Abstract
The pyridine nucleotides, NAD+ and NADH, are coenzymes that provide oxidoreductive power for the generation of ATP by mitochondria. In skeletal muscle, exercise perturbs the levels of NAD+, NADH, and consequently, the NAD+/NADH ratio, and initial research in this area focused on the contribution of redox control to ATP production. More recently, numerous signaling pathways that are sensitive to perturbations in NAD+(H) have come to the fore, as has an appreciation for the potential importance of compartmentation of NAD+(H) metabolism and its subsequent effects on various signaling pathways. These pathways, which include the sirtuin (SIRT) proteins SIRT1 and SIRT3, the poly(ADP-ribose) polymerase (PARP) proteins PARP1 and PARP2, and COOH-terminal binding protein (CtBP), are of particular interest because they potentially link changes in cellular redox state to both immediate, metabolic-related changes and transcriptional adaptations to exercise. In this review, we discuss what is known, and not known, about the contribution of NAD+(H) metabolism and these aforementioned proteins to mitochondrial adaptations to acute and chronic endurance exercise.
Keywords: nicotinamide adenine dinucleotide, sirtuin-1, sirtuin-3, poly(ADP-ribose) polymerase, COOH-terminal binding protein
nicotinamide (nam) adenine dinucleotide [NAD+; initially known as diphosphopyradine nucleotide (DPN+)], is a ubiquitous cellular coenzyme that was first discovered by Arthur Harden and William Young, when they identified a heat-labile fraction of cell-free glucose fermentation containing ATP, Mg2+ and NAD+, which they coined, “cozymase” (78). Our understanding of the role of NAD+ and its reduced form, NADH, in cellular function and metabolism was subsequently expanded by a “who's who” of biochemistry, with researchers such as Hans von Euler-Chelpin, Otto Warburg, Conrad Elvehjem, Arthur Kornberg, Albert Lehninger, and Britton Chance, all making substantial contributions. Four of the aforementioned researchers were awarded the Nobel Prize, with Harden and von Euler-Chelpin sharing the Nobel Prize in 1929 for their work on the fermentation of sugar and fermentative enzymes, which included the identification of the “nucleotide sugar phosphate” NAD+. Subsequently, Warbug demonstrated that NAD+ acted as a carrier of hydrogen and transferred it from one molecule to another, which was key to understanding the metabolic function of NAD+ (128). Ultimately, it was work by Freidkin and Lehninger (55) that showed that NADH was an integral component of ATP production via oxidative phosphorylation. Thus, for many years, the primary cellular function of NAD+ was considered to be its ability to harness energy from glucose, fatty acids, and amino acids in pathways such as glycolysis, β-oxidation, and the citric acid cycle.
In recent years, however, the importance of NAD+ as a central signaling molecule and substrate that can impact numerous fundamental biological processes has come to the fore. Indeed, a remarkable number of regulatory pathways that utilize NAD+ in signaling reactions have been identified, and these cover broad aspects of cellular homeostasis, including functions in energy metabolism, lifespan regulation, DNA repair, apoptosis, and telomere maintenance (11, 12, 84, 97, 190). Thus, while the tissue NAD+/NADH ratio was once thought to be “simply” a balance of the redox state, the complexity of NAD+ metabolism has evolved considerably with the discovery of highly integrated networks of NAD+-consuming pathways and NAD+ biosynthetic and salvage pathways (11, 12, 84, 97, 128, 144, 190). Part of the reason for the renaissance of NAD+ has been the discovery of NAD+-consuming enzymes, particularly sirtuins (SIRT). SIRT1 is the best-described of the seven mammalian sirtuins, and, based on its dependence for NAD+ as a substrate (and therefore its sensitivity to perturbations in NAD+), SIRT1 has been put forth as a key regulator of acute and chronic exercise-mediated mitochondrial adaptations in skeletal muscle (40, 70, 72, 76, 174, 185, 193). In addition, SIRT3 and poly(ADP-ribose) (PAR) polymerases (PARPs), which also use NAD+ as a substrate, have been proposed as important regulators of mitochondrial function and/or biogenesis (40, 76, 125, 174, 185, 193). In this review, our aim is to provide an overview of NAD+ metabolism in skeletal muscle and the changes that occur in NAD+, NADH, and the NAD+/NADH ratio in response to acute and chronic endurance exercise. Our intention is not to discuss the impact of the redox state and NAD+/NADH ratio on cellular bioenergetics and substrate utilization, which is covered in highly informative reviews by others (9, 26, 106, 109, 110). Rather, our goal is to discuss the changes in pyridine nucleotide redox state that occur with exercise in the context of what we know and do not know about the effects of SIRT1, SIRT3, the PARPs, and carboxyl-terminal binding protein (CtBP) on mitochondrial adaptations to exercise in skeletal muscle. It is, of course, difficult to extrapolate the findings from one cell line or tissue type to another, and we acknowledge that we do not discuss many important studies that have contributed to our understanding of NAD+ metabolism and SIRT1, SIRT3, and PARP biology in cell lines and tissue types other than skeletal muscle and muscle cell lines. For a more general and encompassing discussion on NAD+ metabolism and its potential clinical implications, readers are encouraged to read some excellent and comprehensive reviews (see Refs. 11, 12, 84, 97, 128, 144, and 190).
