SHH gene and protein expression. Surprisingly, although SHH transcripts were detected as early as H&H 3, no SHH protein or HH receptors (PTC1, PTC2, HHIP) were detected in the axial midline at this time (Fig. 2D–F; Aglyamova and Agarwala, 2007; Lawson et al., 2001; Pearse et al., 2001). Modest levels of SHH protein were first detected in the cytoplasm of most FP and notochord cells at H&H 8−, although some cells remained SHH-negative at this time (Fig. 2G–G″). SHH expression became stronger by H&H 10 and resembled that of the spinal cord, with strong, cytoplasmic expression along the apical surface of FP cells (Fig. 2H, H′; Chamberlain et al., 2008). Robust and transient expression of PTC1 and GLI1 was seen at the ventral midline between H&H 8− to <H&H 10, with more lateral expression seen at later ages (Fig. 2I; Aglyamova and Agarwala, 2007). Together these data defined a narrow window of time during which HH signaling might accomplish midbrain FP induction.
We used a novel early electroporation paradigm to misexpress SHH-ires-RFP between H&H 4–6 to obtain robust SHH misexpression by H&H 7, immediately prior to the onset of endogenous SHH protein expression (Brown et al., 2012; Eom et al., 2011). Interestingly, while SHH misexpression at this time readily induced ectopic SHH expression and suppressed dorsal expression of LMX1A/B, it failed to induce an LMX1A/B+/SHH+ MFP (Fig. 2J–K′). Thus, early HH activity is not sufficient for midbrain MFP induction, although it is sufficient for LFP induction and suppression of dorsal midbrain fates.
SHH is necessary for midbrain MFP and LFP specification
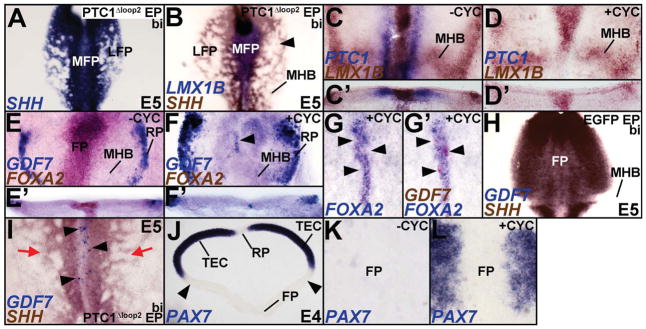
To determine whether HH signaling was necessary for the specification of the MFP, we misexpressed Ptc1Δloop2, a dominant negative regulator of HH signaling, between H&H 7–10 when HH signaling is critically required for midbrain cell fate specification (Fig. 2G–I; Bayly et al., 2007). Bilateral misexpression of Ptc1Δloop2 resulted in a robust disruption of the LFP while leaving LMX1B expression in the MFP largely unperturbed (Fig. 3A, B; Bayly et al., 2007).
Figure 3. SHH is necessary for MFP induction.
(A–B) Broad, bilateral misexpression of PTC1?Δloop2 disrupts the SHH+/LMX1B-negative LFP (B, arrowhead), but does not affect the SHH+/LMX1B+ MFP. Control for 3B can be seen in Figure 4E. (C, D) H&H 3 explants treated with either vehicle (−CYC) or cyclopamine (+CYC) for 24 hours demonstrate that HH blockade suppresses PTC1 expression in LFP, but does not affect LMX1B expression in the MFP. (C′, D′) Cross-sections of the wholemounts shown in C and D. (E, E′) Control explants in flattened wholemount (E) and cross-sectional (E′) view displaying GDF7 (RP) and FOXA2 (FP) expression. (F, F′) Cyclopamine treated explants displaying the loss of FOXA2 and the ectopic induction of GDF7 at the ventral midline (arrowhead). Note that compared to E, E′, the roof plate (RP) expression of GDF7 is expanded in F, F′. (G, G′) Cyclopamine treated explants with mild HH blockade showing ectopic expression of GDF7 (arrowheads) in the ventral midline whereever FOXA2 expression is abolished. Note that G and G′ show the same explant before and after GDF7 staining. Control for G, G′ is shown in Fig. E. (H) Control brain demonstrating that GDF7 is not expressed in the SHH+ FP or MHB. See also, 1N; 7G, H for controls. (I) Bilateral HH blockade results in a disruption of the LFP (red arrows) and a conversion of the MFP into GDF7+ RP (arrowheads). (J) Midbrain cross-section demonstrating that PAX7 expression is absent from the RP and ventral midbrain, but present throughout dorsal midbrain (TEC). (K, L) Control (K) and cyclopamine treated explants (L) shown in wholemount view demonstrating that the ventral expansion of PAX7 into ventral midbrain (K) excludes the ventral midline. The PAX7-negative region between the arrowheads in J was flattened in K, L.
Despite the lack of evidence of an HH signaling cascade prior to H&H 8−, it was possible that low, undetectable levels of early HH activity occurred along the ventral midline and were required for midbrain MFP specification (Ribes et al., 2010). In previous work, we noted that high embryonic lethality was associated with Ptc1Δloop2 electroporations, and despite robust misexpression throughout the midbrain, only a small number of Ptc1Δloop2+ cells were found in the ventral midline (Bayly et al., 2007). Possible reasons for this include the differential cell-affinities of HH+ and HH-negative cells, but exclude apoptosis (Bayly et al., 2007; Lawrence, 1997; Rodriguez and Basler, 1997; Wijgerde et al., 2002).
To obtain more ubiquitous HH blockade at early stages, we therefore exposed early whole embryo explants to cyclopamine, a fast-acting inhibitor of HH signaling (Incardona et al., 1998). Although we initially divided the explants into 3 age groups (H&H 3–5, 6–7, 8–9), these data were pooled since all explants displayed similar FP phenotypes in response to cyclopamine. Bath application of 100 μM cyclopamine for 24 hours in H&H 3–9 explants successfully reduced or abolished the expression of HH pathway/target genes, PTC1, DISP1 and FOXA2 (Fig. 3C–F′; Supplementary Fig. S4A, B; Incardona et al., 1998). However, cyclopamine treatment did not affect LMX1B expression at the ventral midline (n=16/18; Fig. 3C–D′; Pearse et al., 2001). Together, the gain and loss of function experiments reported above suggest that regardless of the age of manipulation, HH activity was necessary, but not sufficient for the full execution of the MFP program.
Since the RP, FP and MHB all express LMX1B, we next asked whether the LMX1B expressing ventral midline retained FP identity following HH blockade (Fig. 1K, L; data not shown; Chizhikov and Millen, 2004). To our surprise, the cyclopamine-treated explants displayed ectopic GDF7 expression in ventral midline cells in which FOXA2 expression had been abolished, suggesting that some FP cells acquired ectopic RP identity in the absence of HH signaling (Fig. 3E–G′). Ptc1Δloop2 electroporations at H&H 7–11 provided additional evidence for the conversion of some FP cells into GDF7+ RP cells (Fig. 3H, I; arrowheads in 3I). With the exception of GDF7, exclusive RP markers that were not also expressed in other midbrain signaling centers were not available (Fig. 1K–Q; Table 1). However, the ectopic ventral midline expression of WNT1 (which is expressed in the RP and MHB, but not the avian FP; Fig. 1M; Fig. 7D) and WNT1+/GDF7+ cells in the ventral midline following Ptc1Δloop2 electroporations further confirmed the transformation of FP cells into RP identity (Supplementary Fig. S5A, B; Fig. 6H).
