Abstract
Francisella tularensis causes the zoonosis tularemia in humans and is one of the most virulent bacterial pathogens. We utilized a global proteomic approach to characterize protein changes in bronchoalveolar lavage fluid from mice exposed to one of three organisms, F. tularensis ssp. novicida, an avirulent mutant of F. tularensis ssp. novicida (F.t. novicida-ΔmglA); and Pseudomonas aeruginosa. The composition of BALF proteins was altered following infection, including proteins involved in neutrophil activation, oxidative stress and inflammatory responses. Components of the innate immune response were induced including the acute phase response and the complement system, however the timing of their induction varied. Francisella tularensis ssp. novicida infected mice do not appear to have an effective innate immune response in the first hours of infection, however within 24 hours they show an upregulation of innate immune response proteins. This delayed response is in contrast to P. aeruginosa infected animals which show an early innate immune response. Likewise, F.t. novicida-ΔmglA infection initiates an early innate immune response, however this response is dimished by 24 hours. Finally, this study identifies several candidate biomarkers, including Chitinase 3-like-1 (CHI3L1 or YKL-40) and peroxiredoxin 1, that are associated with F. tularensis ssp. novicida but not P. aeruginosa infection.
Keywords: innate immunity, Francisella tularensis, proteomics, bronchoalveolar lavage fluid
Introduction
Francisella tularensis is an intracellular, gram-negative coccobacillus that causes tularemia. F. tularensis is classified as a category A bioterrorism agent by the U.S. Centers for Disease Control and Prevention due to its high infectivity, ability to spread via an airborne route, and its capacity to cause significant morbidity and mortality. Inhalation of fewer than 10 bacteria can cause human disease1,2. Humans can acquire tularemia through direct exposure to infected animals or from the bite of a blood-feeding arthropod.
F. tularensis is divided into four subspecies. F. tularensis subsp. tularensis (type A) and F. tularensis subsp. holarctica (type B) are typically associated with human disease. F. tularensis subsp. mediasiatica is of relatively low virulence. Lastly, F. tularensis subsp. novicida (F.t. novicida) is infectious only for immunocompromised humans and lethal to mice at even small doses. A partially attenuated live vaccine strain (LVS) was developed from a type B strain of F.tularensis and is attenuated in humans but virulent in mice. The mouse, a natural host for F. tularensis, serves as an excellent experimental model for tularemia. While all Francisella subspecies are virulent in mice, F.t. novicida has proven useful to investigate the host-pathogen interactions both because of its ease of genetic manipulation and because the pathogenesis of infection in mice is largely similar to human infection3,4.
F. tularensis infection initiates both specific and non-specific host responses. PMNs are recruited to the site of infection within hours, albeit not as vigorously as after infection with other Gram negative bacilli5,6, at least in part because F. tularensis lipopolysaccaride (LPS) is poorly recognized by Toll-like receptor 47. Macrophages and dendritic cells appear to be the major reservoir and site of replication of F. tularensis in the host8,9. Following the infection of macrophage or dendritic cells, virulent strains of F. tularensis actively suppress the ability of these cells to secrete proinflammatory cytokines10–12.
Macrophage receptors mediate phagocytosis and initiate a wide range of responses, including inflammatory and signaling cascades. The specific macrophage receptors responsible for the uptake and survival of F. tularensis are poorly characterized. Recent work demonstrated that the bacterium enters human macrophages by a process of engulfment within asymmetric pseudopod loops. Adherence and uptake by the macrophages is dependent upon complement C3, complement receptors and actin microfilaments13. Further studies indicated a role for complement receptor 3, mannose receptor and surfactant protein A in fostering the association of F. tularensis and human macrophages14. Following bacterium uptake into the macrophage, F.t. novicida and LVS both occupy a membrane-bound compartment within the macrophage that is not fused with lysosomes4. Additionally, growth of LVS in mouse macrophages requires an acidified compartment that facilitates the availability of iron essential for bacterial growth15.
Several virulence factors are required for growth of F. tularensis within macrophages, including macrophage growth locus A (mglA). Disruption of mglA inhibits phagosome escape and decreases virulence in mice16–18. MglA and a related regulator, SspA, positively regulate the expression of genes encoded by the Francisella pathogenicity island (FPI) and additional genes. Previous proteomic studies indicated that MglA influences the expression of F.t. novicida proteins involved in responding to stress, including oxidative stress19.
In this work, a comparative proteomic study of lung lavage fluid was undertaken using a mouse model of inhalation tularemia, induced by Francisella tularensis subspecies novicida. To better understand the initial pulmonary host response to virulent F. tularensis, mice were also exposed to F.t. novicida ΔmglA, an attenuated mutant of F.t. novicida defective for intracellular replication. To place these studies in a broader context of acute Gram negative bacterial infection, mice also were challenged with Pseudomonas aeruginosa, an extracellular pathogen that elicits a rapid and vigorous host response6,20. This work expands on our previous report illustrating that small protein and peptide profiles detected by matrix assisted laser desorption/ionization mass spectrometry (MALDI-MS) could be used to differentiate lavage fluid samples from different exposures. The detected immune cells counts were used to supplement the conclusions from the MALDI-MS analysis. While the MALDI-MS analysis illustrated the potential for using a limited subset of detected proteins for differentiation, the detected masses provided no protein identification and therefore no insights into the different host response pathways involved. To better understand the host response pathways involved, samples from the same study were used in a bottom-up proteomic analysis to compare the protein identities and relative abundance changes between sample treatments. This type of analysis provides more insight into the collection of proteins and indications of host response pathways that are impacted across the proteome of the lung surface.
Materials and Methods
Bacterial strains and growth conditions
Francisella tularensis subspecies novicida U112 (F.t. novicida) and the F. novicida ΔmglA mutant (F.t. novicida ΔmglA) were provided by Dr. Francis Nano (University of Victoria, Canada). Pseudomonas aeruginosa PAK (P. aeruginosa) was obtained from Dr. Steve Lory (Harvard University). Glycerol stocks were inoculated 1:1000 into nutrient broth (TSB-0.1% L-cysteine for F. t. novicida strains and LB for P. aeruginosa) and incubated for 16h at 37°C in a rotating platform incubator at 200 rpm. Bacteria were pelleted by centrifugation at 4° C, washed twice with PBS and resuspended in cold PBS to a concentration of 5 × 109 CFU/ml as estimated by OD540.
Animals
This study was carried out in strict accordance with the recommendations in the Guide for the Care and Use of Laboratory Animals of the National Institutes of Health. The Institutional Animal Care and Use Committee of the University of Washington approved all experimental procedures under protocols 2671-06 and 2671-07. Specific-pathogen-free male C57BL/6 mice were obtained from Jackson Laboratories (Bar Harbor, ME) and were 8 weeks old at the time of study. All animals were housed in laminar flow cages and were permitted “ad libitum” access to sterile food and water. Euthanasia was accomplished with intraperitoneal pentobarbital followed by exsanguination from cardiac puncture and all efforts were made to minimize suffering.
Infection of animals
Mice were exposed to aerosolized bacteria or PBS using a snout-only inhalation system (In-Tox products, Moriarty, NM). Aerosols were generated from MiniHEART hi-flo nebulizers (Westmed, Tucson, AZ) driven at 40 psi. Airflow through the system was maintained at 20 L/min for a 10 minute aerosol exposure, followed by a 5 minute purge with air. Aerosol exposure was conducted over two days following a schedule as outlined in Wunschel et al6. Bacterial deposition in each experiment was determined from quantitative culture of the left lungs harvested from three mice sacrificed immediately after exposure. Measured bacterial depositions (CFU/lung) are as follows (mean ± SEM, n=3): F. t. novicida experiment 1: 4248 ± 1215; F. t. novicida experiment 2: 3030 ± 214; F. t. novicida ΔmglA experiment 1: 3827 ± 815; F. t. novicida ΔmglA experiment 2: 1262 ± 408; and P. aeruginosa 126127 ± 14239. Animals were examined daily for signs of illness, and body surface temperatures were monitored with an infrared thermometer (Raytek, Santa Cruz, CA). No animals developed severe hypothermia (<25°C) or other signs of morbidity warranting euthanasia (ruffled fur, eye crusting, hunched posture, and lack of resistance to handling) during the 48 hour period of observation. Previous studies have shown that the infecting doses of F.t. novicida used in these experiments results in exponential bacterial replication, widespread dissemination of infection, and uniform lethality within 96 hour 18. In contrast, airborne infection with F.t. novicida ΔmglA or P. aeruginosa under the conditions tested results in limited bacterial replication and no mortality 18,20.
BALF collection
Mice were euthanized to harvest bronchoalveolar lavage fluid (BALF) immediately, 4h, 24h, and 48h after inhalation of PBS; 4h, 24h, and 48h after inhalation of F. novicida and F. novicida ΔmglA; and 4h and 24h after inhalation of P. aeruginosa. The trachea was cannulated with a 22g catheter and both lungs were lavaged by instilling 1.0 mL of pre-warmed 0.9% sodium chloride containing 0.6 mM EDTA, which was retrieved after a 30 second dwell time. This procedure was repeated with three more 0.8 ml volumes of saline/EDTA. All four volumes of BALF from each mouse were pooled and centrifuged at 300 × g for 10 minutes at room temperature. The supernatants were harvested, phenylmethylsulfonyl fluoride (PMSF, Sigma-Aldrich, St. Louis, MO) added to give a final concentration of 1 mM, and stored at −80°C.
Precipitation and trypsin digestion of BALF proteins
To concentrate BALF sample proteins prior to trypsin digestion, ice-cold trichloroacetic acid (TCA) was added to the samples to a final concentration of 10%. All samples were incubated at 4°C overnight followed by centrifugation at 14K RCF for 5 minutes. The pellet was washed one time with cold acetone and allowed to dry at room temperature for 5 minutes. The protein pellet was resuspended in 25 uL of denaturing buffer (100 mM ammonium bicarbonate, 8M urea, 2 M thiourea and 5 mM dithiothreitol (DTT) and heated to 60°C for 30 min. The sample was then diluted 4-fold with 100 mM ammonium bicarbonate and CaCl2 was added to 1 mM. Methylated, sequencing-grade trypsin (Promega, Madison, WI) was added at a substrate-to-enzyme ratio of 50:1 (mass:mass) and incubated at 37°C for 15 hours. Sample cleanup was achieved using a 1-mL SPE C17 column (Supelco, Bellefonte, PA). The peptides were eluted from each column with 1 mL of methanol and concentrated via SpeedVac. The samples were reconstituted to 1 µg/µL with 25 mM ammonium bicarbonate and frozen at −20°C until analyzed.
LC-MS/MS and LC-MS Analysis
Peptide samples were analyzed using a custom-built automated high pressure nanocapillary LC system21 coupled on-line to a LTQ-Orbitrap XL mass spectrometer (Thermo Fisher Scientific). The reversed-phase capillary column was prepared by slurry packing 3-µm Jupiter C18 bonded particles (Phenomenex, Torrence, CA) into a 65-cm-long, 75-µm-inner diameter fused silica capillary (Polymicron Technologies, Phoenix, AZ). The mobile phase consisted of 0.2% acetic acid and 0.05% TFA in water (A) and 0.1% TFA in 90% acetonitrile, 10% water (B). After loading 5 µL of peptides onto the column, the mobile phase was held at 100% A for 50 min. Exponential gradient elution was performed by increasing the mobile phase composition from 0 to 55% B over 115 min. The HPLC column was coupled to the mass spectrometer by an in-house manufactured electrospray ionization interface 22. The heated capillary temperature and spray voltage were 200°C and 2.2kV, respectively. Data were acquired for 100 min, beginning 65 min after sample injection (15 min into gradient). Orbitrap spectra (AGC 5×105) were collected from 400–2000 m/z at a resolution of 100K followed by data dependent ion trap MS/MS spectra (AGC 1×104) of the three or six most abundant ions using a collision energy of 35%. A dynamic exclusion time of 60 sec was used to discriminate against previously analyzed ions.
The MS/MS spectra were analyzed using the peptide identification software SEQUEST 23 to independently search all MS/MS spectra against two organism protein FASTA data files: the mouse International Protein Index (IPI) database (version 3.51, released November 5th, 2008) and Francisella tularensis novicida U112 (obtained from NCBI, NC_008601). For each SEQUEST analysis of an MS/MS spectrum (at a given parent ion charge state), only the peptide identification with the highest XCorr value (i.e., the “top ranked hit”) was retained. In addition, the SEQUEST peptide identifications were required to satisfy Washburn-Yates criteria24. Specifically, ΔCn ≥ 0.1 was required and, for each parent ion charge state, XCorr was required to be ≥ 1.9 (+1), 2.2 (+2), 3.75 (≥ +3). The Proteomics Research Information Storage and Management system was used to run SEQUEST, to run software that calculated predicted and observed normalized elution time (NET) values, and to manage the proteomics data in general25.