Where in the Cell Is NAD+?
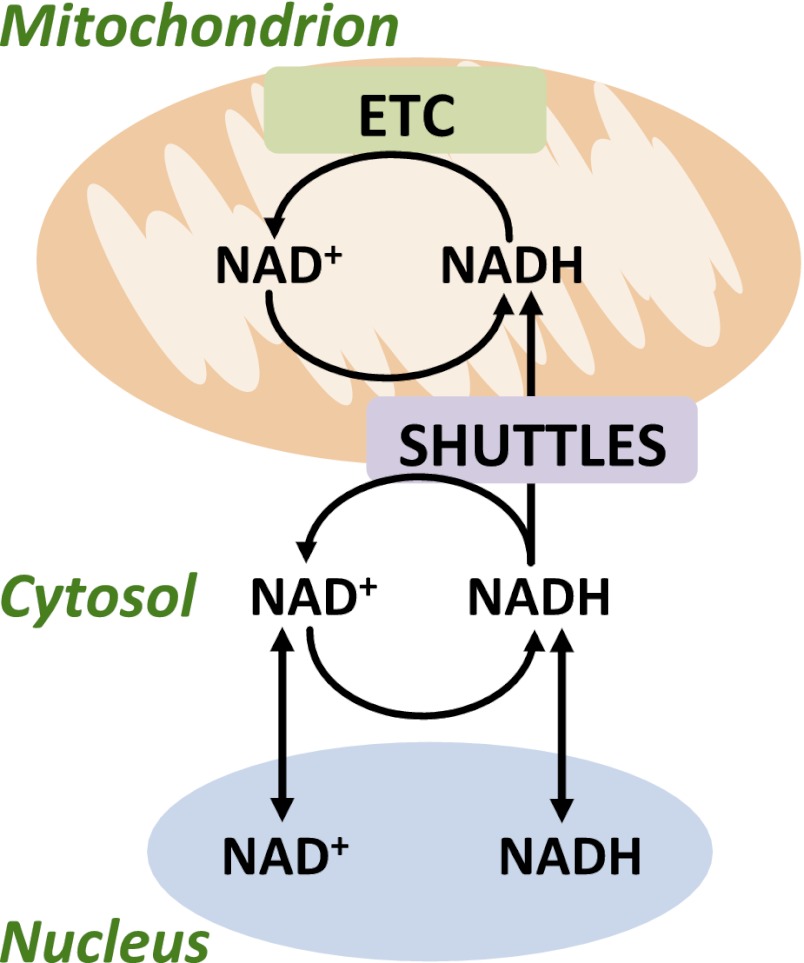
It is broadly accepted that NAD+ is primarily found in three distinct cellular pools: 1) the cytosolic, 2) the mitochondrial, and 3) the nuclear pools. A general overview of the compartmentation of NAD+ and NADH is provided in Fig. 1 and provides a point of reference for the ensuing discussion on NAD+(H) “compartmentation” and their movement into the mitochondria and nucleus. Initial studies used differential centrifugation methods, cell disruption methods, and compounds to modulate mitochondrial NAD+(H) metabolism in order to determine NAD+(H) location. More recently, the compartmentation of NAD+, which was originally suggested by Ragland and Hackett (146), has been extrapolated from the localization of enzymes in the NAD+ consuming, biosynthetic, and salvage pathways and the use of innovative molecular biology techniques (11, 12, 84, 97, 144, 190). Thus, Dölle et al. (43) used the novel PAR Assisted Protein Localization AssaY (PARAPLAY) in HeLa S3 cells, in which they targeted the catalytic domain of PARP1 (which consumes NAD+) to various cellular compartments. The idea behind this method is that if NAD+ is present in the compartment to which PARP1 is targeted, then PAR will accumulate and can be detected by immunocytochemistry (43). Using PARAPLAY, NAD+ was found in mitochondria (specifically the matrix but not intermembrane space) and peroxisomes, and surprisingly to the endoplasmic reticulum (ER) and Gogli complex (43, 112). Cytosolic NAD+ was not detected in that study, most likely due to the fact that PAR glycohydrolase (PARG), which consumes PAR, is most abundant in the cytosol. Little is known about the role of NAD+ and NADH in regulating Golgi complex and ER function, and certainly its function in skeletal muscle is unknown. Furthermore, surprisingly very little is known about nuclear NAD+ levels in general, and to our knowledge nuclear NAD+(H) levels have not been measured in skeletal muscle. Overall, the free cytosolic and nuclear NAD+(H) compartments are traditionally thought to be in equilibrium, with NAD+(H) being able to freely pass through pore complexes in the nuclear membrane (46, 98–103, 187, 190). In Cos7 cells, the free nuclear NAD+ concentration is estimated to be ∼10–100 μM (53, 188), which is comparable to the estimations for the cytosol (∼150 μM) of muscle (42, 119). Thus, in response to exercise, it would be expected that the pyridine redox state in the nucleus reflects changes that occur in the cytosol. The relevance of nuclear NAD+(H) to adaptations to exercise will be covered when discussing SIRT1, PARPs, and CtBP.