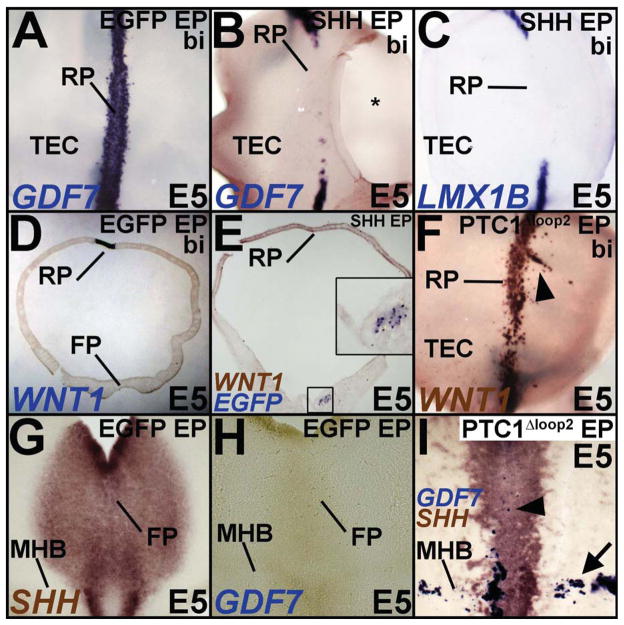
Figure 7.
(A–E) Top down (A–C) and cross-sectional views (D, E) demonstrating that compared to EGFP-electroporated controls (A, D), RP induction is blocked by ventral electroporations of SHH. Inset in E shows magnified view of the boxed region and demonstrates the location of SHH misexpression by EGFP transgene expression. * in B marks a hole cut in dorsal midbrain for irrigation. (F) Ventral PTC1Δloop2 misexpression results in ectopic RP induction (arrowhead). (G–I) Midbrain flatmounts showing that unlike controls (G, H), PTC1Δloop2 misexpression results in reduced SHH expression accompanied by ectopic GDF7 expression in both the ventral midline (arrowhead) and the MHB (arrow).
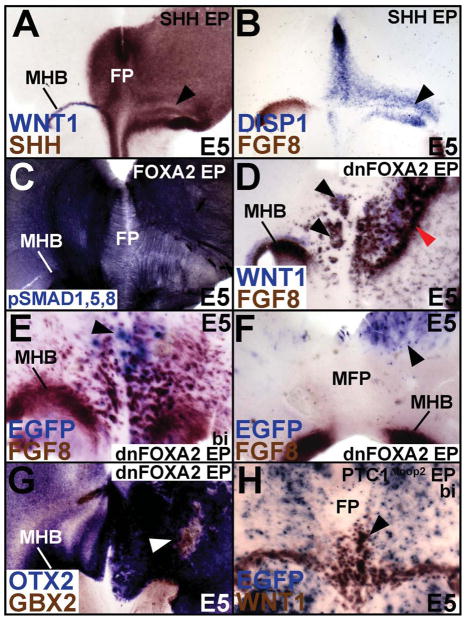
Figure 6. SHH and FOXA2 regulate the signaling centers of the midbrain.
(A, B) Unilateral SHH misexpression (right side) induces the MFP (DISP1+/SHH+) while concurrently suppressing MHB (FGF8+, WNT1+) fates (arrowheads, A, B). (C) FOXA2 misexpression converts the MHB (right side) into an ectopic MFP, which recapitulates the endogenous MFP pattern of axons, identified by phosphorylated-SMAD 1/5/8. (D) dnFOXA2 misexpression can convert the FP into MHB (arrowheads). Note also that the MHB is expanded and shifted rostrally (red arrowhead) on the right, most likely due to ectopic hindbrain induction caudal to it (See also Fig. 6G). (E, F) Ectopic MHB can only be induced by dnFOXA2 misexpression within the FOXA2+ FP (arrowhead, E) and not outside it (arrowhead, F). (G) dnFOXA2 suppresses OTX2+ midbrain fate and upregulates GBX2+ hindbrain fates (arrowhead). (H) Bilateral HH blockade also results in the conversion of caudal MFP into MHB (arrowhead).
To determine whether the ectopic ventral midline expression of GDF7 represented a specific conversion of the MFP to the RP or a generic ventral to dorsal conversion in the absence of HH activity, we examined PAX7 expression, which is absent in the ventral midbrain as well as RP, but otherwise present throughout the dorsal midbrain (Fig. 3J). Interestingly, cyclopamine treatment resulted in ectopic PAX7 expression throughout ventral midbrain except for the LMX1B+ region along the ventral midline (Fig. 3K, L). The presence of GDF7+/WNT1+/LMX1B+/PAX7-negative cells in the MFP demonstrated that in the absence of HH signaling, the ventral midline of the midbrain was specified as RP rather than as MFP. Taken together, these results show that HH signaling is necessary, but not sufficient to execute the full midbrain MFP program (Charrier et al., 2002; Peyrot et al., 2011; Ribes et al., 2010).
FOXA2 induces the full midbrain FP program via HH-dependent and independent mechanisms
FOXA2 is necessary and sufficient to induce MFP and LFP in ventral midbrain
Unlike SHH, FOXA2 mRNA and protein expression are seen at the axial midline of the embryo at H&H 2, prior to the activation HH signaling (Fig. 4A, B; Fig. 2E, G; Ruiz i Altaba et al., 1993). Given our limited understanding of FOXA2 function in regionalizing the amniote FP, we asked whether FOXA2 might induce midbrain MFP (Ang and Rossant, 1994; Norton et al., 2005; Ribes et al., 2010; Sasaki and Hogan, 1993; Sasaki and Hogan, 1994; Strahle et al., 2004). Interestingly FOXA2 electroporations early (H&H 4–6) or late (H&H 7–11) non-autonomously converted ventral midbrain into FP (Fig. 4C; 4D, inset 4D; Supplementary Fig. S6A, B). Unlike SHH however, FOXA2 was sufficient to induce both LFP and MFP markers throughout the midbrain and not just along the MHB (Fig. 4C–F; Fig. 2A–C; Supplementary Fig. S3A–F). Conversely, dominant negative FOXA2 (dnFOXA2) and FOXA2 RNAi electroporations between H&H 4–11 prevented the induction of both the MFP and the LFP (Fig. 4G, H; control for 4H is shown in Fig. 4K; Supplementary Fig. S6A, C).
Figure 4. Foxa2 is necessary and sufficient for midbrain FP specification.
(A, B) FOXA2 mRNA (A, inset A) and protein (B) are expressed in the axial midline of the embryo by H&H 5, prior to HH signaling. A: Whole embryo, top down view. Inset, A: Section taken from this embryo at the level indicated by the black line. (C) Unilateral misexpression of FOXA2-ires-EGFP (right) is sufficient to non-autonomously upregulate SHH. (D) Unilateral misexpression of FOXA2-ires-EGFP (right) results in DISP1 upregulation both along (arrow) and away (red arrowhead) from the MHB. Boxed region in D is magnified in the inset to demonstrate the non-autonomous induction of DISP1 (blue) by FOXA2-ires-EGFP (brown) misexpressing cells. (E, F). EGFP (E) and bilateral FOXA2 misexpression (F) demonstrating that ectopic FOXA2 can induce an LMX1B+/SHH+ MFP away from the MHB (arrowhead, F). (G, H) Bilateral dnFOXA2 electroporation (identified by EGFP detection in H) is sufficient to block LFP (arrow, G) and MFP (arrowheads, G, H) induction. Arrow in H points to the non-cell autonomous expansion of DISP1 following FOXA2 knockdown. (I, J) Unilateral PTC1Δloop2 and FOXA2 co-electroporation demonstrates that FOXA2-mediated induction of LMX1B is HH-independent. Note that the cells ectopically inducing LMX1B also express GDF7 and adopt a RP fate (arrowheads) in the absence of HH. The same wholemount is displayed in G and H before and after GDF7 detection. (K, L) Compared to controls (K), FOXA2 RNAi results in the suppression of MFP (compare the lines in K and L, marking the mediolateral extent of DISP1) as well as the ectopic induction of MFP in lateral midbrain (arrowhead, L).