The high resolution mass spectral datasets were analyzed using the accurate mass and elution time (AMT) tag approach26. The peak lists for each analysis were matched against a mass tag database constructed from the LC-MS/MS analyses of the BALF samples from this study, combined with LC-MS/MS analyses from our previous proteomic BALF studies27,28. The estimated false-positive rate of the peptide identifications included in the AMT tag database was ≤5% and was obtained by searching a decoy database of the reversed IPI database.29.
Features from the LC-MS analyses (i.e., m/z peaks deconvolved of isotopic and charge state effects and then correlated by mass and NET) were matched to the AMT tag database to identify peptides. The mass deisotoping and alignment process was performed using Decon2LS30, and the matching process was performed using VIPER31 (http://ncrr.pnl.gov/software;). The data were examined in a 2-D fashion to identify groups of features from adjacent spectra comprising a peptide peak, computing a corresponding median monoisotopic mass, central NET, and abundance estimate by summing the intensities of the MS peaks that comprise the entire LC-Orbitrap MS feature. Peptide abundance data were further processed to remove peptides identified with low confidence, using a uniqueness filter that required that the peptide have a SLiC value of 0.5 or greater and a DelSLiC of 0.2 or greater32.
Statistical Analysis
The identification of significant peptides was evaluated on the Log10 abundance. Given the small sample sizes, a non-parametric version of ANOVA called Kruskal-Wallis was used to assess if there is a significant difference between the measured abundance across groups; peptides with no measured abundance value for a sample are treated as a missing value and ignored for the purpose of quantitative statistical data analysis (Kruskal-Wallis). However, missing peptide abundance data can also indicate differential expression, for example when a peptide is completely missing from a treatment it may demonstrate the suppression of the expression of a specific protein. This type of qualitative change in peptide/protein expression will not be identified by an ANOVA-type analysis. To address this issue, a modified Chi-square test, called a G-test, was used to determine if the amount of missing data in any group is less than expected by chance33. Using either the Kruskal-Wallis or the G-test, 1480 peptides were identified with a q-value of 0.05 or less.
A peptide occurrence filter based upon the minimum number of observations required for statistical analysis was employed to remove peptides with little information content33. Following the occurrence filter the number of peptides evaluated was 2175. An additional filter requiring at least two unique peptides per protein further reduced the peptide list to 2009 peptides. The 2009 peptides were used for protein inference; the peptide abundance measurements were rolled up to the corresponding protein abundances using RRollup within DAnTE (http://nccr.pnl.gov/software/ 34. Peptide redundancy, mapping to more than one protein, was reduced either with Protein Prophet35 or manually by selecting known proteins over similar but less completely annotated entries appearing as predicted proteins or hypothetical proteins.
Ingenuity Pathway Analysis
Differentially regulated proteins identified were further analyzed using Ingenuity Pathway Analysis (IPA; Ingenuity Systems, Mountain View, CA; http://www.ingenuity.com webcite). IPA was used to interpret the differentially expressed proteins in terms of predominant canonical pathways. The Ingenuity Pathways Knowledge Base (IKB) is a regularly updated curated database consisting of interactions between different proteins collected from the scientific literature. IPA uses this database to construct protein interaction clusters that involve direct and indirect interactions, physical binding interactions, enzyme-substrate relationships, and cis-trans relationships in transcriptional control. To infer significant signaling pathways from our global proteomic results, the whole dataset containing protein identifiers and corresponding ratiometric values (TreatmentGroup: Control Group) were uploaded to IPA. A fold change of 2.0 was required to identify focus proteins whose expression was significantly regulated. These focus proteins were overlaid onto a global molecular network developed from information contained in the Ingenuity Pathways Knowledge Base. The significance values for canonical pathways were determined by using the right-tailed Fisher’s Exact Test to calculate a p-value determining the probability that the association between the proteins in the dataset and the canonical pathway was explained by chance alone. This analysis was done by comparing the number of focus proteins that participate in a given function or pathway relative to the total number of occurrences of these proteins in all functional/pathway annotations stored in the ingenuity pathway knowledge base (IPKB).
Results
Mice (young male C57BL/6) were exposed to either PBS (control mice) or one of three organisms: virulent strains of F. t. novicida and P. aeruginosa, and an avirulent F. novicida mutant for the transcriptional regulator mglA (F.t. novicida ΔmglA). BALF was collected from control animals at 0, 4-, 24- and 48-hours following exposure to PBS. Likewise, BALF was collected 4-, 24- and 48-hours following infection with the attenuated (Mgla) and the F.t. novicida strains and at 4- and -24 hours for the P. aeruginosa treated mice, resulting in eight treatment groups and four control groups (Table 1). In previous work, aliquots of these BALF samples were used in MALDI MS studies to identify patterns of peptides and small proteins that could differentiate between samples6. In this work, we have extended these studies to identify protein alterations in the BALF samples in response to bacterial exposures. To achieve this, BALF proteins were precipitated and digested with trypsin, and analyzed by high resolution MS. The resultant peptide list was filtered (see methods) resulting in a list of 2009 peptides, corresponding to 239 proteins. This number of identified proteins is lower than is typically identified in either plasma or lung tissue and likely reflects the decreased protein complexity in BALF compared with lung tissue or plasma27,36–38. The averaged relative abundances for these peptides and proteins along with the ratiometric data against the control samples are listed in Supplemental Tables 1 and 2, respectively. Bacterial proteins were not detected in any of the control or infected samples, possibly because they are present at too low of a concentration to be detected by ESI-MS or they are removed in the BALF preparation steps.
Table 1.
Experimental design and correlation score analysis within treatment groups
| Treatment Group |
Median Correlation (r) |
Number of animals/group |
|---|---|---|
| CTLa | 0.83 | 16 |
| MGLA4 | 0.82 | 4 |
| MGLA24 | 0.79 | 4 |
| MGLA48 | 0.76 | 4 |
| FTN 4 | 0.74 | 4 |
| FTN24 | 0.80 | 4 |
| FTN48 | 0.89 | 4 |
| PA4 | 0.90 | 4 |
| PA24 | 0.95 | 4 |
Control animals were exposed to phosphate buffered saline
To evaluate the variability between biological replicates, we analyzed the correlation coefficients of the log10 peptide abundance across the biological replicates within each of the nine treatment/time group combinations (Table 1). The median correlation values ranged from 0.75 to 0.89 for the control, F.t. novicida- and the MGLA-exposed treatment groups/time groups. The correlation coefficients for the two P. aeruginosa treatment groups were higher, 0.90 and 0.95 for the 4- and the 24-hour times respectively. Based on the correlation analysis across biological replicates all analyzed samples were retained for analysis.
BALF protein changes in F.t. novicida-exposed mice
Overall, at the earliest time studied following F.t. novicida infection, a small number of proteins in the BALF are upregulated, however this number increases dramatically as the infection progresses (Supplemental Table 2). At four hours only 11% (27/239) of proteins from F.t. novicida-exposed animals are upregulated two-fold or more versus PBS-control animals. Of the 27 proteins upregulated at four hours following F.t. novicida inhalation, 12 are oxidative stress response proteins, including aldehyde dehydrogenase (ALDH1a7), retinal dehydrogenase 1 (ALDH1a1), peroxiredoxin-1 (PRDX1), Llactate dehydrogenase A chain (LDHA), L-lactate dehydrogenase B chain (LDHB), superoxide dismutase (SOD1), glutathione S-transferase P1 (GSTP1), glutathione S-transferase A3 (GSTS3), glutathione S-transferase Mu1 (GSTM1), glutathione S-transferase Mu2 (GSTM2), and glutathione S-transferase A4 (GSTA4).
Table 2.
Top four and selected Ingenuity canonical pathways assigned to >2 fold up/down-regulated abundance changes relative to controls
| Treatment Group |
Pathway Name | p-value | #proteins/ total in IPA pathway |
Proteins |
|---|---|---|---|---|
| Mgla04 | ||||
| Acute Phase Response Signaling |
2.0E-22 | 21/169 | TTR, HPX, FTL, C3, APOH, APOA2, AHSG, CP, SERPINA3, C5, ORM1, HRG, F2, PLG, HP, TF, C4A, SERPINA1, FGB, FGA, A2M |
|
| NRF2-mediated Oxidative Stress Response |
2.5E-11 | 13/178 | GSTA3, GSTM1, FTL, GSTP1, SOD1, GSTM5, PRDX1, ACTB, GSTO1, GSTA4, ACTA1, CBR1, FTH1 |
|
| Coagulation System | 1.4E-09 | 7/38 | PLG, SERPINC1, SERPINA1, FGB, FGA, A2M, F2 | |
| Aryl Hydrocarbon Receptor Signaling |
1.5E-07 | 9/150 | TGM2, GSTA3, GSTM1, GSTP1, ALDH1A1, GSTM5, GSTA4, HSP90AA1, GSTO1 |
|
| Complement System | 1.3E-06 | 5/33 | CFD, C3, C4A, C8B, C5 | |
| Mgla24 | ||||
| Acute Phase Response Signaling |
2.9E-08 | 8/169 | TTR, HPX, ITIH4, AHSG, C4A, C5, A2M, ORM1 | |
| Glutathione Metabolism | 1.5E-03 | 3/91 | GSTP1,GSTM5,PRDX6 | |
| NRF2-mediated Oxidative Stress Resp. |
1.7E-03 | 4/178 | GSTP1, GSTM5, PRDX1, CBR1 | |
| Stilbene, Coumarine and Lignin Biosynthesis |
4.0E-03 | 2/77 | PRDX1,PRDX6 | |
| Complement System | 4.3E-03 | 2/33 | C4A, C5 | |
| Coagulation System | 5.1E-03 | 2/38 | SERPINC1,A2M | |
| Mgla48 | ||||
| Acute Phase Response | 8.7E-04 | 4/169 | APOA1, SERPINA3, A2M, HRG | |
| Hepatic Fibrosis/Hepatic Stellate Cell Act. |
4.3E-03 | 3/128 | MYH6, MYH9, A2M | |
| Germ Cell-Sertoli Cell Junction Signaling |
7.4E-03 | 3/156 | TUBA1A, TUBB2C, A2M | |
| Tight Junction Signaling | 7.4E-03 | 3/156 | MYH6, MYH9, SPTAN1 | |
| Ftn04 | ||||
| Glutathione Metabolism | 4.4E-09 | 7/91 | GSTA3, GSTM1, GSTP1, PGD, GSTM5, GSTA4, PRDX6 |
|
| NRF2-mediated Oxidative Stress Resp. |
6.9E-08 | 8/178 | GSTA3, GSTM1, GSTP1, SOD1, GSTM5, GSTA4, PRDX1,CBR1 |
|
| Glycolysis/Gluconeogen. | 8.5E-08 | 7/132 | GPI, ALDH1A1, ALDH1A7, PGAM1, TPI1, LDHA, LDHB |
|
| Aryl Hydrocarbon Receptor Signaling |
3.2E-07 | 7/150 | TGM2, GSTA3, GSTM1, GSTP1, ALDH1A1, GSTM5, GSTA4 |
|
| Acute Phase Response | 1.0E-06 | 7/169 | APOH, APCS, ITIH4, CFB, FGA, A2M, HRG | |
| Complement System | 1.6E-04 | 3/33 | CFD, CFB, C8B | |
| Ftn24 | ||||
| Acute Phase Response Signaling |
3.2E-16 | 16/169 | TTR, HPX, FTL, C3, APOA1, APOA2, AHSG, C9, CP, SERPINA3, PLG, TF, ITIH4, CFB, SERPINA1, FGA |
|
| NRF2-mediated Oxidative Stress Resp. |
2.9E-07 | 9/178 | GSTM1, FTL, GSTP1, SOD1, GSTM5, PRDX1, ACTB,ACTA1,FTH1 |
|
| Complement System | 7.4E-07 | 5/33 | C3, C9, CFB, C8B, C8G | |
| Actin Cytoskeleton Signaling | 1.8E-06 | 9/223 | MYH10, MYH6, PFN1, MYL12B, ACTB, MYH11, GSN, KNG1,ACTA1 |
|
| Ftn48 | ||||
| Acute Phase Response Signaling |
1.6E-25 | 25/169 | RAC2, FTL, APOH, APOA1, APOA2, C9, CP, SERPINA3,ORM1, HRG, F2, FGG, SERPIND1, ITIH4, SERPINA1, FGB, HPX, TTR, C3, AHSG, SERPINF2, PLG, HP, TF, A2M |
|
| Coagulation System | 5.0E-12 | 9/39 | PLG, SERPINC1, SERPINA1, FGB, A2M, SERPINF2, FGG, F2,SERPIND1 |
|
| Actin Cytoskeleton Signaling | 1.0E-09 | 14/223 | MYH10, RAC2, ACTR2, PFN1, CFL1, ACTB, MYH11, GSN, F2,MYH9, VCL, KNG1, ACTA1, MSN |
|
| Complement System | 1.7E-07 | 6/33 | CFD, C3, C9, C8B, C6, C8G | |
| Regulation of Actin-based Motility by Rho . |
3.5E-06 | 7/83 | RAC2, ACTR2, PFN1, CFL1, ACTB, GSN, ACTA1 | |
| NRF2-mediated Oxidative Stress Resp |
3.7E-04 | 7/178 | GSTM1, FTL, GSTP1, ACTB, TXN, ACTA1, FTH1 | |
| PA04 | ||||
| Acute Phase Response Signaling |
2.5E-24 | 22/169 | TTR, HPX, C3, APOH, APOA1, APOA2, C9, AHSG, CP, SERPINA3, SERPINF2, ORM1, HRG, F2, SERPIND1, PLG, ALB, TF, SERPINA1, FGB, FGA, A2M |
|
| Coagulation System | 3.2E-13 | 9/38 | PLG, SERPINC1, SERPINA1, FGB, FGA, A2M, SERPINF2, F2, SERPIND1 |
|
| Complement System | 2.8E-08 | 6/33 | CFD, C3, C9, C7, C8B, C8G | |
| Actin Cytoskeleton Signaling | 3.7E-08 | 11/223 | MYH10, MYH6, PFN1, CFL1, ACTB, MYH9, MYH11, GSN, KNG1, ACTA1, F2 |
|
| NRF2-mediated Oxidative Stress Resp. |
3.1E-03 | 5/178 | GSTP1, SOD1, ACTB, ACTA1, CBR1 | |
| PA24 | ||||
| Acute Phase Response Signaling |
5.0E-28 | 25/169 | FTL, APOH, APOA1, APOA2, C9, CP, SERPINA3, ORM1, HRG, F2, FGG, SERPIND1, APCS, SERPINA1, FGB, HPX, TTR, C3, AHSG, C5, SERPINF2, PLG, ALB, TF, A2M |
|
| Coagulation System | 7.9E-13 | 9/38 | PLG, SERPINC1, SERPINA1, FGB, A2M, SERPINF2, FGG, F2, SERPIND1 |
|
| Complement System | 2.0E-11 | 8/33 | CFD, C3, C9, C7, C8B, C5, C6, C8G | |
| Actin Cytoskeleton Signaling | 7.6E-10 | 13/223 | MYH10, ACTR2, MYH6, PFN1, ACTB, MYH11, GSN, F2, MYL12B, MYH9, VCL, KNG1, ACTA1 |
|
| NRF2-mediated Oxidative Stress Resp. |
2.3E-02 | 4/178 | FTL, ACTB, ACTA1, FTH1 | |
By 24 and 48 hours, 37% (89/241) and 54% (131/241) of BALF proteins are upregulated in F.t. novicida-exposed mice. Proteins with the largest increases in abundance at 24 hours post-exposure include myeloperoxidase (MPO; 78 fold), S100a9 (23 fold), Lactotransferrin (LTF; 18 fold), and neutrophil gelatinase-associated protein (LCN2; 8 fold). Proteins showing the greatest increases at 48 hour post-exposure include S100-A9 (193 fold), MPO (166 fold), Histone H4 (107 fold), LTF (76 fold), Transketolase (45 fold), SERPIND1 (44 fold), LCN2 (37 fold), S100A8 (17 fold) and Chitinase-3-like protein 3 (CHI3L3, also referred to as Ym1; 13 fold).