Fig. 1.
Compartmentation of NAD+ and NADH in skeletal muscle. NAD+ and NADH move freely across pores in the nuclear membrane, and as such, the cytosolic and nuclear compartment concentrations of NAD+ and NADH are thought to be comparable. In the cytosol, NADH is generated by glycolysis. Because mitochondria are impermeable to NADH, transfer of these reducing equivalents occurs via a variety of shuttles including the glycerol 3-phosphate shuttle, malate-aspartate shuttle, lactate shuttle, and the NADH/cytochrome c electron transport shuttle, as described in the text. Depending on the shuttle, NADH is produced. The cytosolic/nuclear NAD+ pool is replenished when NADH is converted back to NAD+ in the reactions of the aforementioned shuttles, including the conversion of pyruvate to lactate. NAD+ levels in nuclear, cytosolic, and mitochondrial compartments are also replenished via specific de novo and salvage pathways that are discussed in the text and overviewed in Fig. 2. Within mitochondria, NADH is oxidized to NAD+ in the electron transport chain (ETC).
NAD+ and NADH Concentrations in Skeletal Muscle at Rest
While PARAPLAY provides qualitative insight into the location of NAD+, determining the precise concentration of NAD+ in various compartments remains challenging. Typically, absolute concentrations of NAD+ and NADH have been calculated using biochemical and extraction methods, whereas the metabolite indicator method (MIM) has been used to extrapolate the “free” cytoplasmic and mitochondrial NAD+/NADH ratio by measuring the concentrations of specific cytoplasmic and mitochondrial redox couples. The MIM carries a number of assumptions, such as the selected dehydrogenase reaction being a near-equilibrium reaction and that the reaction occurs in one cellular compartment, at pH 7.0 (63, 107, 179). In skeletal muscle, the most common application of the MIM is calculation of the cytosolic free NAD+/NADH ratio via measurement of lactate and pyruvate levels, based on the lactate dehydrogenase (LDH) reaction (107, 179). The mitochondrial free NAD+/NADH ratio can be determined by measuring the concentrations of glutamate, α-ketoglutarate, and NH3 and is based on the glutamate dehydrogenase (GDH) reaction (107, 179), although GDH activity is low in skeletal muscle (10, 179).
In resting human muscle, total NAD+ and NADH concentrations are estimated to be ∼1.5–1.9 and ∼0.08–0.20 mmol/kg dry wt muscle, respectively (62, 80, 93, 154, 155, 159, 160). Based on the approximate volumes of distributions of mitochondria, the extramitochondrial space (i.e., cytosol) and their mass fractions [i.e., %cell volume: cytosol = 90% and mitochondria = 10% (50)], Cabrera and colleagues (42, 119) estimate the total, mitochondrial, and cytosolic compartment concentrations in skeletal muscle for NAD+ and NADH, respectively, to be approximately as follows: total, 0.45 and 0.05 mmol/kg cell wet wt; cytosol, 0.15 and 0.00028 mmol/kg cytosolic wet wt; mitochondria, 3.15 and 0.5 mmol/kg mitochondrial wet wt [Note: to convert to dry weight muscle, multiply by ∼4.2 (145)]. Thus, the NAD+/NADH ratio in resting skeletal muscle is estimated to be much higher in the cytosol (∼540) compared with mitochondria (∼6.3), and overall, greater than ∼95% of cellular NADH is estimated to be in the mitochondrial compartment. The nucleus comprises ∼1% of muscle cell volume (50), and considering that the nuclear-to-cytosolic NAD+(H) levels are considered to be in equilibrium, the nuclear NAD+ and NADH concentrations would be estimated to be comparable to the aforementioned values for the cytosol. Although higher than estimates in other cells [NAD+, ∼10–100 μM; NADH, ∼130 nM (53, 188)], considering the high density of mitochondria and metabolic turnover of skeletal muscle, these approximations seem reasonable.