FOXA2 cannot ventralize the FP without turning on SHH
We next determined the extent to which the FOXA2-mediated induction of MFP depended upon HH signaling. Joint electroporations of FOXA2 and Ptc1Δloop2 mimicked FOXA2 misexpression by inducing LMX1B expression throughout ventral midbrain, demonstrating that LMX1B induction by FOXA2 is HH-independent (Fig. 4I, J). However, in contrast to FOXA2 misexpression alone, ectopic LMX1B+ puncta induced by FOXA2/Ptc1Δloop2 co-electroporations also expressed GDF7, an exclusive marker of midbrain RP (Fig. 4I, J, arrowheads). Thus, following joint FOXA2 misexpression and HH blockade, cells ectopically expressing LMX1B adopt a RP, rather than a FP identity. We conclude that FOXA2 is necessary for midbrain MFP and LFP induction, and is necessary and sufficient to induce MFP-specific genes in the absence of SHH. By contrast, HH activity is required for inducing HH-pathway genes (DISP1, PTC1, FOXA2), without which the ventral midline cannot acquire a specific MFP identity (Fig. 3C–I). Thus the HH pathway is required for LFP specification and the ventralization of the MFP, while HH-dependent and independent activities of FOXA2 are required for the execution of the full midbrain FP program.
FOXA2 is necessary to restrict MFP induction to the ventral midline of the midbrain
A HH-dependent lateral expansion of netrin1 and nkx2.2a is seen in fish foxa2 (monorail) mutants, suggesting a role for FOXA2 in limiting HH activity laterally and preventing the lateral expansion of FP (Norton et al., 2005). In agreement with these observations, dnFOXA2 and FOXA2 RNAi-electroporated midbrains demonstrated a non-autonomous induction of MFP in lateral midbrain (n=8; arrow, Fig. 4H, arrowhead, Fig. 4L). Interestingly, such non-autonomous induction of the MFP was not seen in embryos electroporated with Ptc1Δloop2, suggesting that it was likely to be HH-dependent (Fig. 3B). Thus, in addition to its role in inducing the MFP and LFP, FOXA2 also plays a role in restricting the MFP domain to the ventral midline of the midbrain, as suggested by mouse Foxa2 enhancer analyses and fish foxa2 mutants (Norton et al., 2005; Sasaki et al., 1997).
FOXA2, but not SHH, is sufficient for specifying the full spinal FP program
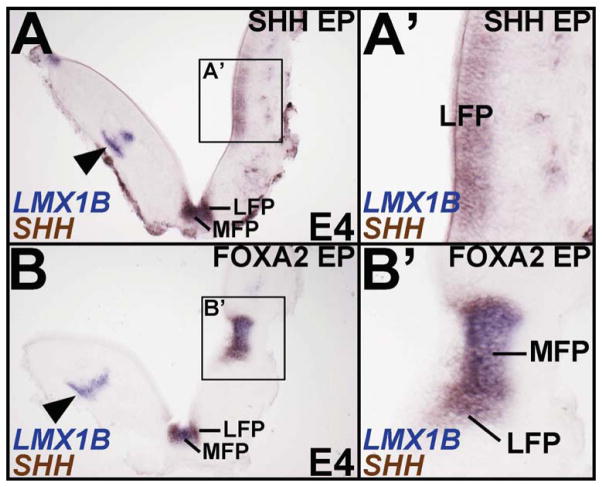
In the chick spinal cord, a notochord transplant can induce a complete FP, while ectopic SHH can only induce the LFP (Charrier et al., 2002). Since FOXA2 is expressed in the spinal FP and notochord, we next asked whether SHH and FOXA2 might play different roles in spinal FP specification as they do in the midbrain. Like the midbrain, we noted the presence of an LMX1B+/SHH+ MFP and LMX1B-negative/SHH+ LFP in the spinal cord (Fig. 5A). Unilateral electroporation of SHH in the H&H 10 rostral spinal cord suppressed dorsal fates (e.g. dI5 interneurons, arrowhead; Fig. 5A, A′) and readily induced ectopic LMX1B-negative/SHH+ LFP (Fig. 5A, A′). However, it did not induce an LMX1B+/SHH+ MFP (n= 10/10; Fig. 5A, A′). By contrast, FOXA2 misexpression induced both ectopic MFP and LFP, in addition to recapitulating the characteristic FP morphology (Fig. 5B, B′). Taken together, our observations suggest that FOXA2, but not SHH, is sufficient to execute the entire FP program in the midbrain and spinal cord.
Figure 5. FOXA2 is sufficient for MFP and LFP induction in the spinal cord.
Boxed areas in A and B are magnified in A′ and B′, respectively. (A, A′) Unilateral SHH misexpression (right side) is sufficient to induce a SHH+ LFP (boxed area, A; A′), but fails to induce a SHH+/LMX1B+ MFP. (B, B′) FOXA2 misexpression (right side) in the spinal cord is sufficient to induce both the MFP and LFP (boxed area, B; B′), accompanied by a FP-like morphology. Note that unlike the control side (arrowheads, A, B) FOXA2 and SHH both suppress LMX1gB+ dorsal dI5 interneurons.
SHH and FOXA2 regulate the specification of all midbrain signaling centers
The midbrain-hindbrain boundary
In Fig. 2 and 4, we demonstrated that ectopic SHH and FOXA2 can induce an MFP along the MHB, although how this occurs was not explored (Figs. 2A–C; 4D). FOXA2 or SHH-mediated induction of MFP markers along the MHB resulted in a concurrent suppression of the MHB markers, FGF8 and WNT1 (Fig. 6A, B; Supplementary Fig. S5C). To determine whether a functional MFP formed along the MHB, we asked whether the ectopic MFP provided axon guidance cues, a known function of the FP (Charron et al., 2003). Immuno-labeling by phosphorylated-SMAD 1/5/8 (a readout of canonical BMP signaling) allows for the identification of multiple axon trajectories in the midbrain, either attracted or repelled by FP signals (Fig. 6C; Fedtsova et al., 2008). Interestingly, FOXA2-mediated induction of the MFP along the MHB resulted in a complete recapitulation of axon trajectories seen at the MFP, while repressing those seen at the MHB (Fig. 6C, right side). Thus, FOXA2 and SHH suppress the MHB program and convert it into a functional MFP, which can redirect axon trajectories.
By contrast, dnFOXA2 resulted in a massive induction of WNT1 and FGF8, converting the FP into MHB and the midbrain into hindbrain (black arrowheads, Fig. 6D, E and red arrowhead, Fig. 6D; see also Fig. 6G; Agarwala and Ragsdale, 2002; Liu et al., 1999). We noted that this MHB induction was non-autonomous, but required that the dnFOXA2 electroporations were targeted to the ventral midline (Fig. 6E, F). By contrast, no induction of FGF8 occurred when dnFOXA2 was misexpressed outside the FOXA2 domain (Fig. 6F). We conclude that the non-autonomous induction of WNT1 and FGF8 throughout the midbrain was an indirect consequence of converting the MFP into the MHB and of expanding the endogenous MHB.
FGF8 misexpression is known to convert the midbrain into hindbrain by inducing GBX2 (Liu et al., 1999). Indeed, dnFOXA2 cell-autonomously upregulated GBX2 and suppressed OTX2, converting midbrain into hindbrain, while non-autonomously upregulating OTX2 (Fig. 6G; data not shown). HH blockade by PTC1Δloop2 replicated the effects of dnFOXA2 and also converted the MFP into MHB (Fig. 6H; Bayly et al., 2007). Together, these data suggest that FOXA2 and HH signaling are involved in determining whether a midbrain signaling center will take on MHB or MFP fates.