A subset of proteins (29/241) were decreased greater than two-fold at the earliest times following F.t. novicida-infection, however many of these proteins were transiently decreased and by 48 hours post infection only 11 proteins continue to be downregulated by more than two-fold compared with proteins from PBS-control animals. Transiently decreased proteins included Lcn2, Histidine-rich glycoprotein (HRG), galectin-3 (LGALS3) and alpha-enolase (ENO1). Persistently downregulated proteins throughout F.t. novicida-infection include carbonic anhydrase2 (CAR2), annexin A4 (ANXA4), and uteroglobin (SCGB1a1).
Infection enriched IPA pathways
To identify canonical pathways that were significantly altered in infected animals compared with PBS-control animals, Ingenuity Pathway Analysis (IPA) was performed. IPA contains lists of canonical pathways and associated proteins. The relative abundance data of BALF proteins statistically altered during infection, was uploaded into IPA and the protein list filtered to identify proteins that were up- or down-regulated relative to PBS exposed controls by at least two-fold. The filtered proteins, referred to as the focus proteins, were overlaid onto a global molecular network developed from information contained in the Ingenuity Pathways Knowledge Base. Additionally, IPA determines a ratio of the number of focus molecules to overall molecules in each pathway. The results from this pathway analysis are summarized in Table 2 and Figure 1.
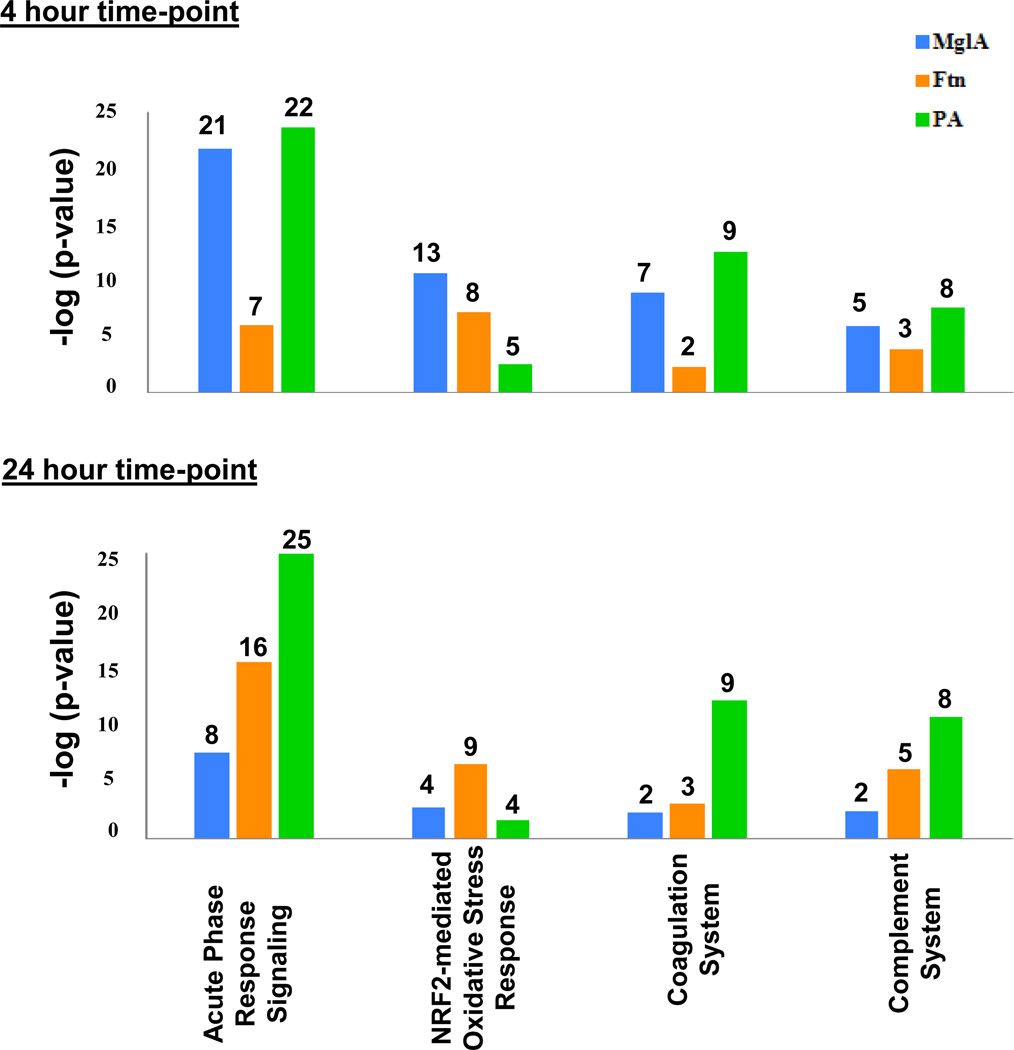
Figure 1.
Canonical pathway analysis of infected mice. Proteins that were found to be significantly different in infected versus control animals were analyzed by Ingenuity Pathway Analysis (IPA). The top canonical pathways are shown. The y-axis indicates the negative log of p-values while the number above each bar corresponds to the number of proteins identified by IPA in each pathway. IPA includes 169, 178, 38, and 33 proteins in the Acute Phase Response Signaling, the NRF2-mediated Oxidative Stress Response, Coagulation System, and the Complement System pathways respectively.
In general, BALF proteins associated with innate immunity pathways were highly represented in the list of significantly altered canonical pathways. Innate immunity provides immediate defense against infection utilizing major mechanisms such as the recognition of pathogens by pattern recognition receptors, the recruitment of immune cells, activation of the complement cascade, activation of the acute phase proteins, induction of cytokine production and the activation of the adaptive immune system. Several of these pathways are represented in IPA as the acute phase response signaling pathway, the complement system, and an oxidative stress response pathway, NRF2-mediated Oxidative Stress Response pathway. These pathways were significantly altered in all three infections, however there were important differences in the kinetics of activation of the pathways. Figure 1 summarizes the overall trends of the identified innate immunity pathways. These differences are discussed further below and the proteins associated with these pathways identified both by IPA and references in the literature are listed in Table 3 (acute phase proteins and the subset of complement proteins), Table 4 (oxidative stress response proteins), and Table 5 (additional innate immune response proteins). To identify trends, the protein abundance ratio (treatment group/ control group) from all the identified proteins from each pathway was averaged. The averages ranged from less than a one-fold change to greater than three-fold changes between the treatment group and the control PBS group.
Table 3.