Relevant to the redox state and covalent activation of NAD+- or NADH-dependent signaling proteins is the fact that most cellular NAD+ and NADH is bound to proteins (13, 54, 171, 176, 179, 180). This makes it quantitatively difficult to determine the free NAD+ and NADH levels (and the free NAD+/NADH ratio), which ultimately represent the metabolically active forms of these coenzymes. Measurement of free NAD+(H) levels is further complicated by the fact that NADH binds proteins more firmly than NAD+ (54, 171, 180). It should be noted, however, that studies in rat hippocampus using time-resolved fluorescence and anisotropy decay suggest the ratio of free-to-bound NADH to be ∼0.78 (175). Whether this is the case in skeletal muscle is unknown. Based on the MIM for LDH, in resting skeletal muscle the free cytosolic NADH level is estimated to be ∼0.5–1.5% of total cytosolic NADH (158).
In skeletal muscle, NAD+ levels are highest in mitochondria (42, 119); thus, by extension one might infer that oxidative skeletal muscle (with a greater abundance of mitochondria) would have overall higher NAD+ levels compared with glycolytic muscle. Supporting this notion, in human resting muscle, NAD+ concentration is positively correlated with the percentage of slow-twitch fibers (62). However, in rat soleus and extensor digitorum longus (EDL) muscles, no differences in NAD+ levels were noted, although differences in the degree of reduction of the NAD+ couple were found (i.e., higher NAD+ levels in soleus vs. EDL mitochondria), which may be indicative of the differing metabolic characteristics of these muscles (158).
Changes in NAD+ and NADH Concentrations and the NAD+/NADH Ratio in Muscle During Exercise
Animal studies.
Early studies by Britton Chance and colleagues (27, 28, 33) and others (61, 87, 88), typically in amphibian muscle, used fluorescence-based methods (128, 129) to demonstrate that NADH levels decrease (and thus NAD+ levels increase) during muscle contraction. With respect to mammalian muscle, Jobsis and Stainsby (89) used the same technique to study NADH oxidation in the gastrocnemius-plantaris and gracilis muscle groups in dogs and found that low-intensity (5 Hz) and tetanic contractions increased NAD+ levels. By manipulating the ability of mitochondria to oxidize NADH, they concluded that the increase in tissue NAD+ primarily occurs inside mitochondria (89). In contrast to studies that show that NAD+ increases with contraction, Duboc et al. (44) reported an increase in NADH during tetanic contractions in soleus and EDL muscles of the rat. A limitation of the fluorometric technique used in these studies is that it does not provide quantitative assessment of NAD+, NADH, and the NAD+/NADH ratio. Addressing this limitation, Edington et al. (48) measured NAD+ biochemically and estimated the NAD+/NADH ratio using the MIM method (using the lactate/pyruvate and β-hydroxybutyrate/acetoacetate ratios). Thus, in untrained and trained rats, cytosolic and mitochondrial NAD+ concentrations, as well as the NAD+/NADH ratio, were increased by low-intensity muscle contraction of the gastrocnemius-plantaris muscles. As one would expect, the increase in the mitochondrial NAD+/NADH ratio during the same absolute exercise was lower in trained rats (47, 48). In the soleus and EDL muscles of the rat, twitch or tetanic contractions increased NAD+ levels (as measured by decreased NADH fluorescence) during contraction (178). Supporting this notion, studies in insect and canine muscle using the MIM method [based on the glutamate dehydrogenase (GDH) reaction] found that the mitochondrial NAD+/NADH ratio is increased during exercise at a variety of exercise intensities (34, 135, 152, 153, 181). Chronic low-frequency (10 Hz) stimulation of the rat tibialis anterior muscle also increased NAD+ levels after 15 min of contraction, and the NAD+/NADH ratio was significantly increased for up to 24 h of stimulation (65). In mice, swimming exercise increased muscle NAD+ levels (23), and in rats, endurance exercise training resulted in a sustained (as samples were measured 2 days after the last exercise bout) increase in NAD+ levels in gastrocnemius muscle of young and old rats (104). However, an increase in NAD+ and the NAD+/NADH ratio during exercise is not a universal finding. In one study, NADH increased and the NAD+/NADH ratio decreased during flight in insect muscle (77), whereas in mouse muscle no change in NAD+ levels at the end of running exercise was found, although an increase 3 h after exercise was noted (22). In addition, in electrically stimulated canine muscle (gastrocnemius-plantaris muscles), cytoplasmic NAD+ levels were reduced during exercise (64), whereas in electrically-stimulated (5 Hz) soleus muscle, no change in NAD+ levels was found (167).