The roof plate
We showed above that HH mediated-ventralization of the midbrain determined whether the LMX1B+ ventral midline expressed MFP or RP fates (Fig. 3E–I; Fogel et al., 2008). We explored the inter-convertibility of MFP and RP further with SHH gain and loss of function experiments. Ventral SHH misexpression suppressed the specification of a GDF7+/WNT1+/LMX1B+ RP (Fig. 7A–E). Interestingly, very small ventral electroporations of SHH were sufficient to non-autonomously suppress RP specification (Fig. 7D, E, inset E). Conversely, ventral electroporations of Ptc1Δloop2resulted in an expansion and ectopic induction of the midbrain RP, a phenotype similar to the RP expansion noted in Shh−/− mice (Fig. 7F; Fogel et al., 2008). Ventral FOXA2 blockade mimicked the effects of HH blockade by expanding the endogenous midbrain RP and by ectopically inducing RP in dorsal midbrain (Supplementary Fig. S5D).
Surprisingly, suppression of HH activity along the MHB also resulted in a conversion of the MHB into GDF7+ RP cells (Fig. 7G–I, arrow in 7I). Together, these results suggest that SHH plays a role in repressing the RP program dorsally and along the MHB. In its absence the RP expands and the MHB and the MFP take on RP identity. These results suggest that HH is required for the specification and/or maintenance of all midbrain signaling centers, including the FP, RP and the MHB.
Discussion
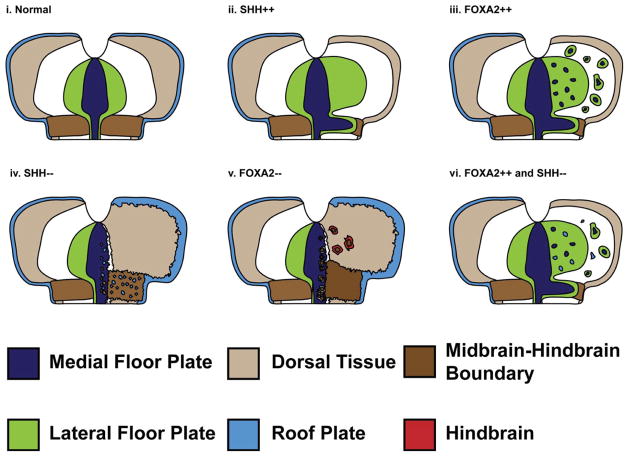
Our results suggest that SHH is both necessary and sufficient for LFP induction, but it is only necessary, and not sufficient for inducing the MFP program (Odenthal et al., 2000; Strahle et al., 2004). By contrast, FOXA2 is necessary and sufficient to specify the entire FP and does so in a HH-dependent and independent manner (Fig. 8). We have also identified a novel function for HH signaling in determining the identity of midbrain signaling centers, with critical consequences for 3-dimensional midbrain patterning and for midbrain and hindbrain identity (Fig. 8).
Figure 8. Cartoon summarizing the role of SHH and FOXA2 in patterning the MFP, MHB and RP.
(i) Schematic depicting the normal spatial relationships among the RP (light blue), MFP (dark blue), LFP (green), MHB (dark brown) and dorsal midbrain (beige). (ii) Unilateral SHH misexpression (right side) ectopically induces LFP and can induce MFP along the MHB while concurrently suppressing MHB fates. Dorsal cell fates, including RP are also suppressed. (iii) Unilateral FOXA2 misexpression can ectopically induce MFP along the MHB as well as away from the MHB. (iv) In the absence of HH, the MFP can be converted into the MHB or the RP. (v) The loss of FOXA2 converts the ventral midbrain and the MFP into the MHB. As a consequence of ectopic MHB induction, midbrain fate (OTX2+) is suppressed (not shown) and hindbrain fate (GBX2+, red) is induced. (vi) FOXA2 overexpression combined with HH blockade results in ectopic RP induction throughout ventral midbrain.
The role of FOXA2 and SHH in FP specification
The role of SHH in MFP specification has been difficult to ascertain, with evidence ranging from an absolute requirement for HH signaling in the mouse, to a role in MFP maintenance in the fish (Blaess et al., 2006; Fedtsova et al., 2008; Fogel et al., 2008; Odenthal et al., 2000; Perez-Balaguer et al., 2009). In this study, we examined the precise requirement for HH signaling in midbrain MFP specification and show that it is required for ventralizing the MFP by turning on HH pathway and target genes (e.g., SHH, FOXA2, DISP1). However, it is insufficient for executing the full MFP program because it cannot turn on MFP-specific genes such as DISP1, MN-CAD, LMX1B, LMX1A and BMP7. We also show that HH blockade experiments between H&H 3–21 result in a loss of LFP, but do not eliminate all MFP markers (Bayly et al., 2007). Thus our data suggest that while HH signaling is necessary for MFP and LFP specification, it is only sufficient for LFP, but not MFP specification.
In this and previous work, we have also addressed the issue of the timing and duration of HH requirement (Bayly et al., 2007). Some studies have suggested that FP induction is accomplished at presomitic stages by a brief, but intense pulse of SHH (Patten et al., 2003; Ribes et al., 2010). Interestingly, we and others have shown that although SHH transcripts are present at the axial midline by H&H 3, the HH signal transduction machinery (PTC1, PTC2, HHIP, GLI1) is not present until H&H 5 (Aglyamova and Agarwala, 2007; Pearse et al., 2001). Surprisingly, the onset of SHH protein expression in the axial midline occurs later, at H&H 8−, when strong GLI1 and PTC1, transcriptional targets and readouts of HH signaling, are also transiently seen at the FP (H&H 8 to <H&H 10; Aglyamova and Agarwala, 2007; Pearse et al., 2001).
This post-somitic expression of SHH proteins in the chick is in contrast to the presomitic SHH expression reported in the mouse, and suggests that HH-mediated FP specification in the midbrain may occur at different time points in these species (Blaess et al., 2006; Fogel et al., 2008; Marti et al., 1995; Placzek and Briscoe, 2005; Ribes et al., 2010). Furthermore, neither the midbrain FP nor the subjacent notochord expresses the high levels of SHH seen at spinal cord levels (Ribes et al., 2010). However, it is still possible that midbrain FP patterning requires early and low/undetectable levels of the HH signaling emanating from the node or notochord (Jeong and McMahon, 2005; Placzek and Briscoe, 2005). Even so, HH signaling is still only necessary for the induction of HH-target/pathway genes at the ventral midline and not for the induction or blockade of MFP-specific markers such as LMX1B and BMP7. Thus the principal role of HH signaling in MFP patterning appears to be its ability to ventralize the FP. The precise molecular mechanisms underlying midbrain ventralization have not been extensively studied, but are likely to include the induction of HH pathway genes (DISP1, PTC1) and the exclusion of dorsal markers (PAX7) from the ventral neural tube (Bayly et al., 2007; Ericson et al., 1997; Fogel et al., 2008).
HH-independent mechanisms in the ventral midbrain patterning have also been noted in the Shh−/− and Smo−/− mice (Fogel et al., 2008). These mutants do not express Gli1 and Ptc1 in the FP and the entire ventral midbrain is dorsalized (Pax7+) by E9 (Blaess et al., 2006; Fogel et al., 2008). Although these results could be interpreted to signify an absolute requirement for HH signaling in the midbrain FP, a Pax7-negative ventral midbrain region is initially specified in the mouse, but it cannot be maintained beyond E9 without HH signaling (Blaess et al., 2006; Chiang et al., 1996; Fedtsova et al., 2008; Fogel et al., 2008). Taken together, these data may reconcile the apparent differences in chick and mouse midbrain FP specification by suggesting that the FP can be initiated in the absence of HH signaling, but requires HH signaling for its ventralization in both amniotes. A full comparison between the chick and mouse FP however would require knowing whether MFP-specific gene induction also occurs in the Shh−/− and Smo−/− mice, but these data are currently not available.