Acute phase proteins
| Description | Gene Name |
Ratiometric- ratioed to PBS Control | |||||||
|---|---|---|---|---|---|---|---|---|---|
| Mgla 4hr | Mgla 24hr | Mgla 48hr | Ftn 4hr | Ftn 24hr | Ftn 48hr | PA 4hr | PA 24hr | ||
| Complement proteins | |||||||||
| Complement component 3 | C3 | ↑ 2.9a | ↓1.6 | ↓1.4 | ↓1.1 | ↑5.0 | ↑14.3 | ↑6.2 | ↑20 |
| Complement C4-b | C4b | ↑ 2.8 | ↓2.0 | 1.0 | ↑1.4 | ↓2 | ↑1.1 | ↓1.1 | |
| Complement C5 | C5 | ↑3.1 | ↓2.4 | ↓1.4 | ↑1.2 | ↑1.5 | ↑1.2 | ↑1.6 | ↑2.6 |
| Complement component c8 alpha chain | C8a | +b | + | + | + | ||||
| Complement component C8 beta chain | C8b | ↑6.3 | ↑6.1 | ↑4.3 | ↑6.4 | ↑5.2 | ↑22 | ||
| Complement component C8 gamma chain |
C8g | ↑2.4 | ↑3.9 | ↑4.8 | ↑12 | ||||
| Complement component 9 | C9 | ↓1.8 | ↓1.7 | ↑1.4 | ↑4.8 | ↑4.2 | ↑8.3 | ↑7.5 | |
| Complement factor B (fragment) | Cfb | ↓1.9 | ↓1.5 | ↓2.0 | ↓5.0 | ↓2.5 | ↑1.1 | ↓1.4 | ↑1.6 |
| Isoform 1 of complement factor d | Cfd | ↑4.1 | ↓2.0 | ↓2.3 | ↑2.6 | ↑1.5 | ↑3.4 | ↑4.4 | ↑6.1 |
| Complement factor I | Cfi | + | + | + | + | ||||
| Coagulation proteins | |||||||||
| Coagulation factor XII | F12 | + | + | + | |||||
| Prothrombin | F2 | ↑4.2 | ↓1.3 | ↓1.4 | ↑1.3 | ↑2.4 | ↑3.0 | ↑3.6 | |
| Fibrinogen, alpha polypeptide isoform 2 | Fga | ↓4.3 | ↓1.5 | ↓5 | ↓10 | ↓1.4 | ↓2.5 | ↓1.4 | |
| Fibrinogen beta chain | Fgb | ↑ 2.8 | ↑2.0 | ↑7.0 | ↑2.7 | ↑8.1 | |||
| Fibrinogen gamma chain | Fgg | ||||||||
| Plasma kallikrein | Klkb1 | + | + | + | |||||
| Plasminogen | Plg | ↑ 3.1 | ↑1.1 | ↑1.5 | ↑3.2 | ↑5.7 | ↑4.8 | ↑17 | |
| Antithrombin-III | Serpinc1 | ↑2.4 | ↓2.9 | ↓1.7 | ↓1.1 | ↑2.0 | ↑3.2 | ↑3.5 | ↑7.2 |
| Heparin cofactor 2 | Serpind1 | ↑44 | ↑22 | ↑28.8 | |||||
| Alpha-2-antiplasmin | Serpinf2 | ↑1.6 | ↓1.3 | ↑1.1 | ↑1.6 | ↑3.6 | ↑3.7 | ↑7.4 | |
| Protease inhibitor proteins | |||||||||
| Inter-alpha-trypsin inhibitor H1 | Itih1 | + | |||||||
| Inter-alpha-trypsin inhibitor H2 | Itih2 | + | + | + | + | ||||
| Inter-alpha-trypsin inhibitor H3 | Itih3 | + | + | ||||||
| Inter alpha-trypsin inhibitor H4 | Itih4 | ↑1.2 | ↑1.4 | ↓2 | ↓2 | ↑2.3 | ↓2 | ↑1.1 | |
| Alpha-1-antitrypsin 1-2 | Serpina1b | ↑ 2.5 | ↓1.7 | ↓1.4 | ↑1.1 | ↑2.5 | ↑2.9 | ↑3.6 | ↑6.6 |
| Serine (or cysteine) proteinase inhibitor, clade a, member 1c |
Serpina1c | ↑2.5 | ↓1.5 | ↓1.3 | ↑1.1 | ↑2.5 | ↑2.9 | ↑3.5 | ↑6.7 |
| Alpha-1-antitrypsin 1-5 | Serpina1e | ↑2.4 | ↓1.8 | ↓1.4 | ↑1.1 | ↑2.4 | ↑2.9 | ↑3.4 | ↑6.5 |
| Serine protease inhibitor A3c | Serpina3c | ↑2.4 | ↓1.2 | ↑2.1 | ↓1.3 | ↑2.3 | ↑2.3 | ↑3.3 | ↑9.9 |
| Serine protease inhibitor A3f | Serpina3f | ↑ 1.9 | ↓1.6 | ↓1.1 | ↑1.1 | ↑3.5 | ↑7.3 | ↑4.2 | ↑4.3 |
| Serine protease inhibitor A3k. | Serpina3k | ↑1.8 | ↓2.3 | ↓1.3 | ↓1.1 | ↑2.1 | ↑1.9 | ↑3.0 | ↑7.0 |
| Serine protease inhibitor A3n | Serpina3n | ↑2 | ↓1.2 | ↑2.1 | ↓1.3 | ↑2.2 | ↑2.7 | ↑2.8 | ↑2.5 |
| Serpin B6 | Serpinb6a | ↓ 3.3 | ↓2.5 | ↓2.0 | ↓2.5 | ↓3.3 | ↓10 | ↓1.1 | |
| Miscellaneous acute phase proteins | |||||||||
| Alpha-2-HS-glycoprotein | Ahsg | ↑ 2.7 | ↓2.2 | ↓1.4 | ↑1.1 | ↑2.4 | ↑2.9 | ↑3.8 | ↑8.2 |
| Albumin | Alb | ↑1.7 | ↓1.6 | ↓1.4 | ↓1.1 | ↑1.7 | ↑1.8 | ↑2.8 | ↑4.9 |
| Serum amyloid p-component | Apcs | ↑1.3 | ↑1.3 | ↑1.2 | ↓2.5 | ↓1.3 | ↓1.1 | ↓1.3 | ↓2.5 |
| Apolipoprotein A-II | Apoa2 | ↑5.1 | ↓1.7 | ↑1.3 | ↑2.0 | ↑2.4 | ↑6.3 | ↑2.2 | |
| Apolipoprotein A-IV | Apoa4 | ↑ 1.5 | ↓1.1 | 1.0 | ↑1.2 | ↑1.9 | ↑1.3 | ↑3.6 | ↑4.9 |
| Ceruloplasmin | Cp | ↑ 2.5 | ↓1.1 | ↓1.7 | ↑1.2 | ↑2.7 | ↑6.7 | ↑3.3 | ↑9.0 |
| Vitamin D-binding protein | Gc | ↑ 1.9 | ↓2.3 | ↓1.3 | ↑1.2 | ↑2.3 | ↑7.1 | ↑3.9 | ↑9.9 |
| Haptoglobin | Hp | ↑ 2.2 | ↓1.7 | ↓1.7 | ↓1.1 | ↑1.9 | ↑4.8 | ↑1.6 | ↑1.6 |
| Hemopexin | Hpx | ↑ 3.3 | ↓2 | ↓1.4 | ↑1.3 | ↑2.7 | ↑10 | ↑4.8 | ↑13 |
| Histidine-rich glycoprotein | Hrg | ↑2.5 | ↓5 | ↓2 | ↑1.8 | ↑2.3 | ↑2.5 | ↑3.5 | |
| Murinoglobulin-1 | Mug1 | ↑ 1.8 | ↓1 | ↓3.3 | ↑1.1 | ↑4.6 | ↑7.2 | ↑8.6 | ↑41 |
| Alpha-1-acid glycoprotein 1 | Orm1 | ↑ 2.7 | ↓2.1 | ↓1.7 | ↓1.1 | ↑1.4 | ↑4.9 | ↑2.2 | ↑3.4 |
| Alpha-1-acid glycoprotein 2 | Orm2 | ↑ 3.8 | ↓2.5 | ↓1.7 | ↓1.4 | ↑1.5 | ↑3.4 | ↑1.6 | ↑2.5 |
| Alpha-2-macroglobulin | Pzp | ↓ 2 | ↓4.7 | ↓5 | ↑2.1 | ↑1.3 | ↑2.2 | ↑2.3 | ↑10 |
| Serotransferrin | Trf | ↑2.7 | ↓1.6 | ↓1.4 | ↑1.1 | ↑2.5 | ↑3.5 | ↑3.6 | ↑6.0 |
| Transthyretin | Ttr | ↑2.3 | ↓2.1 | ↓1.4 | 1.0 | ↑2.2 | ↑2.7 | ↑3.5 | ↑3.5 |
| Vitronectin. | Vtn | + | + | + | + | + | |||
Arrows indicate increasing or decreasing abundance relative to control. Blue indicates decreased expression and pink indicates increased expression. Lightest shading indicates a 2- to 5-fold change; medium shading indicates a 5- to 10-fold change; and darkest shading indicates a >10-fold change.
+ indicates the protein is detected in this treatment group, but not in the control treatment group
Table 4.
Oxidative Stress Proteins
| Description | Gene Name |
Ratiometric-ratioed to PBS Control | |||||||
|---|---|---|---|---|---|---|---|---|---|
| Mgla 4hr | Mgla 24hr | Mgla 48hr | Ftn 4hr | Ftn 24hr | Ftn 48hr | PA 4hr | PA 24hr | ||
| Actin, alpha skeletal muscle | Acta1 | ↑3.4a | ↑1.1 | ↓1.4 | ↑1.4 | ↑2.8 | ↑12 | ↑2.8 | ↑7.7 |
| Actin, cytoplasmic 1 | Actb | ↑3.3 | 1.0 | ↓1.4 | ↑1.3 | ↑3.0 | ↑13 | ↑2.9 | ↑8.0 |
| Retinal dehydrogenase 1 | Aldh1a1 | ↑8.5 | ↑2.3 | 1.0 | ↑4.3 | ↑2.9 | ↓1.2 | 1.0 | ↑1.5 |
| Aldehyde dehydrogenase, cytosolic 1 | Aldh1a7 | ↑6.3 | ↑1.9 | ↓1.1 | ↑3.3 | ↑2.9 | ↓1.2 | ↑1.2 | ↑1.6 |
| Carbonyl reductase 2 | Cbr2 | ↑7 | ↑3.2 | ↓2 | ↑2.5 | ↑1.2 | ↓1.2 | ↓2.5 | ↑1.2 |
| Ferritin heavy chain | Fth1 | ↑2 | ↓1.3 | ↓1.4 | ↑1.8 | ↑2.2 | ↑6.9 | ↓1.3 | ↑2.7 |
| Ferritin light chain 1 | Ftl1 | ↑1.5 | ↓1.7 | ↓2 | ↑1.2 | ↑2.4 | ↑3.9 | ↓1.4 | ↓2.5 |
| glutathione peroxidase 3 | Gpx3 | ↑1.5 | ↓1.4 | ↓1.1 | ↑1.6 | ↑2.4 | ↑2.7 | ↑3.2 | ↑7.2 |
| Glutathione S-transferase A3 | Gsta3 | ↑5.2 | ↑1.6 | ↑1.6 | ↑2.0 | ↑1.6 | ↓1.4 | ↑1.1 | ↓2 |
| Glutathione S-transferase A4 | Gsta4 | ↑6.2 | ↑1.2 | ↑1.2 | ↑5.2 | ↓1.1 | |||
| Glutathione S-transferase Mu1 | Gstm1 | ↑5.9 | ↑2.2 | ↓1.1 | ↑3.5 | ↑2.3 | ↑1.9 | ↑1.1 | ↓1.1 |
| Glutathione S-transferase Mu 2 | Gstm2 | ↑4 | ↑1.7 | ↑1.1 | ↑2.7 | ↑2.3 | ↑2.2 | ↑1.2 | ↓1.1 |
| Glutathione transferase Omega-1 | Gsto1 | ↑5.2 | ↑1.2 | ↓1.7 | ↑1.7 | ↑1.7 | ↓1.7 | ↓1.4 | ↓2 |
| Glutathione S-transferase P 1 | Gstp1 | ↑2.9 | ↑2.6 | ↓2 | ↑2.0 | ↑2.6 | ↑2.6 | ↑4.0 | |
| L-Lactate dehydrogenase A chain | Ldha | ↑5.9 | ↑1.9 | ↓2 | ↑3.9 | ↑2.9 | ↑11 | ↑3.4 | ↑3.9 |
| L-Lactate dehydrogenase B chain | Ldhb | ↑6.9 | ↑1.2 | ↑1.2 | ↑8.0 | ↑2.0 | ↑1.5 | ||
| Lysozyme C-2 | Lyz2 | ↑1.9 | ↓1.7 | ↓2 | ↓1.1 | ↑1.3 | ↓1.1 | ↓1.4 | ↓1.4 |
| Myeloperoxidase | Mpo | ↑31 | ↑78 | ↑166 | ↑48 | ||||
| Serum Paraoxonase/Arylesterase 1 | Pon1 | + | + | ||||||
| Peroxiredoxin-1 | Prdx1 | ↑7.4 | ↑2.8 | ↑1.1 | ↑5.7 | ↑2.3 | ↑1.9 | ↑1.5 | ↓1.7 |
| Peroxiredoxin-5 | Prdx5 | ↑2.5 | ↑1.8 | ↑1.5 | ↑1.9 | ↑6.8 | ↑2.7 | ↑1.6 | |
| Peroxiredoxin-6 | Prdx6 | ↑3.3 | ↑2.2 | 1.0 | ↑2.4 | ↑2.3 | ↑1.4 | ↑1.9 | ↓1.4 |
| Superoxide dismutase | Sod1 | ↑4.8 | ↑1.2 | ↓1.4 | ↑2.6 | ↑2.1 | ↑1.4 | ↑2.3 | ↑1.1 |
| Thioredoxin | Txn1 | ↑1.3 | ↓1.3 | ↑1.3 | ↓1.1 | ↑3.0 | ↑1.8 | ↓2 | |
Arrows indicate increasing or decreasing abundance relative to control. Blue indicates decreased expression and pink indicates increased expression. Lightest shading indicates a 2- to 5-fold change; medium shading indicates a 5- to 10-fold change; and darkest shading indicates a >10-fold change.
Table 5.