FOXA2 is both necessary and sufficient for turning on SHH in the midbrain, but accomplishes the ventral midline induction of LMX1B and BMP7 in a HH-independent manner. Its ability to induce LMX1B, a critical regulator of mDA neuron induction, may account for its greater potency in inducing mDA neurons (Andersson et al., 2006; Lin et al., 2009; Nakatani et al., 2010; Smidt et al., 2000; Yan et al., 2011). Whether FOXA2 interacts with the Nodal pathway in the chick midbrain is currently under study in our laboratory. Previous studies suggest that joint misexpression of Nodal and SHH is sufficient to induce “area a” cells to a FP fate, but whether FP subregions differentially depend upon Nodal and/or SHH signaling is not known (Patten et al., 2003). Our preliminary data suggest that multiple TGFβ ligands and their essential effectors (phosphorylated SMAD 2, 3) are active throughout the midbrain and are essential early regulators of FOXA2 expression in the FP (Amarnath et al., unpublished observations). Given their widespread distribution, any specific action of Nodal/TGFβ signaling within the FP is likely to be mediated via regulation of FOXA2 at the axial midline.
Since SHH can induce FOXA2, why is SHH not as potent as FOXA2 in MFP induction? One reason for this is that SHH protein is not expressed in the axial midline at early stages when FOXA2 protein expression is clearly detected (this study; Ruiz i Altaba et al., 1993). Since onset of LMX1B and BMP7 mRNA expression at the ventral midline occurs prior to SHH protein expression and FOXA2 can induce these and other MFP markers in a HH-independent manner, FOXA2 is likely to initiate the MFP program in the midbrain and not SHH (Amarnath et al., unpublished data; Yuan and Schoenwolf, 1999). However, once expressed, SHH must rapidly take control of FOXA2 expression since cyclopamine-treated explants collected at H&H 9, one stage (~90 min) after the onset of SHH protein expression, display severely reduced FOXA2 (Fig. 3F). This short time however is not sufficient to turn down LMX1B expression in such explants, despite the critical role of FOXA2 in its induction (Fig. 4A–H; Supplementary Fig. S6A–C).
The role of SHH and FOXA2 in regulating midbrain’s signaling centers
Signaling centers that pattern a given tissue are known to cross-regulate each other. Such crosstalk is a key to establishing a 3-dimensionally correct tissue pattern and has been noted between the signaling centers of the limb (Benazet and Zeller, 2009; Duboc and Logan, 2011). Evidence has also recently accumulated that the modulation of one of midbrain’s signaling centers affects the patterning or maintenance of other signaling centers (Aoto et al., 2002; Basson et al., 2008; Blaess et al., 2006; Blaess et al., 2008; Fogel et al., 2008).
A clear role for SHH in the maintenance of the MHB and suppression of the RP program has been established in the chick and/or mouse (Bayly et al., 2007; Blaess et al., 2006; Fogel et al., 2008). Ventral blockade of SHH and FOXA2 cause an expansion and ectopic induction of RP in dorsal midbrain in both chick and mouse, while their overexpression suppresses RP induction (data not shown; Fogel et al., 2008). Although this could be interpreted as a generic role for SHH in ventralization, our previous data demonstrate specific up and downregulation of dorsal midbrain genes by SHH (Fogel et al., 2008). Whether SHH regulates RP specification directly or indirectly is not known, although a transient, early wave of SHH activity floods and then recedes from the dorsal neural tube, and HH effectors, GLI1-GLI3 are all expressed dorsally (Aglyamova and Agarwala, 2007; Chamberlain et al., 2008). Finally, multiple HH-binding proteins and pathway members, which could potentially transduce HH signals (e.g., Megalin, BOC), are also expressed in dorsal midbrain (Aglyamova and Agarwala, 2007).
In this study, we show that ventral midline cells can become MFP or RP cells, depending on whether HH signaling is present or absent. Why only a handful of MFP cells take on the RP phenotype is not understood. Given the mosaic nature of FP gene expression, one possibility is that only a subset of LMX1B+ MFP cells are competent to turn on RP fates in the absence of HH signaling. Alternatively, we and others have noted the scarcity of HH-negative cells at the ventral midline following attempts to block HH signaling in this region (Bayly et al., 2007; Briscoe et al., 2001; Wijgerde et al., 2002). This may be due to the differential cell-affinity based segregation of HH-negative and HH+ cells as noted in the fly (Rodriguez and Basler, 1997). In this scenario, only a few HH-negative cells would remain at the ventral midline, although all of these would turn on RP fates in the absence of HH signaling.
The midbrain-hindbrain boundary
Previous studies suggest that although SHH is not required for MHB induction, it is required for maintaining its spatial coherence and signaling properties (Aoto et al., 2002; Bayly et al., 2007; Blaess et al., 2006; Blaess et al., 2008). In the mouse, the absence of SHH results in a progressive loss of FGF8 and a near complete loss of WNT1 in the MHB by E12.5 (Blaess et al., 2006; Fogel et al., 2008). Interestingly, GLI1-GLI3 and Megalin are all expressed within the chick MHB at various time points, and MHB regulation by GLI3 has been clearly demonstrated (Aglyamova and Agarwala, 2007; Aoto et al., 2002; Blaess et al., 2008). Here we extend these observations to show that overexpression of SHH blocks MHB induction by converting the MHB into a MFP fate. Together these results show that the MHB is sensitive to both increased and decreased levels of HH signaling, being converted to MFP in the presence of excess SHH and losing its signaling properties when SHH is downregulated.
More surprisingly, the absence of HH signaling can also convert the MFP into the MHB. In such experiments, the MHB can also be converted into a GDF7+/WNT1+ RP. Fate mapping and lineage analyses suggest a common set of precursors for the MHB and the RP, which might explain their inter-convertibility in our experiments (Alexandre and Wassef, 2003; Zervas et al., 2005). To our knowledge, no common RP/MFP or MFP/MHB precursors have been described. However, as we demonstrate in Fig. 1K, L, all 3 signaling centers are physically contiguous and cell exchange among them is therefore feasible. We also note that all midbrain signaling centers express a common set of developmental control genes (BMP7, LMX1B). Although the significance of BMP7 expression in all 3 signaling centers is not understood, LMX1B is clearly involved in FP, MHB and RP specification (Adams et al., 2000; Chizhikov and Millen, 2004; Matsunaga et al., 2002; Millen et al., 2004; O’Hara et al., 2005; Yan et al., 2011). Thus, LMX1B may be a key regulator of midbrain signaling center induction and may specify the MFP, MHB or RP depending upon the specific cellular context. If so, a key component of that cellular context would be the presence or absence of SHH.
Supplementary Material
Highlights.
We identify medial and lateral subdivisions in avian midbrain and spinal floor plate
These subdivisions differentially require FOXA2 and SHH for their induction
Only FOXA2 induces the full floor plate via SHH independent and dependent mechanisms
The medial region shares gene expression with other midbrain signaling centers
Midbrain signaling centers are interconvertible and depend on SHH for their identity >
Acknowledgments
We thank Drs. J. Briscoe, O. Conneely, M. Goulding, C. Goridis, A. Graham, B. Hogan, B. Houston, T. Jessell, J. Lahti, S. Lamont, J. Lewis, G. Martin, A. McMahon, C. Ragsdale, H. Sasaki, C. Tabin, B. Vogelstein, M. Wassef for DNA reagents and J. Gross, T. Shimogori and J. Eberhardt for critical comments. This research was supported by a grant from NIH-NINDS to SA and a graduate student continuing fellowship awarded to RB from the University of Texas, Austin.
Abbreviations
- bi
bilaterally electroporated
- CYC
cyclopamine
- DI
diencephalon
- E
embryonic day
- EP
electroporated
- FP
floor plate
- HH
hedgehog
- H&H
Hamburger & Hamilton
- HB
hindbrain
- hn
Henson’s Node
- LFP
lateral floor plate
- MB
midbrain
- me
mesendoderm
- MFP
medial floor plate
- MHB
midbrain-hindbrain boundary
- ne
neurectoderm
- RP
roof plate
- TEC
tectum
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Adams KA, Maida JM, Golden JA, Riddle RD. The transcription factor Lmx1b maintains Wnt1 expression within the isthmic organizer. Development. 2000;127:1857–67. doi: 10.1242/dev.127.9.1857. [DOI] [PubMed] [Google Scholar]
- Afonso C, Henrique D. PAR3 acts as a molecular organizer to define the apical domain of chick neuroepithelial cells. J Cell Sci. 2006;119:4293–304. doi: 10.1242/jcs.03170. [DOI] [PubMed] [Google Scholar]
- Agarwala S, Ragsdale CW. A role for midbrain arcs in nucleogenesis. Development. 2002;129:5779–88. doi: 10.1242/dev.00179. [DOI] [PubMed] [Google Scholar]
- Agarwala S, Sanders TA, Ragsdale CW. Sonic hedgehog control of size and shape in midbrain pattern formation. Science. 2001;291:2147–50. doi: 10.1126/science.1058624. [DOI] [PubMed] [Google Scholar]
- Aglyamova GV, Agarwala S. Gene expression analysis of the hedgehog signaling cascade in the chick midbrain and spinal cord. Dev Dyn. 2007;236:1363–73. doi: 10.1002/dvdy.21146. [DOI] [PubMed] [Google Scholar]
- Alexandre P, Wassef M. The isthmic organizer links anteroposterior and dorsoventral patterning in the mid/hindbrain by generating roof plate structures. Development. 2003;130:5331–8. doi: 10.1242/dev.00756. [DOI] [PubMed] [Google Scholar]
- Andersson E, Tryggvason U, Deng Q, Friling S, Alekseenko Z, Robert B, Perlmann T, Ericson J. Identification of intrinsic determinants of midbrain dopamine neurons. Cell. 2006;124:393–405. doi: 10.1016/j.cell.2005.10.037. [DOI] [PubMed] [Google Scholar]
- Ang SL, Rossant J. HNF-3 beta is essential for node and notochord formation in mouse development. Cell. 1994;78:561–74. doi: 10.1016/0092-8674(94)90522-3. [DOI] [PubMed] [Google Scholar]
- Aoto K, Nishimura T, Eto K, Motoyama J. Mouse GLI3 regulates Fgf8 expression and apoptosis in the developing neural tube, face, and limb bud. Dev Biol. 2002;251:320–32. doi: 10.1006/dbio.2002.0811. [DOI] [PubMed] [Google Scholar]
- Basson MA, Echevarria D, Ahn CP, Sudarov A, Joyner AL, Mason IJ, Martinez S, Martin GR. Specific regions within the embryonic midbrain and cerebellum require different levels of FGF signaling during development. Development. 2008;135:889–98. doi: 10.1242/dev.011569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bayly RD, Ngo M, Aglyamova GV, Agarwala S. Regulation of ventral midbrain patterning by Hedgehog signaling. Development. 2007;134:2115–24. doi: 10.1242/dev.02850. [DOI] [PubMed] [Google Scholar]
- Benazet JD, Zeller R. Vertebrate limb development: moving from classical morphogen gradients to an integrated 4-dimensional patterning system. Cold Spring Harb Perspect Biol. 2009;1:a001339. doi: 10.1101/cshperspect.a001339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blaess S, Bodea GO, Kabanova A, Chanet S, Mugniery E, Derouiche A, Stephen D, Joyner AL. Temporal-spatial changes in Sonic Hedgehog expression and signaling reveal different potentials of ventral mesencephalic progenitors to populate distinct ventral midbrain nuclei. Neural Dev. 2011;6:29. doi: 10.1186/1749-8104-6-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blaess S, Corrales JD, Joyner AL. Sonic hedgehog regulates Gli activator and repressor functions with spatial and temporal precision in the mid/hindbrain region. Development. 2006;133:1799–809. doi: 10.1242/dev.02339. [DOI] [PubMed] [Google Scholar]
- Blaess S, Stephen D, Joyner AL. Gli3 coordinates three-dimensional patterning and growth of the tectum and cerebellum by integrating Shh and Fgf8 signaling. Development. 2008;135:2093–103. doi: 10.1242/dev.015990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Briscoe J, Chen Y, Jessell TM, Struhl G. A hedgehog-insensitive form of patched provides evidence for direct long-range morphogen activity of sonic hedgehog in the neural tube. Mol Cell. 2001;7:1279–91. doi: 10.1016/s1097-2765(01)00271-4. [DOI] [PubMed] [Google Scholar]
- Brown CY, Eom DS, Amarnath S, Agarwala S. A simple technique for early in vivo electroporation of E1 chick embryos. Dev Dyn. 2012;241:545–52. doi: 10.1002/dvdy.23747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Catala M, Ziller C, Lapointe F, Le Douarin NM. The developmental potentials of the caudalmost part of the neural crest are restricted to melanocytes and glia. Mech Dev. 2000;95:77–87. doi: 10.1016/s0925-4773(00)00349-x. [DOI] [PubMed] [Google Scholar]
- Chamberlain CE, Jeong J, Guo C, Allen BL, McMahon AP. Notochord-derived Shh concentrates in close association with the apically positioned basal body in neural target cells and forms a dynamic gradient during neural patterning. Development. 2008;135:1097–106. doi: 10.1242/dev.013086. [DOI] [PubMed] [Google Scholar]
- Charrier JB, Lapointe F, Le Douarin NM, Teillet MA. Dual origin of the floor plate in the avian embryo. Development. 2002;129:4785–96. doi: 10.1242/dev.129.20.4785. [DOI] [PubMed] [Google Scholar]
- Charron F, Stein E, Jeong J, McMahon AP, Tessier-Lavigne M. The morphogen sonic hedgehog is an axonal chemoattractant that collaborates with netrin-1 in midline axon guidance. Cell. 2003;113:11–23. doi: 10.1016/s0092-8674(03)00199-5. [DOI] [PubMed] [Google Scholar]
- Chiang C, Litingtung Y, Lee E, Young KE, Corden JL, Westphal H, Beachy PA. Cyclopia and defective axial patterning in mice lacking Sonic hedgehog gene function. Nature. 1996;383:407–13. doi: 10.1038/383407a0. [DOI] [PubMed] [Google Scholar]
- Chizhikov VV, Millen KJ. Control of roof plate development and signaling by Lmx1b in the caudal vertebrate CNS. J Neurosci. 2004;24:5694–703. doi: 10.1523/JNEUROSCI.0758-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dal-Pra S, Thisse C, Thisse B. FoxA transcription factors are essential for the development of dorsal axial structures. Dev Biol. 2011;350:484–95. doi: 10.1016/j.ydbio.2010.12.018. [DOI] [PubMed] [Google Scholar]
- Duboc V, Logan MP. Regulation of limb bud initiation and limb-type morphology. Dev Dyn. 2011;240:1017–27. doi: 10.1002/dvdy.22582. [DOI] [PubMed] [Google Scholar]