Additional innate immunity proteins
| Description | Gene Name |
Ratiometric- ratioed to Control | |||||||
|---|---|---|---|---|---|---|---|---|---|
| Mgla 4hr | Mgla 24hr | Mgla 48hr | Ftn 4hr | Ftn 24hr | Ftn 48hr | PA 4hr | PA 24hr | ||
| Cathelin-related antimicrobial peptide |
Camp | +b | + | + | + | ||||
| Monocyte differentiation antigen CD14 |
Cd14 | + | + | + | + | ||||
| CD177 antigen | Cd177 | + | + | ||||||
| Coronin-1a | Coro1a | + | + | + | |||||
| Resistin-like alpha (Fizz1) | FIZZ1 | ↑5.2a | ↑12.8 | ||||||
| Neutrophil gelatinase-associated lipocalin (NGAL) |
Lcn2 | ↑1.9 | ↓2 | ↑8.8 | ↑37 | ↑2.7 | ↑18 | ||
| Galectin-3 | Lgals3 | ↑1.6 | ↓3.6 | ↑1.5 | ↑3.5 | ||||
| Lactotransferrin | Ltf | ↑18 | ↑6.3 | ↑5.7 | ↑28 | ||||
| Long palate, lung and nasal epithelium carcinoma-associated protein 1 |
Lplunc1 | ↑3.0 | ↑1.1 | 1.0 | ↑2.7 | ↑3.2 | ↑2.0 | ||
| Lysozyme C-2 | Lyz2 | ↑1.9 | ↓1.7 | ↓2 | ↓1.1 | ↑1.3 | ↓1.1 | ↓1.4 | ↓1.4 |
| Matrix metalloproteinase 9 - gelatinase |
Mmp9 | + | + | + | + | ||||
| Myeloperoxidase | Mpo | ↑31 | ↑78 | ↑166 | ↑48 | ||||
| Neutrophilic granule protein | Ngp | + | + | + | + | ||||
| Protein plunc | Plunc | ↑4.1 | ↓10 | ↑1.1 | ↑3.3 | ↑3.2 | ↑1.6 | ||
| Leukocyte elastase inhibitor a | Serpinb1a | + | + | + | |||||
| Plasma protease c1 inhibitor | Serping1 | + | + | + | |||||
| Pulmonary surfactant-associated protein A |
Sftpa1 | ↑4.1 | ↓1.4 | ↓1.6 | ↑1.1 | ↑1.3 | ↑1.2 | ↑1.4 | ↓2 |
| Pulmonary surfactant-associated protein D |
Sftpd | ↑3.1 | ↓2 | ↓1.6 | ↑1.1 | ↑2.4 | ↑3.6 | ↑1.7 | ↑1.1 |
| Chitinase-3-like protein 3 (Ym1) | YM1 / Chi3l3 | ↑1.9 | ↓1.7 | ↓1.4 | ↓1.3 | ↑3.5 | ↑13 | ↑1.6 | ↑2.6 |
Arrows indicate increasing or decreasing abundance relative to control. Blue indicates decreased expression and pink indicates increased expression. Lightest shading indicates a 2- to 5-fold change; medium shading indicates a 5- to 10-fold change; and darkest shading indicates a >10- fold change.
+ indicates the protein is detected in this treatment group, but not in the control treatment group
Acute phase proteins
The acute phase proteins are plasma proteins produced in the liver with plasma concentrations that either increase (positive acute phase protein) or decrease (negative acute phase protein) in response to inflammation39. Animals exposed to P. aeruginosa develop an early and robust acute phase response (Table 2 and 3). The most significantly altered canonical pathway in P. aeruginosa -exposed animals was the acute phase response signaling pathway at 4 hours (p-value = 2.5×10−24) and at 24 hours (p-value = 5.0×10-28) following infection. Additionally, at the earliest time, 22 proteins were upregulated out of a total of 169 proteins documented in the IPA acute phase response pathway (Table 2). At 24 hours, 25 proteins were associated with this pathway and 23 of these proteins were upregulated while Serum amyloid p-component and Complement factor b were downregulated.
Similarly, F.t. novicida-ΔmglA exposed mice produced an initial acute phase response (p-value = 2.0×10−22) within the first four hours of infection with 21 proteins altered, 19 of which are upregulated and 2 downregulated (Complement component 9 and factor b) (Figure 1 and Table 2 and 3). However, in F.t. novicida-ΔmglA exposed mice this early response is dampened by 24-hours post-infection, with only one acute phase response protein (Itih4) continuing to show a 1.5-fold increased expression and 7 acute phase response proteins showing a 2-fold decrease in expression. Furthermore, by 48 hours post-F.t. novicida-ΔmglA exposure the acute phase response signaling pathway, while still significantly altered (p-value 8.7×10−4), has only four proteins altered by two-fold or greater, with only one protein upregulated (SERPINA3n).
In contrast, F.t. novicida-exposed animals show little initial acute phase response at the four hour time point. However, by 24 hours post-exposure the acute phase response pathway is significantly altered (p-value=3.2×10−16) with 13 proteins upregulated and three proteins down-regulated (Fibrinogen, α isoform 2, Inter β-trypsin inhibitor H4 and SERPINB6). This robust response continues through the 48 hour time point (p-value = 1.6×10−25) with 25 proteins demonstrating an upregulation of at least two-fold.
One acute phase protein, murinoglobulin-1 (MUG-1), a mouse alpha-macroglobulin, is increased in abundance in F.t. novicida- and especially P. aeruginosa-exposed mice. Murinoglobulin 1 increased in P. aeruginosa -exposed animals as early as four hours following infection. The level of MUG-1 continued to increase in P. aeruginosa-exposed animals, reaching a 40-fold increase over MUG-1 levels in control animals at 48 hours after infection (Table 3). F.t. novicida exposed animals did not show an increase in MUG-1 expression until 24 hours but this increase continued till at least 48 hours after exposure. The other alpha-macroglobulin identified in this study, PZP, demonstrated a modest increase, compared to MUG-1, with the largest increase being a ten-fold increase in abundance at the 48-hour time point in mice infected with P. aeruginosa.
Complement system
The complement proteins are primarily synthesized in the liver and utilized by the innate immune response to trigger inflammation, the recruitment of inflammatory cells, and the opsonization of pathogens for subsequent destruction. A number of complement proteins are increased in abundance in mice infected with either F.t. novicida or P. aeruginosa, including C3, C4b, C5, C8a, C9, Cfb, Cfd and Cfi (Table 2 and 3). The complement system pathway (IPA) is significantly altered in most of the treatment groups, however the timing of this alteration follows a pattern similar to the acute phase protein pathway (Figure 1 and Table 2). Again, in the P. aeruginosa-exposed animals this pathway is increased at both 4 (p-value = 2.8×10−8) and 24 hours (p-value = 2.0×10−11) with 6 and 8 proteins increased by two-fold or more, respectively. Similarly, the F.t. novicida-ΔmglA exposed mice demonstrate an altered complement system (p-value = 1.3×10−6) at four hours with 5 proteins from this pathway upregulated. However, by 24 hours post-infection the pathway is significantly downregulated (p-value = 4.3×10−3). By 48 hours, the complement pathway continues to be downregulated, however it is no longer downregulated to a significant extent (p-value = 0.081).
Again, similar to acute phase response pathway, the complement system is delayed in its activation in F.t. novicida- relative to P. aeruginosa - infected animals. While the pathway is significantly altered at 4 hours, (p-value = 1.6×10−4) only two proteins are up regulated (C8b ↑6.1, Cfd ↑2.6) and one is downregulated (Cfb ↓4.7) in the mice infected with F.t. novicida (Table 3). However, by the 24 hours the complement system is more strongly altered in F.t. novicida infected animals (p-value = 7.4×10−7) and continues to be altered at 48 hours after infection (p-value = 1.7×10−7) with 5 proteins (C3 ↑5, C8b ↑, C8g↑2.4, C9↑4.8, Cfb↓2.6) and 6 proteins (C3↑14.3, C8b↑6.4, C8g↑3.9, C9↑4.2, Cfd↑3.4, C4b↓2) affected respectively. One of the complement proteins, C3, is increased more sharply in the F.t. novicida-exposed mice compared with the other complement proteins (↑14.3 at 48 hour time point).
The oxidative stress response
The innate immune system utilizes reactive oxygen and nitrogen to damage pathogens. Neutrophils, along with macrophages, activate a respiratory burst, responsible for the generation of hydrogen peroxide, free radicals and hypochlorite. The production of respiratory burst compounds activates the oxidative stress responses in the surrounding tissue including the cellular antioxidants superoxide dismutase, glutathione peroxidase, paraoxonase, aldehyde dehydrogenases and glutathione-S transferases. IPA includes the canonical pathway, NRF2-mediated Oxidative Stress Response, a pathway composed of proteins induced by the promoter nuclear factor-erythroid 2-related factor 2 (NRF2), which binds to the antioxidant response elements (ARE) within the promoter of a number of antioxidant enzymes and activates their transcription.
Mice infected with F.t. novicida-ΔmglA show a strong early response to oxidative stress as indicated by the significantly altered NRF2-mediated Oxidative Stress Response (p-value = 2.5×10−11) and the number of oxidative response proteins upregulated (Figure 1 and Table 2) at just four hours following infection. Indeed at this early time 13 proteins are upregulated in the F.t. novicida- ΔmglA mice as compared to the 8 proteins in F.t. novicida-exposed and just 5 proteins in P. aeruginosa -exposed animals. This strong oxidative stress response in mice exposed to F.t. novicida-ΔmglA is still affected by 24 hours following exposure, albeit to a lesser extent (p-value = 1.7×10−3) and is no longer significantly affected by 48 hours post-infection (p-values = 0.36).
The NRF2-mediated Oxidative Stress Response pathway is also significantly altered four hours following F.t. novicida- and P. aeruginosa - infection (p-values = 6.9×10−8 and 3.1×10−3, respectively) however with fewer proteins affected compared with the F.t. novicida-ΔmglA mice, as discussed above. In contrast to F.t. novicida- ΔmglA exposed mice, the majority of the oxidative stress response proteins in F.t. novicida- and P. aeruginosa -exposed mice continue to be upregulated 24- and 48- hours following exposure. Results from IPA support this conclusion and show that F.t. novicida- and P. aeruginosa - exposed mice significantly alter the NRF2-mediated Oxidative Stress Response pathway at 24 hrs (p-value: 2.9×10−7 and 2.3×10−2, respectively) (Figure 1 and Table 2). The NRF2-mediated Oxidative Stress Response pathway is elevated only in the F.t. novicida-exposed mice at 48 hrs (p-value: 3.7×10−4).
A number of oxidative stress response proteins are not represented in the NRF2-mediated Oxidative Stress Response pathway including, MPO, lysozyme c-2 (LYZ2), peroxiredoxin-5 (PRDX5), and peroxiredoxin-6 (PRDX6) (Table 4). These proteins by and large follow the trends outlined above (i.e. increased at early times with modulation of the response as the infection progresses), however the abundance level of MPO does not modulate over the early course of the infection, rather this protein continues to increase throughout the course of the F.t. novicida and P. aeruginosa infections, perhaps reflecting the degree of neutrophil infiltration.
Additional innate immune proteins including neutrophil granule proteins
Neutrophils are the most abundant polymorphonuclear leukocytes (PMN) in the blood and function as the first line of defense in the innate immune response. Indeed, measurement of PMNs in these samples showed that BALF from P. aeruginosa-infected animals had increased recovery of PMNs as early as 4 hours after exposure whereas BALF from F.t. novicida-infected animals did not show measurable numbers of PMNs until 24 hours after infection and BALF from F.t. novicidaΔmglA-infected mice contained no PMNs through 48h after infection6. PMN carry high concentrations of proteins, including both proteases and antibacterial compounds, within several different neutrophil granules. The contents of the neutrophil granules are released, following the mobilization and extravasation of PMN to the site of infection. A number of neutrophil-associated and additional proteins important in innate immunity were detected in the BALF samples (Table 5). The results from some of these proteins such as the alpha-macroglobulins, the surfactant proteins, LTF, MPO, CD14, S100A8 and -A9, and galactin-3 are highlighted in the following paragraphs. Alpha-macroglobulins are proteinase inhibitors and have roles in modulating inflammatory responses40. In mice there are two main alpha-macroglobulins, alpha-2-macroglobulin (PZP or A2M) and MUG-1, both of which act as proteinase inhibitors and carriers of cytokines and growth factors41. MUG-1 expression was increased in both F.t. novicida and P. aeruginosa infected animals, although with differing kinetics (Table 3). The other alpha-macroglobulin identified in this study, PZP, binds to and removes the active forms of the gelatinase proteins, MMP2 and MMP942. PZP demonstrated a modest increase, compared to MUG-1, with the largest increase being a ten-fold increase in abundance at the 48-hour time point in mice infected with P. aeruginosa (Table 3). The delayed expression of MUG-1 and relative lack of PZP in F.t. novicida infection relative to P. aeruginosa indicates a possible increase in MMP activity. This finding mirrors a previous study that showed that an increase in expression of MMP-9 is important for F. tularensis pathogenesis2.
There is a sharp increase in the expression of LTF following both F.t. novicida and P. aeruginosa infections, although the increase is delayed following F.t. novicida infection (Table 5). LTF is an iron-binding glycoprotein important to the innate immune response with unique bacteriocidal effects both because of its ability to bind and sequester iron and to bind lipopolysaccaride. LTF is a component of the secondary granules of neutrophils and is released in infected tissue and blood during the immune response43. There is recent evidence indicating that LTF may also contribute to acquired immunity44,45.
Myeloperoxidase (MPO) is highly abundant in neutrophil granulocytes and stored in the neutrophil’s azurophilic granules. It catalyses the formation of oxidants such as hypochlorous acid as part of the neutrophil’s respiratory burst46. MPO is sharply upregulated twenty-four hours following F.t. novicida and P. aeruginosa infection (Table 5) in good agreement with neutrophil recruitment occurring at these time points6. By twenty four hours following infection the protein abundance for this protein has increased 78-fold in F.t. novicida-infected mice and 48-fold in P. aeruginosa -infected mice. The peptide data at four hours following infection is incomplete (only one peptide identified) making it difficult to determine MPO protein levels early in F.t. novicida infection. Interestingly, MPO is not detected in F.t. novicida-ΔmglA infected mice, consistent with the absent neutrophil response to this organism 6.
Neutrophil gelatinase-associated lipocalin showed an increase in the samples from mice twenty four hours after exposure to F.t. novicida, and P. aeruginosa relative to control samples (Table 5). Neutrophilic granule protein (Ngp) was detected only in the F.t. novicida- and P. aeruginosa exposed samples and not in F.t. novicida-ΔmglA-exposed or control samples. Likewise, the leukocyte elastase inhibitor protein (SERPINB1a) was detected only in BALF collected from F.t. novicida- and P. aeruginosa- exposed animals.
Neutrophils carry a high concentration of specific serine proteases within their granules including Neutrophil elastase (ELA2), cathepsin G and proteinase-3. Only one peptide of ELA2 was detected in the BALF samples (data not shown). The ELA2 peptide is detected in mice exposed to F.t. novicida at the 24- (2 of 4 samples), and 48-hour time points (4 of 4 samples) and P. aeruginosa at the 24-hour time point (1 of 4 samples). No other serine proteases (cathepsin G and proteinase-3) were detected in the mouse BALF samples.
However, a number of serine protease inhibitors were identified including the Leukocyte elastase inhibitor A (SERPINB1a). Leukocyte inhibitors have been described to have pleiotropic functions including activation of innate immunity and antimicrobial activity47. Twelve SERPINB1a peptides were detected in BALF from F.t. novicida- and P. aeruginosa -infected animals, however these peptides were not detected in a single PBS-control or F.t. novicida-ΔmglA sample (Supplemental Table 1). All twelve SERPINB1a peptides were detected in BALF samples isolated 48 hours following F.t. novicida-exposure indicating a high expression level of this protein relative to the other samples.
CD14 is constitutively expressed primarily on the surface of monocytes, macrophages and neutrophils as membrane CD14. The membrane CD14 is important in the recognition of LPS by macrophages and appears to play an early role in F. tularensis infection48. In addition to the membrane form, a soluble form of CD14 is abundant in plasma and other fluids either from macrophage shedding of the membrane form, or by secretion from the liver. CD14 is not observed in BALF from control mice, likely reflecting a low abundance of the protein in normal lung lining fluid. However, CD14 is observed in samples from F.t. novicida and P. aeruginosa infected mice and is upregulated in F.t. novicida mice relative to P. aeruginosa mice by more than 2-fold (Table 5 and Supplemental Table 1 and 2). This increased expression is continued through the 48 hour time point. Indeed the protein is detected in all four samples from the 48 hour time point, whereas, in the earlier time point and both the P. aeruginosa time points, CD14 is detected in only one of the four samples from these treatment groups. The CD14 increase in BALF from F.t. novicida infected animals could be due to an influx into the lung of the soluble serum form of CD14 or it is shed from leukocytes or macrophages in the lung49,50. Previous work indicated a decrease in the CD14 mRNA level in F.t. novicida-infected monocytes51.
There are four surfactant-associated proteins (SP) found in the lung, SP-A, -B, -C and –D. Two of the surfactant-associated proteins, SP-A and SP-D, are important mediators of the innate immunity response including opsonization of bacteria and the regulation of inflammatory proteins52,53. SP-B and SP-C are required for proper biophysical function of the lung and are important in spreading surfactant over the lung surface. SP-A, -B, and –D were all detected in BALF samples from all treatment groups of infected mice (Table 5 and Supplemental Tables). SP-A and –D are upregulated four hours following infection with F.t. novicida-ΔmglA, however the level of these proteins is decreased by 24 hours and remains decreased through the 48 hour time point. The SP-A protein concentration is largely unchanged during the course of a F.t. novicida infection. However, SP-A levels decrease 2-fold at the 24 hour time point during P. aeruginosa infection in agreement with previous work54. SP-D levels are increased throughout the course of an F.t. novicida infection, however they remain largely unchanged during P. aeruginosa infection. SP-B was also detected in the BALF samples and found to decrease in abundance during the course of all three infections, F.t. novicida-ΔmglA, F.t. novicida, and P. aeruginosa, compared with PBS-controls. Whether the increased expression of SP-D indicates an immunomodulatory role for this protein following F.t. novicida infection or some other role during its infection is unknown at this time. The final surfactant-associated protein, SP-C, is not detectable in any of the BALF samples.
Galectin-3 (LGALS3) is a lectin-binding protein that is secreted by several cell types including monocytes, macrophages and epithelial cells55. Galectin-3 is implicated in several innate immune responses including neutrophil activation and adhesion, pro-inflammatory activity, chemoattraction of monocytes/macrophages, and neutrophil apoptosis (for review see Henderson et al56). Additionally, Sato et al57 demonstrated that LGALS3 levels increased in mouse lung following Streptococcal pneumoniae infection. In this study, two peptides corresponding to galactin-3 were identified (Supplemental Table 1). The LGALS3 peptides were identified at a low frequency in samples from control (1/16 samples), F.t. novicida-ΔmglA- (2/4 samples at 4 hour time point), and certain time points of F.t. novicida- (1/4 samples at 4- and 24- hour time points) infected animals. However, by 48 hours following F.t. novicida-infection, the LGALS3 peptides are seen in all 4 of the BALF samples analyzed and the protein demonstrates a 3.5 fold increase in abundance relative to control samples (Table 5). Interesting, LGALS3 peptides were not identified in BALF samples from animals infected with P. aeruginosa.
S100A8 and S100A9 are small calcium-binding proteins, possessing pro-inflammatory properties including the ability to increase the expression of pro-inflammatory chemokines58. They are the most abundant proteins in neutrophils, comprising 40% of the cytosolic protein content59. Additionally they are involved in the chemotaxis and accumulation of neutrophils and macrophages60,61. S100A8 and S100A9 proteins are both significantly upregulated during P. aeruginosa and F.t. novicida infections, however the proteins are not detectable in F.t. novicidaΔmglA infected BALF samples (Table 5). S100A9 is significantly upregulated, increasing by 23-fold and 62-fold in F.t. novicida- and P. aeruginosa -infected samples compared to control samples, respectively, at 24 hours post-exposure. By 48 hours post-exposure this protein has increased almost 200-fold in F.t. novicida infected animals compared with control animals. S100A8 is increased more moderately showing a 6-fold increase in P. aeruginosa -infected animals (24-hour time point) and a 17-fold increase in F.t. novicida-infected animals (48-hour time point). These results are in good agreement with previous findings showing the upregulation of S100A9 during F.t. novicida infection in mice62–64 and S100A8 in humans65.
Studies have identified a number of reliable markers expressed by alternatively activated macrophages including FIZZ1 (Retnla) and YM1 (also known as Chitinase 3-like-3)66. In this study, 21 peptides corresponding to YM1 were identified with a majority of the peptides identified across all samples and treatment groups (Supplemental Table 1). The protein abundance level is not altered to a large extent early in infection (4 hour time point). However, 24- and 48-hours following F. t. novicida infection, YM1 is increased significantly, 3.5- and 12.7-fold respectively (Table 5). YM1 levels increase 2.6-fold twenty-four hours following P. aeruginosa exposure. Only two peptides corresponding to FIZZ1 were identified, however these peptides were both increased 24- and 48-hours following F.t. novicida infection only (Table 5).
An analysis of the MS data reveals a trend in the BALF protein abundance data collected from F.t. novicida-exposed and P. aeruginosa -exposed mice, namely that many detectable proteins increase in abundance beginning shortly after exposure, continuing through the last time that samples were collected. The mice exposed to the attenuated strain of F.t. novicida do not share this increase in mass-spectrometric identified peptide abundance. It is important to note that for each sample, the same amount of peptide was loaded onto the mass spectrometer. Mass spectrometry predominantly identifies medium and high abundance proteins and lacks sufficient sensitivity to identify low abundance proteins. Therefore, a possible explanation for the observed trend is that during F.t. novicida and P. aeruginosa infection there is an influx into the lung of moderate and highly abundant proteins (i.e. plasma proteins such as albumin and acute phase proteins) from the surrounding tissue resulting in an overrepresentation of these proteins in BALF from these animals.
Biomarker assessment
To identify potential protein biomarkers specific to F.t. novicida infection the Biomarker filter in IPA was utilized. Proteins that were 2-fold up- or downregulated in samples collected 24 hours following F.t. novicida- and P. aeruginosa -infection were included in this analysis. An additional filter was used that required two or more peptides from a protein to be observed in at least 75% of the samples from either F.t. novicida or P. aeruginosa treatment groups. Thirteen candidate biomarkers were identified that were common between the two treatment groups: APOA1, B2M, CLU, CP, FTL, GSN, LCN2, MPO, PLG, S100A9, SERPINA, TTR, and VIM. Four candidate biomarkers were identified that were unique to the F.t. novicida treatment group: ALDH1a1, CHI3L1, FGA, and PRDX1.
Discussion
Understanding the host response to aerosol pathogen exposure and infection is an important first step to determining markers useful for early diagnoses. In this work, a mouse model for F. tularensis novicida, was used to examine the early responses to infection. Aerosol exposure of a mouse to Francisella tularensis novicida was used to study the protein profile variations from 4 up to 48 hours post exposure. These variations were compared to infection with an attenuated F. novicida strain as well as to infection with P. auriginosa, a pathogen known to elicit an intense inflammatory response.
Previous work illustrated that patterns of protein or metabolites could be used to differentiate exposures6. By contrast, the current study identifies a broad profile of host proteins detected in the lavage fluid. Protein identification and quantitation provide insight into different pathways involved in response to each infection that is not possible to obtain through pattern comparison alone. A number of innate immunity pathways were altered in the three infections including the acute phase proteins, the complement and coagulation systems and the oxidative stress response, however the kinetics of these alterations varied across the three infections. F.t. novicida infected mice do not appear to efficiently activate an innate immune response in the first hours of infection. However, within 24 hours and continuing at least until 48 hours after infection, the BALF of F.t. novicida-infected mice show a sharp upregulation of the innate immune response proteins. A number of these responses, including the upregulation of complement proteins, coagulation proteins and S100A9 protein have been cited as contributing to severe sepsis in the late phase of lethal murine infection with F.t. novicida63,64,67.
Francisella novicida’s delayed activation of the innate immune response is in contrast to mice infected with F.t. novicida-ΔmglA, which demonstrate an innate immune response within the first four hours of infection despite the fact that it replicates much more slowly in vivo than F. t. novicida18. This early innate immune response becomes attenuated by 24 hours and 48 hours following infection. This observation is consistent with the possibility that MglA mediates the active early suppression of the host innate immune response in mice infected with wild type F.t. novicida. Pseudomonas aeruginosa also demonstrate an innate immune response within the first four hours of infection. The pathology of P. aeruginosa infection is well known to be an inflammatory response in contrast to the necrosis induced by F.t. novicida68. The contrasting pathologies make the comparison between the two exposures important for full analysis of response pathways involved from their associated protein markers.
Activated macrophages secrete inflammatory mediators and kill intracellular pathogens. However, there are multiple pathways to macrophage activation resulting in classically activated macrophages and alternatively activated macrophages69. Recent evidence indicates that Francisella tularensis infection may be capable of inducing alternative activation of macrophages in mouse lungs70,71. It has been further proposed that the accumulation of necrotic cell debris during Francisella infection is responsible for the induction of alternative macrophage differentiation. Studies have identified a number of reliable markers expressed by alternatively activated macrophages including FIZZ1 (resistin-like alpha) and YM1 (also known as Chitinase 3-like-3)66. It was found that YM1 levels increased significantly following both F.t. novicida and P.aeruginosa exposure while FIZZ1 levels increased only in F.t. novicida exposed animals. These results are in good agreement with studies demonstrating increased YM1 and FIZZ1 mRNA levels during Francisella macrophage infection70,71 providing further evidence of alternatively activated macrophages during Francisella infection in vivo.
Several BALF candidate protein biomarkers specific to early F.t. novicida infection were identified including Chi3L1 and Prdx1. These markers showed a greater than 2-fold increase in F.t. novicida-infected animals but not P. aeruginosa -infected animals and may provide useful diagnostic biomarkers of early Francisella aerosol exposure. CHI3L1, also known as YKL-40 (humans) or BRP-39 (mouse), binds chitin however it lacks actual chitinase activity. CHI3L1 is expressed by a range of cell types including macrophages, neutrophils and epithelial cells (reviewed in72). Its exact function remains unclear however it appears to play a role in inflammation and is elevated in a number of inflammatory conditions including asthma, idiopathic pulmonary fibrosis, and inflammatory bowel disease73,74. YKL-40 mRNA expression is strongly induced during the maturation of monocytes to macrophages. Additionally, YKL-40 is stored in neutrophil granules and released during neutrophil activation. In this study, compared to control samples CHI3L1 shows an upregulation of 3.6 fold in F.t. novicida samples at 24 hours after infection whereas it is increased only 1.8 fold in P. aeruginosa infected samples.