- Echelard Y, Epstein DJ, St-Jacques B, Shen L, Mohler J, McMahon JA, McMahon AP. Sonic hedgehog, a member of a family of putative signaling molecules, is implicated in the regulation of CNS polarity. Cell. 1993;75:1417–30. doi: 10.1016/0092-8674(93)90627-3. [DOI] [PubMed] [Google Scholar]
- Eom DS, Amarnath S, Fogel JL, Agarwala S. Bone morphogenetic proteins regulate neural tube closure by interacting with the apicobasal polarity pathway. Development. 2011;138:3179–88. doi: 10.1242/dev.058602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ericson J, Rashbass P, Schedl A, Brenner-Morton S, Kawakami A, van Heyningen V, Jessell TM, Briscoe J. Pax6 controls progenitor cell identity and neuronal fate in response to graded Shh signaling. Cell. 1997;90:169–80. doi: 10.1016/s0092-8674(00)80323-2. [DOI] [PubMed] [Google Scholar]
- Fedtsova N, Quina LA, Wang S, Turner EE. Regulation of the development of tectal neurons and their projections by transcription factors Brn3a and Pax7. Dev Biol. 2008;316:6–20. doi: 10.1016/j.ydbio.2007.12.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferri AL, Lin W, Mavromatakis YE, Wang JC, Sasaki H, Whitsett JA, Ang SL. Foxa1 and Foxa2 regulate multiple phases of midbrain dopaminergic neuron development in a dosage-dependent manner. Development. 2007;134:2761–9. doi: 10.1242/dev.000141. [DOI] [PubMed] [Google Scholar]
- Fogel JL, Chiang C, Huang X, Agarwala S. Ventral specification and perturbed boundary formation in the mouse midbrain in the absence of Hedgehog signaling. Dev Dyn. 2008;237:1359–72. doi: 10.1002/dvdy.21536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray SD, Dale JK. Notch signalling regulates the contribution of progenitor cells from the chick Hensen’s node to the floor plate and notochord. Development. 2010;137:561–8. doi: 10.1242/dev.041608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamburger V, Hamilton HL. A series of normal stages in the development of the chick embryo. J Morphol. 1951;88:49–92. doi: 10.1002/aja.1001950404. [DOI] [PubMed] [Google Scholar]
- Hatta K, Kimmel CB, Ho RK, Walker C. The cyclops mutation blocks specification of the floor plate of the zebrafish central nervous system. Nature. 1991;350:339–41. doi: 10.1038/350339a0. [DOI] [PubMed] [Google Scholar]
- His W. Geschichte des Gehirns, sowie der centralen und peripherischen Nervenbahnen beim menschlichen embryo. Abd d math phys Kl d Konigl Saschsischen Gesellschaft d Wiss. 1888:341–392. [Google Scholar]
- Hynes M, Ye W, Wang K, Stone D, Murone M, Sauvage F, Rosenthal A. The seven-transmembrane receptor smoothened cell-autonomously induces multiple ventral cell types. Nat Neurosci. 2000;3:41–6. doi: 10.1038/71114. [DOI] [PubMed] [Google Scholar]
- Incardona JP, Gaffield W, Kapur RP, Roelink H. The teratogenic Veratrum alkaloid cyclopamine inhibits sonic hedgehog signal transduction. Development. 1998;125:3553–62. doi: 10.1242/dev.125.18.3553. [DOI] [PubMed] [Google Scholar]
- Jacob J, Ferri AL, Milton C, Prin F, Pla P, Lin W, Gavalas A, Ang SL, Briscoe J. Transcriptional repression coordinates the temporal switch from motor to serotonergic neurogenesis. Nat Neurosci. 2007;10:1433–9. doi: 10.1038/nn1985. [DOI] [PubMed] [Google Scholar]
- Jeong J, McMahon AP. Growth and pattern of the mammalian neural tube are governed by partially overlapping feedback activities of the hedgehog antagonists patched 1 and Hhip1. Development. 2005;132:143–54. doi: 10.1242/dev.01566. [DOI] [PubMed] [Google Scholar]
- Joksimovic M, Anderegg A, Roy A, Campochiaro L, Yun B, Kittappa R, McKay R, Awatramani R. Spatiotemporally separable Shh domains in the midbrain define distinct dopaminergic progenitor pools. Proc Natl Acad Sci U S A. 2009;106:19185–90. doi: 10.1073/pnas.0904285106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kingsbury BF. The extent of the floor plate of His and its significance. J Comp Neurol. 1920:113–135. [Google Scholar]
- Kittappa R, Chang WW, Awatramani RB, McKay RD. The foxa2 gene controls the birth and spontaneous degeneration of dopamine neurons in old age. PLoS Biol. 2007;5:e325. doi: 10.1371/journal.pbio.0050325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawrence PA. Developmental biology. Straight and wiggly affinities. Nature. 1997;389:546–7. doi: 10.1038/39188. [DOI] [PubMed] [Google Scholar]
- Lawson A, Colas JF, Schoenwolf GC. Classification scheme for genes expressed during formation and progression of the avian primitive streak. Anat Rec. 2001;262:221–6. doi: 10.1002/1097-0185(20010201)262:2<221::AID-AR1019>3.0.CO;2-F. [DOI] [PubMed] [Google Scholar]
- Lin W, Metzakopian E, Mavromatakis YE, Gao N, Balaskas N, Sasaki H, Briscoe J, Whitsett JA, Goulding M, Kaestner KH, Ang SL. Foxa1 and Foxa2 function both upstream of and cooperatively with Lmx1a and Lmx1b in a feedforward loop promoting mesodiencephalic dopaminergic neuron development. Dev Biol. 2009;333:386–96. doi: 10.1016/j.ydbio.2009.07.006. [DOI] [PubMed] [Google Scholar]
- Liu A, Losos K, Joyner AL. FGF8 can activate Gbx2 and transform regions of the rostral mouse brain into a hindbrain fate. Development. 1999;126:4827–38. doi: 10.1242/dev.126.21.4827. [DOI] [PubMed] [Google Scholar]
- Marti E, Takada R, Bumcrot DA, Sasaki H, McMahon AP. Distribution of Sonic hedgehog peptides in the developing chick and mouse embryo. Development. 1995;121:2537–47. doi: 10.1242/dev.121.8.2537. [DOI] [PubMed] [Google Scholar]
- Matsunaga E, Katahira T, Nakamura H. Role of Lmx1b and Wnt1 in mesencephalon and metencephalon development. Development. 2002;129:5269–77. doi: 10.1242/dev.129.22.5269. [DOI] [PubMed] [Google Scholar]
- Millen KJ, Millonig JH, Hatten ME. Roof plate and dorsal spinal cord dl1 interneuron development in the dreher mutant mouse. Dev Biol. 2004;270:382–92. doi: 10.1016/j.ydbio.2004.03.008. [DOI] [PubMed] [Google Scholar]
- Momose T, Tonegawa A, Takeuchi J, Ogawa H, Umesono K, Yasuda K. Efficient targeting of gene expression in chick embryos by microelectroporation. Dev Growth Differ. 1999;41:335–44. doi: 10.1046/j.1440-169x.1999.413437.x. [DOI] [PubMed] [Google Scholar]
- Nakatani T, Kumai M, Mizuhara E, Minaki Y, Ono Y. Lmx1a and Lmx1b cooperate with Foxa2 to coordinate the specification of dopaminergic neurons and control of floor plate cell differentiation in the developing mesencephalon. Dev Biol. 2010;339:101–13. doi: 10.1016/j.ydbio.2009.12.017. [DOI] [PubMed] [Google Scholar]