Another potential protein biomarker to early F.t. novicida infection, Peroxiredoxin 1 (PRDX1), functions as a regulator of hydrogen peroxide signaling and as a protein chaperone75. PRDX1 is elevated in various cancers and is secreted by non-small cell lung cancer cells. Recent work indicates that PRDX1 may act as a danger signal interacting with TLR4 and stimulating the release of proinflammatory cytokines76. The level of PRDX1 is increased 2.3 fold in BALF from mice infected with F.t. novicida 24 hours post-infection whereas it is decreased 1.7 fold at the same time point in P. aeruginosa infected samples. Furthermore, this protein may serve as a candidate biomarker even at 4 hours post-infection as it is increased 5.7 fold in F.t. novicida infected samples and only 1.5 fold in P. aeruginosa infected samples.
In conclusion, it is clear that numerous mouse host protein levels are altered in response to pulmonary infection with F.t. novicida and that an innate immune response is initially delayed followed by a sharp upregulation of many of the innate immune response proteins. Several BALF candidate protein biomarkers specific to early F.t. novicida infection were identified including Chi3L1 and Prdx1. These markers showed a greater than 2-fold increase in F.t. novicida-infected animals but not P. aeruginosa -infected animals and may provide useful diagnostic biomarkers of early Francisella aerosol exposure. Broad differences in the host pathways involved during response provide a clearer understanding of the immunopathogenesis of this infection. This combination of highly sensitive mass spectrometric measurements and methods for interpreting large proteomic datasets is essential for host response pathway elucidation. A comprehensive view of immunopathogenesis pathways is necessary if protein biomarkers are going to be useful in a clinical setting.
Supplementary Material
Acknowledgements
Portions of this work were funded by the U.S. Department of Energy through the Environmental Biomarkers Initiative at Pacific Northwest National Laboratory (PNNL), by NIH/NIAID grant 3U54AI057141, NIH/NHLBI grant HL098067, and by the NIH National Center for Research Resources (RR18522, RR030249). Some of the experimental work was performed in the Environmental Molecular Science Laboratory, a national scientific user facility sponsored by the Department of Energy's Office of Biological and Environmental Research located at PNNL. The authors thank Mark Pelletier for expert technical support. PNNL is operated by Battelle for the U.S. Department of Energy under contract (AC06-76RLO 1830).
References
- 1.McCrumb FR. Aerosol Infection of Man with Pasteurella Tularensis. Bacteriol Rev. 1961;25:262–267. doi: 10.1128/br.25.3.262-267.1961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Malik M, Bakshi CS, McCabe K, Catlett SV, Shah A, Singh R, Jackson PL, Gaggar A, Metzger DW, Melendez JA, Blalock JE, Sellati TJ. Matrix metalloproteinase 9 activity enhances host susceptibility to pulmonary infection with type A and B strains of Francisella tularensis. J Immunol. 2007;178:1013–1020. doi: 10.4049/jimmunol.178.2.1013. [DOI] [PubMed] [Google Scholar]
- 3.Hollis DG, Weaver RE, Steigerwalt AG, Wenger JD, Moss CW, Brenner DJ. Francisella philomiragia comb. nov. (formerly Yersinia philomiragia) and Francisella tularensis biogroup novicida (formerly Francisella novicida) associated with human disease. J Clin Microbiol. 1989;27:1601–1608. doi: 10.1128/jcm.27.7.1601-1608.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Anthony LD, Burke RD, Nano FE. Growth of Francisella spp. in rodent macrophages. Infect Immun. 1991;59:3291–3296. doi: 10.1128/iai.59.9.3291-3296.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hajjar AM, Harvey MD, Shaffer SA, Goodlett DR, Sjostedt A, Edebro H, Forsman M, Bystrom M, Pelletier M, Wilson CB, Miller SI, Skerrett SJ, Ernst RK. Lack of in vitro and in vivo recognition of Francisella tularensis subspecies lipopolysaccharide by Toll-like receptors. Infect Immun. 2006;74:6730–6738. doi: 10.1128/IAI.00934-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wunschel D, Webb-Robertson B-J, Frevert CW, Skerrett S, Beagley N, Willse A, Colburn H, Antolick K. Differentiation of gram-negative bacterial aerosol exposure using detected markers in bronchial-alveolar lavage fluid. PLoS One. 2009;4:e7047. doi: 10.1371/journal.pone.0007047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Duenas AI, Aceves M, Orduna A, Diaz R, Sanchez Crespo M, Garcia-Rodriguez C. Francisella tularensis LPS induces the production of cytokines in human monocytes and signals via Toll-like receptor 4 with much lower potency than E. coli LPS. Int Immunol. 2006;18:785–795. doi: 10.1093/intimm/dxl015. [DOI] [PubMed] [Google Scholar]
- 8.Fortier AH, Slayter MV, Ziemba R, Meltzer MS, Nacy CA. Live vaccine strain of Francisella tularensis: infection and immunity in mice. Infection and immunity. 1991;59:2922–2928. doi: 10.1128/iai.59.9.2922-2928.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bosio CM, Dow SW. Francisella tularensis induces aberrant activation of pulmonary dendritic cells. J Immunol. 2005;175:6792–6801. doi: 10.4049/jimmunol.175.10.6792. [DOI] [PubMed] [Google Scholar]
- 10.Telepnev M, Golovliov I, Grundstrom T, Tarnvik A, Sjostedt A. Francisella tularensis inhibits Toll-like receptor-mediated activation of intracellular signalling and secretion of TNFalpha and IL-1 from murine macrophages. Cell Microbiol. 2003;5:41–51. doi: 10.1046/j.1462-5822.2003.00251.x. [DOI] [PubMed] [Google Scholar]
- 11.Bosio CM, Bielefeldt-Ohmann H, Belisle JT. Active suppression of the pulmonary immune response by Francisella tularensis Schu4. J Immunol. 2007;178:4538–4547. doi: 10.4049/jimmunol.178.7.4538. [DOI] [PubMed] [Google Scholar]
- 12.Chase JC, Celli J, Bosio CM. Direct and indirect impairment of human dendritic cell function by virulent Francisella tularensis Schu S4. Infect Immun. 2009;77:180–195. doi: 10.1128/IAI.00879-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Clemens DL, Lee BY, Horwitz MA. Francisella tularensis enters macrophages via a novel process involving pseudopod loops. Infect Immun. 2005;73:5892–5902. doi: 10.1128/IAI.73.9.5892-5902.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Balagopal A, MacFarlane AS, Mohapatra N, Soni S, Gunn JS, Schlesinger LS. Characterization of the receptor-ligand pathways important for entry and survival of Francisella tularensis in human macrophages. Infect Immun. 2006;74:5114–5125. doi: 10.1128/IAI.00795-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fortier AH, Leiby DA, Narayanan RB, Asafoadjei E, Crawford RM, Nacy CA, Meltzer MS. Growth of Francisella tularensis LVS in macrophages: the acidic intracellular compartment provides essential iron required for growth. Infect Immun. 1995;63:1478–1483. doi: 10.1128/iai.63.4.1478-1483.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Baron GS, Nano FE. MglA and MglB are required for the intramacrophage growth of Francisella novicida. Mol Microbiol. 1998;29:247–259. doi: 10.1046/j.1365-2958.1998.00926.x. [DOI] [PubMed] [Google Scholar]
- 17.Lauriano CM, Barker JR, Yoon SS, Nano FE, Arulanandam BP, Hassett DJ, Klose KE. MglA regulates transcription of virulence factors necessary for Francisella tularensis intraamoebae and intramacrophage survival. Proc Natl Acad Sci U S A. 2004;101:4246–4249. doi: 10.1073/pnas.0307690101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.West TE, Pelletier MR, Majure MC, Lembo A, Hajjar AM, Skerrett SJ. Inhalation of Francisella novicida Delta mglA causes replicative infection that elicits innate and adaptive responses but is not protective against invasive pneumonic tularemia. Microbes Infect. 2008;10:773–780. doi: 10.1016/j.micinf.2008.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Guina T, Radulovic D, Bahrami AJ, Bolton DL, Rohmer L, Jones-Isaac KA, Chen J, Gallagher LA, Gallis B, Ryu S, Taylor GK, Brittnacher MJ, Manoil C, Goodlett DR. MglA regulates Francisella tularensis subsp. novicida (Francisella novicida) response to starvation and oxidative stress. Journal of bacteriology. 2007;189:6580–6586. doi: 10.1128/JB.00809-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Skerrett SJ, Wilson CB, Liggitt HD, Hajjar AM. Redundant Toll-like receptor signaling in the pulmonary host response to Pseudomonas aeruginosa. Am J Physiol Lung Cell Mol Physiol. 2007;292:L312–L322. doi: 10.1152/ajplung.00250.2006. [DOI] [PubMed] [Google Scholar]
- 21.Livesay EA, Tang K, Taylor BK, Buschbach MA, Hopkins DF, LaMarche BL, Zhao R, Shen Y, Orton DJ, Moore RJ, Kelly RT, Udseth HR, Smith RD. Fully automated four-column capillary LC-MS system for maximizing throughput in proteomic analyses. Anal Chem. 2008;80:294–302. doi: 10.1021/ac701727r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Smith RD, Anderson GA, Lipton MS, Pasa-Tolic L, Shen Y, Conrads TP, Veenstra TD, Udseth HR. An accurate mass tag strategy for quantitative and high-throughput proteome measurements. Proteomics. 2002;2:513–523. doi: 10.1002/1615-9861(200205)2:5<513::AID-PROT513>3.0.CO;2-W. [DOI] [PubMed] [Google Scholar]
- 23.Eng JK, Mccormack AL, Yates JR. An Approach to Correlate Tandem Mass-Spectral Data of Peptides with Amino-Acid-Sequences in a Protein Database. J Am Soc Mass Spectr. 1994;5:976–989. doi: 10.1016/1044-0305(94)80016-2. [DOI] [PubMed] [Google Scholar]
- 24.Washburn MP, Wolters D, Yates JR. Large-scale analysis of the yeast proteome by multidimensional protein identification technology. Nat Biotechnol. 2001;19:242–247. doi: 10.1038/85686. [DOI] [PubMed] [Google Scholar]
- 25.Kiebel GR, Auberry KJ, Jaitly N, Clark DA, Monroe ME, Peterson ES, Tolic N, Anderson GA, Smith RD. PRISM: a data management system for high-throughput proteomics. Proteomics. 2006;6:1783–1790. doi: 10.1002/pmic.200500500. [DOI] [PubMed] [Google Scholar]
- 26.Zimmer JSD, Monroe ME, Qian W-J, Smith RD. Advances in proteomics data analysis and display using an accurate mass and time tag approach. Mass Spectrom Rev. 2006;25:450–482. doi: 10.1002/mas.20071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pounds JG, Flora JW, Adkins JN, Lee KM, Rana GSJB, Sengupta T, Smith RD, McKinney WJ. Characterization of the mouse bronchoalveolar lavage proteome by micro-capillary LC-FTICR mass spectrometry. J Chromatogr B Analyt Technol Biomed Life Sci. 2008;864:95–101. doi: 10.1016/j.jchromb.2008.01.044. [DOI] [PubMed] [Google Scholar]
- 28.Jin S, Daly DS, Springer DL, Miller JH. The effects of shared peptides on protein quantitation in label-free proteomics by LC/MS/MS. J Proteome Res. 2008;7:164–169. doi: 10.1021/pr0704175. [DOI] [PubMed] [Google Scholar]
- 29.Qian W-J, Liu T, Monroe ME, Strittmatter EF, Jacobs JM, Kangas LJ, Petritis K, Camp DG, Smith RD. Probability-based evaluation of peptide and protein identifications from tandem mass spectrometry and SEQUEST analysis: the human proteome. J Proteome Res. 2005;4:53–62. doi: 10.1021/pr0498638. [DOI] [PubMed] [Google Scholar]
- 30.Jaitly N, Mayampurath A, Littlefield K, Adkins JN, Anderson GA, Smith RD. Decon2LS: An open-source software package for automated processing and visualization of high resolution mass spectrometry data. BMC Bioinformatics. 2009;10:87. doi: 10.1186/1471-2105-10-87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Monroe ME, Tolic N, Jaitly N, Shaw JL, Adkins JN, Smith RD. VIPER: an advanced software package to support high-throughput LC-MS peptide identification. Bioinformatics. 2007;23:2021–2023. doi: 10.1093/bioinformatics/btm281. [DOI] [PubMed] [Google Scholar]