- Norton WH, Mangoli M, Lele Z, Pogoda HM, Diamond B, Mercurio S, Russell C, Teraoka H, Stickney HL, Rauch GJ, Heisenberg CP, Houart C, Schilling TF, Frohnhoefer HG, Rastegar S, Neumann CJ, Gardiner RM, Strahle U, Geisler R, Rees M, Talbot WS, Wilson SW. Monorail/Foxa2 regulates floorplate differentiation and specification of oligodendrocytes, serotonergic raphe neurones and cranial motoneurones. Development. 2005;132:645–58. doi: 10.1242/dev.01611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Hara FP, Beck E, Barr LK, Wong LL, Kessler DS, Riddle RD. Zebrafish Lmx1b.1 and Lmx1b.2 are required for maintenance of the isthmic organizer. Development. 2005;132:3163–73. doi: 10.1242/dev.01898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Odenthal J, van Eeden FJ, Haffter P, Ingham PW, Nusslein-Volhard C. Two distinct cell populations in the floor plate of the zebrafish are induced by different pathways. Dev Biol. 2000;219:350–63. doi: 10.1006/dbio.1999.9589. [DOI] [PubMed] [Google Scholar]
- Ono Y, Nakatani T, Sakamoto Y, Mizuhara E, Minaki Y, Kumai M, Hamaguchi A, Nishimura M, Inoue Y, Hayashi H, Takahashi J, Imai T. Differences in neurogenic potential in floor plate cells along an anteroposterior location: midbrain dopaminergic neurons originate from mesencephalic floor plate cells. Development. 2007;134:3213–25. doi: 10.1242/dev.02879. [DOI] [PubMed] [Google Scholar]
- Patten I, Kulesa P, Shen MM, Fraser S, Placzek M. Distinct modes of floor plate induction in the chick embryo. Development. 2003;130:4809–21. doi: 10.1242/dev.00694. [DOI] [PubMed] [Google Scholar]
- Pearse RV, 2nd, Vogan KJ, Tabin CJ. Ptc1 and Ptc2 transcripts provide distinct readouts of Hedgehog signaling activity during chick embryogenesis. Dev Biol. 2001;239:15–29. doi: 10.1006/dbio.2001.0430. [DOI] [PubMed] [Google Scholar]
- Perez-Balaguer A, Puelles E, Wurst W, Martinez S. Shh dependent and independent maintenance of basal midbrain. Mech Dev. 2009;126:301–13. doi: 10.1016/j.mod.2009.03.001. [DOI] [PubMed] [Google Scholar]
- Peyrot SM, Wallingford JB, Harland RM. A revised model of Xenopus dorsal midline development: differential and separable requirements for Notch and Shh signaling. Dev Biol. 2011;352:254–66. doi: 10.1016/j.ydbio.2011.01.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Placzek M, Briscoe J. The floor plate: multiple cells, multiple signals. Nat Rev Neurosci. 2005;6:230–40. doi: 10.1038/nrn1628. [DOI] [PubMed] [Google Scholar]
- Rastegar S, Albert S, Le Roux I, Fischer N, Blader P, Muller F, Strahle U. A floor plate enhancer of the zebrafish netrin1 gene requires Cyclops (Nodal) signalling and the winged helix transcription factor FoxA2. Dev Biol. 2002;252:1–14. doi: 10.1006/dbio.2002.0837. [DOI] [PubMed] [Google Scholar]
- Ribes V, Balaskas N, Sasai N, Cruz C, Dessaud E, Cayuso J, Tozer S, Yang LL, Novitch B, Marti E, Briscoe J. Distinct Sonic Hedgehog signaling dynamics specify floor plate and ventral neuronal progenitors in the vertebrate neural tube. Genes Dev. 2010;24:1186–200. doi: 10.1101/gad.559910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez I, Basler K. Control of compartmental affinity boundaries by hedgehog. Nature. 1997;389:614–8. doi: 10.1038/39343. [DOI] [PubMed] [Google Scholar]
- Ruiz i Altaba A, Prezioso VR, Darnell JE, Jessell TM. Sequential expression of HNF-3 beta and HNF-3 alpha by embryonic organizing centers: the dorsal lip/node, notochord and floor plate. Mech Dev. 1993;44:91–108. doi: 10.1016/0925-4773(93)90060-b. [DOI] [PubMed] [Google Scholar]
- Sasaki H, Hogan BL. Differential expression of multiple fork head related genes during gastrulation and axial pattern formation in the mouse embryo. Development. 1993;118:47–59. doi: 10.1242/dev.118.1.47. [DOI] [PubMed] [Google Scholar]
- Sasaki H, Hogan BL. HNF-3 beta as a regulator of floor plate development. Cell. 1994;76:103–15. doi: 10.1016/0092-8674(94)90176-7. [DOI] [PubMed] [Google Scholar]
- Sasaki H, Hui C, Nakafuku M, Kondoh H. A binding site for Gli proteins is essential for HNF-3beta floor plate enhancer activity in transgenics and can respond to Shh in vitro. Development. 1997;124:1313–22. doi: 10.1242/dev.124.7.1313. [DOI] [PubMed] [Google Scholar]
- Schauerte HE, van Eeden FJ, Fricke C, Odenthal J, Strahle U, Haffter P. Sonic hedgehog is not required for the induction of medial floor plate cells in the zebrafish. Development. 1998;125:2983–93. doi: 10.1242/dev.125.15.2983. [DOI] [PubMed] [Google Scholar]
- Schoenwolf GC, Sheard P. Fate mapping the avian epiblast with focal injections of a fluorescent-histochemical marker: ectodermal derivatives. J Exp Zool. 1990;255:323–39. doi: 10.1002/jez.1402550309. [DOI] [PubMed] [Google Scholar]
- Smidt MP, Asbreuk CH, Cox JJ, Chen H, Johnson RL, Burbach JP. A second independent pathway for development of mesencephalic dopaminergic neurons requires Lmx1b. Nat Neurosci. 2000;3:337–41. doi: 10.1038/73902. [DOI] [PubMed] [Google Scholar]
- Strahle U, Lam CS, Ertzer R, Rastegar S. Vertebrate floor-plate specification: variations on common themes. Trends Genet. 2004;20:155–62. doi: 10.1016/j.tig.2004.01.002. [DOI] [PubMed] [Google Scholar]
- Wijgerde M, McMahon JA, Rule M, McMahon AP. A direct requirement for Hedgehog signaling for normal specification of all ventral progenitor domains in the presumptive mammalian spinal cord. Genes Dev. 2002;16:2849–64. doi: 10.1101/gad.1025702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan CH, Levesque M, Claxton S, Johnson RL, Ang SL. Lmx1a and lmx1b function cooperatively to regulate proliferation, specification, and differentiation of midbrain dopaminergic progenitors. J Neurosci. 2011;31:12413–25. doi: 10.1523/JNEUROSCI.1077-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye W, Shimamura K, Rubenstein JL, Hynes MA, Rosenthal A. FGF and Shh signals control dopaminergic and serotonergic cell fate in the anterior neural plate. Cell. 1998;93:755–66. doi: 10.1016/s0092-8674(00)81437-3. [DOI] [PubMed] [Google Scholar]
- Yuan S, Schoenwolf GC. The spatial and temporal pattern of C-Lmx1 expression in the neuroectoderm during chick neurulation. Mech Dev. 1999;88:243–7. doi: 10.1016/s0925-4773(99)00185-9. [DOI] [PubMed] [Google Scholar]
- Zervas M, Blaess S, Joyner AL. Classical embryological studies and modern genetic analysis of midbrain and cerebellum development. Curr Top Dev Biol. 2005;69:101–38. doi: 10.1016/S0070-2153(05)69005-9. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.