- 32.Anderson KK, Monroe ME, Daly DS. Estimating probabilities of peptide database identifications to LC-FTICR-MS observations. Proteome Sci. 2006;4:1. doi: 10.1186/1477-5956-4-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Webb-Robertson BJ, McCue LA, Waters KM, Matzke MM, Jacobs JM, Metz TO, Varnum SM, Pounds JG. Combined Statistical Analyses of Peptide Intensities and Peptide Occurrences Improves Identification of Significant Peptides from MS-Based Proteomics Data. J Proteome Res. 2010 doi: 10.1021/pr1005247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Polpitiya AD, Qian W-J, Jaitly N, Petyuk VA, Adkins JN, Camp DG, Anderson GA, Smith RD. DAnTE: a statistical tool for quantitative analysis of -omics data. Bioinformatics. 2008;24:1556–1558. doi: 10.1093/bioinformatics/btn217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nesvizhskii AI, Keller A, Kolker E, Aebersold R. A statistical model for identifying proteins by tandem mass spectrometry. Anal Chem. 2003;75:4646–4658. doi: 10.1021/ac0341261. [DOI] [PubMed] [Google Scholar]
- 36.Ventura CL, Higdon R, Hohmann L, Martin D, Kolker E, Liggitt HD, Skerrett SJ, Rubens CE. Staphylococcus aureus elicits marked alterations in the airway proteome during early pneumonia. Infect Immun. 2008;76:5862–5872. doi: 10.1128/IAI.00865-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gharib SA, Nguyen E, Altemeier WA, Shaffer SA, Doneanu CE, Goodlett DR, Schnapp LM. Of mice and men: comparative proteomics of bronchoalveolar fluid. Eur Respir J. 2010;35:1388–1395. doi: 10.1183/09031936.00089409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Schnapp LM, Donohoe S, Chen J, Sunde DA, Kelly PM, Ruzinski J, Martin T, Goodlett DR. Mining the acute respiratory distress syndrome proteome: identification of the insulin-like growth factor (IGF)/IGF-binding protein-3 pathway in acute lung injury. Am J Pathol. 2006;169:86–95. doi: 10.2353/ajpath.2006.050612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gabay C, Kushner I. Acute-phase proteins and other systemic responses to inflammation. N Engl J Med. 1999;340:448–454. doi: 10.1056/NEJM199902113400607. [DOI] [PubMed] [Google Scholar]
- 40.Umans L, Serneels L, Overbergh L, Stas L, Van Leuven F. alpha2-macroglobulin- and murinoglobulin-1-deficient mice. A mouse model for acute pancreatitis. Am J Pathol. 1999;155:983–993. doi: 10.1016/s0002-9440(10)65198-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Correia Soeiro MN, Paiva MM, Waghabi M, Meirelles MN, Lorent K, Araujo-Jorge TC, Van Leuven F. Differential expression of mRNA coding for the alpha-2-macroglobulin family and the LRP receptor system in C57BL/6J and C3H/HeJ male mice. Cell Struct Funct. 2001;26:161–167. doi: 10.1247/csf.26.161. [DOI] [PubMed] [Google Scholar]
- 42.Nagase H, Itoh Y, Binner S. Interaction of alpha 2-macroglobulin with matrix metalloproteinases and its use for identification of their active forms. Ann N Y Acad Sci. 1994;732:294–302. doi: 10.1111/j.1749-6632.1994.tb24744.x. [DOI] [PubMed] [Google Scholar]
- 43.Ward PP, Mendoza-Meneses M, Park PW, Conneely OM. Stimulus-dependent impairment of the neutrophil oxidative burst response in lactoferrin-deficient mice. Am J Pathol. 2008;172:1019–1029. doi: 10.2353/ajpath.2008.061145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Actor JK, Hwang SA, Kruzel ML. Lactoferrin as a natural immune modulator. Curr Pharm Des. 2009;15:1956–1973. doi: 10.2174/138161209788453202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Legrand D, Elass E, Carpentier M, Mazurier J. Lactoferrin: a modulator of immune and inflammatory responses. Cell Mol Life Sci. 2005;62:2549–2559. doi: 10.1007/s00018-005-5370-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.van der Veen BS, de Winther MP, Heeringa P. Myeloperoxidase: molecular mechanisms of action and their relevance to human health and disease. Antioxidants & redox signaling. 2009;11:2899–2937. doi: 10.1089/ars.2009.2538. [DOI] [PubMed] [Google Scholar]
- 47.Fitch PM, Roghanian A, Howie SE, Sallenave JM. Human neutrophil elastase inhibitors in innate and adaptive immunity. Biochem Soc Trans. 2006;34:279–282. doi: 10.1042/BST20060279. [DOI] [PubMed] [Google Scholar]
- 48.Chase JC, Bosio CM. The presence of CD14 overcomes evasion of innate immune responses by virulent Francisella tularensis in human dendritic cells in vitro and pulmonary cells in vivo. Infect Immun. 2010;78:154–167. doi: 10.1128/IAI.00750-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lin SM, Frevert CW, Kajikawa O, Wurfel MM, Ballman K, Mongovin S, Wong VA, Selk A, Martin TR. Differential regulation of membrane CD14 expression and endotoxin-tolerance in alveolar macrophages. Am J Respir Cell Mol Biol. 2004;31:162–170. doi: 10.1165/rcmb.2003-0307OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Senft AP, Korfhagen TR, Whitsett JA, Shapiro SD, LeVine AM. Surfactant protein-D regulates soluble CD14 through matrix metalloproteinase-12. J Immunol. 2005;174:4953–4959. doi: 10.4049/jimmunol.174.8.4953. [DOI] [PubMed] [Google Scholar]
- 51.Butchar JP, Cremer TJ, Clay CD, Gavrilin MA, Wewers MD, Marsh CB, Schlesinger LS, Tridandapani S. Microarray analysis of human monocytes infected with Francisella tularensis identifies new targets of host response subversion. PLoS One. 2008;3:e2924. doi: 10.1371/journal.pone.0002924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kuroki Y, Takahashi M, Nishitani C. Pulmonary collectins in innate immunity of the lung. Cell Microbiol. 2007;9:1871–1879. doi: 10.1111/j.1462-5822.2007.00953.x. [DOI] [PubMed] [Google Scholar]
- 53.Pastva AM, Wright JR, Williams KL. Immunomodulatory roles of surfactant proteins A and D: implications in lung disease. Proc Am Thorac Soc. 2007;4:252–257. doi: 10.1513/pats.200701-018AW. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Beatty AL, Malloy JL, Wright JR. Pseudomonas aeruginosa degrades pulmonary surfactant and increases conversion in vitro. Am J Respir Cell Mol Biol. 2005;32:128–134. doi: 10.1165/rcmb.2004-0276OC. [DOI] [PubMed] [Google Scholar]
- 55.Sato S, Hughes RC. Regulation of secretion and surface expression of Mac-2, a galactoside-binding protein of macrophages. J Biol Chem. 1994;269:4424–4430. [PubMed] [Google Scholar]
- 56.Henderson NC, Sethi T. The regulation of inflammation by galectin-3. Immunol Rev. 2009;230:160–171. doi: 10.1111/j.1600-065X.2009.00794.x. [DOI] [PubMed] [Google Scholar]
- 57.Sato S, Ouellet N, Pelletier I, Simard M, Rancourt A, Bergeron MG. Role of galectin-3 as an adhesion molecule for neutrophil extravasation during streptococcal pneumonia. J Immunol. 2002;168:1813–1822. doi: 10.4049/jimmunol.168.4.1813. [DOI] [PubMed] [Google Scholar]
- 58.Foell D, Wittkowski H, Vogl T, Roth J. S100 proteins expressed in phagocytes: a novel group of damage-associated molecular pattern molecules. J Leukoc Biol. 2007;81:28–37. doi: 10.1189/jlb.0306170. [DOI] [PubMed] [Google Scholar]
- 59.Hessian PA, Edgeworth J, Hogg N. MRP-8 and MRP-14, two abundant Ca(2+)-binding proteins of neutrophils and monocytes. J Leukoc Biol. 1993;53:197–204. [PubMed] [Google Scholar]
- 60.Nacken W, Roth J, Sorg C, Kerkhoff C. S100A9/S100A8: Myeloid representatives of the S100 protein family as prominent players in innate immunity. Microsc Res Tech. 2003;60:569–580. doi: 10.1002/jemt.10299. [DOI] [PubMed] [Google Scholar]
- 61.Ryckman C, Vandal K, Rouleau P, Talbot M, Tessier PA. Proinflammatory activities of S100: proteins S100A8, S100A9, and S100A8/A9 induce neutrophil chemotaxis and adhesion. Journal of immunology (Baltimore, Md : 1950) 2003;170:3233–3242. doi: 10.4049/jimmunol.170.6.3233. [DOI] [PubMed] [Google Scholar]
- 62.Andersson H, Hartmanova B, Kuolee R, Ryden P, Conlan W, Chen W, Sjostedt A. Transcriptional profiling of host responses in mouse lungs following aerosol infection with type A Francisella tularensis. J Med Microbiol. 2006;55:263–271. doi: 10.1099/jmm.0.46313-0. [DOI] [PubMed] [Google Scholar]
- 63.Mares CA, Ojeda SS, Morris EG, Li Q, Teale JM. Initial delay in the immune response to Francisella tularensis is followed by hypercytokinemia characteristic of severe sepsis and correlating with upregulation and release of damage-associated molecular patterns. Infect Immun. 2008;76:3001–3010. doi: 10.1128/IAI.00215-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Sharma J, Li Q, Mishra BB, Pena C, Teale JM. Lethal pulmonary infection with Francisella novicida is associated with severe sepsis. J Leukoc Biol. 2009;86:491–504. doi: 10.1189/jlb.1208728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Andersson H, Hartmanova B, Back E, Eliasson H, Landfors M, Naslund L, Ryden P, Sjostedt A. Transcriptional profiling of the peripheral blood response during tularemia. Genes Immun. 2006;7:503–513. doi: 10.1038/sj.gene.6364321. [DOI] [PubMed] [Google Scholar]
- 66.Raes G, Noel W, Beschin A, Brys L, de Baetselier P, Hassanzadeh GH. FIZZ1 and Ym as tools to discriminate between differentially activated macrophages. Dev Immunol. 2002;9:151–159. doi: 10.1080/1044667031000137629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Sharma J, Mares CA, Li Q, Morris EG, Teale JM. Features of sepsis caused by pulmonary infection with Francisella tularensis Type A strain. Microb Pathog. 2011;51:39–47. doi: 10.1016/j.micpath.2011.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Twenhafel NA, Alves DA, Purcell BK. Pathology of inhalational Francisella tularensis spp. tularensis SCHU S4 infection in African green monkeys (Chlorocebus aethiops) Vet Pathol. 2009;46:698–706. doi: 10.1354/vp.08-VP-0302-T-AM. [DOI] [PubMed] [Google Scholar]
- 69.Mosser DM, Edwards JP. Exploring the full spectrum of macrophage activation. Nat Rev Immunol. 2008;8:958–969. doi: 10.1038/nri2448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Mares CA, Sharma J, Li Q, Rangel EL, Morris EG, Enriquez MI, Teale JM. Defect in efferocytosis leads to alternative activation of macrophages in Francisella infections. Immunol Cell Biol. 2010 doi: 10.1038/icb.2010.81. [DOI] [PubMed] [Google Scholar]
- 71.Shirey KA, Cole LE, Keegan AD, Vogel SN. Francisella tularensis live vaccine strain induces macrophage alternative activation as a survival mechanism. J Immunol. 2008;181:4159–4167. doi: 10.4049/jimmunol.181.6.4159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Lee CG, Da Silva C, Dela Cruz CS, Ahangari F, Ma B, Kang MJ, He CH, Takyar S, Elias JA. Role of Chitin, Chitinase/Chitinase-Like Proteins in Inflammation, Tissue Remodeling, and Injury. Annu Rev Physiol. 2010 doi: 10.1146/annurev-physiol-012110-142250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Ober C, Tan Z, Sun Y, Possick JD, Pan L, Nicolae R, Radford S, Parry RR, Heinzmann A, Deichmann KA, Lester LA, Gern JE, Lemanske RF, Jr, Nicolae DL, Elias JA, Chupp GL. Effect of variation in CHI3L1 on serum YKL-40 level, risk of asthma, and lung function. N Engl J Med. 2008;358:1682–1691. doi: 10.1056/NEJMoa0708801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Wu J, Kobayashi M, Sousa EA, Liu W, Cai J, Goldman SJ, Dorner AJ, Projan SJ, Kavuru MS, Qiu Y, Thomassen MJ. Differential proteomic analysis of bronchoalveolar lavage fluid in asthmatics following segmental antigen challenge. Mol Cell Proteomics. 2005;4:1251–1264. doi: 10.1074/mcp.M500041-MCP200. [DOI] [PubMed] [Google Scholar]
- 75.Wood ZA, Schroder E, Robin Harris J, Poole LB. Structure, mechanism and regulation of peroxiredoxins. Trends Biochem Sci. 2003;28:32–40. doi: 10.1016/s0968-0004(02)00003-8. [DOI] [PubMed] [Google Scholar]
- 76.Riddell JR, Wang XY, Minderman H, Gollnick SO. Peroxiredoxin 1 stimulates secretion of proinflammatory cytokines by binding to TLR4. J Immunol. 2010;184:1022–1030. doi: 10.4049/jimmunol.0901945. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.