Human studies.
In human muscle, the effects of exercise on NAD+ levels and the NAD+/NADH ratio are largely the opposite of those found in animal studies. Muscle NAD+ levels were decreased with exercise at 65% and 100% of maximal oxygen uptake (V̇o2 max), and although increased muscle water accounted for ∼73% of this decrease, NAD+ levels were still reduced when assessed on a dry weight basis (62). The first studies to quantitatively measure both NAD+ and NADH levels in human muscle at rest and during exercise were conducted by Dr. Kent Sahlin and colleagues (80, 93, 154, 155, 160). During maximal exercise and submaximal isometric contractions, NADH increased ∼140% above resting levels, whereas there was no significant change in NAD+ levels (80, 155). In contrast, no change in total muscle NADH concentration was noted throughout exercise at 75% V̇o2 max (157), whereas NADH and the cytosolic NAD+/NADH ratio were decreased during exercise at 50% V̇o2 max (93). Similar to this, a number of studies found that the cytosolic NAD+/NADH ratio is reduced during exercise (66, 141), although the magnitude of reduction is lower after exercise training (141). Exercise intensity appears to be an important contributor to the differences in measured NAD+(H) and NAD+/NADH ratio during exercise in animal versus human studies. For example, NADH decreased (and the cytosolic NAD+/NADH ratio was unchanged) from resting values during exercise at 40% V̇o2 max, but both NAD+ and the cytosolic NAD+/NADH ratio were increased above resting values at 75% and 100% V̇o2 max (160). Moreover, a series of in silico studies (that distill the NAD+ and NADH information from some of the aforementioned papers) predict that whole tissue, cytosolic, and mitochondrial NAD+/NADH ratios are reduced during exercise at 60% V̇o2 max (119) but are increased during exercise at a lower intensity (65 W) (21, 41). Interestingly, estimation of the mitochondrial redox state during exercise in human muscle using the MIM method estimated that the free NAD+/NADH ratio is significantly increased at 75% and 100% V̇o2 max (63).
Summary.
There are conflicting results in both animal and human studies as to whether or not exercise increases or decreases NAD+, NADH, and the NAD+/NADH ratio. There are many reasons that may underlie these differences, including training state, intensity of contraction, duration of exercise, time point of measurement during exercise, the analytical technique used to measure NAD+(H) and the NAD+/NADH ratio (e.g., fluorometric, biochemical, and MIM method), and the compartment that was measured (whole tissue, mitochondrial, or cytosolic). From a more “big picture” perspective, because the majority of change in muscle NADH levels with exercise is presumed to occur within the mitochondrial compartment, a large increase in NADH during exercise would correspond to a decreased redox potential, which could be inhibitory on mitochondrial oxidative enzymes and limit TCA cycle flux (63). The simplest explanation for this would be a “backing up” of the electron transport chain (ETC) due to limitations in the capacity to oxidize NADH. This is supported by the findings that elevated total muscle NADH concentrations decrease to resting levels during recovery from high-intensity exercise (80, 155). Alternatively, an increase in the mitochondrial redox potential would be expected to facilitate generation of NADH by increasing the availability of NAD+ for pyruvate dehydrogenase and the various dehydrogenase reactions of the TCA cycle and β-oxidation (63). In muscle, measurement and extrapolation of NAD+(H) metabolism during exercise is further complicated by the fact that muscle is comprised of subsarcolemmal and intermyofibrillar mitochondria, which are known to have different capacities for substrate oxidation (32, 108, 184). Whether NAD+(H) kinetics during exercise is different within these mitochondrial populations is unknown, and it is likely that fluorometric studies of NAD+(H) metabolism with contraction reflect changes in the subsarcolemmal compartment and not the “whole” muscle. Considering these results and unresolved questions as a whole, it is clear that a major gap in our understanding of NAD+(H) metabolism during exercise is that no study has directly measured the free NAD+ and NADH levels or the subcellular localization and compartmentation of NAD+(H) metabolism. Such analysis is clearly very technically challenging and will likely require the use of advanced techniques such as HPLC and matrix-assisted laser desorption/ionization (MALDI)-mass spectrometry in combination with tissue fractionation methods or two-photon excitation microscopy (139, 162, 182, 188). Ultimately, measuring the free NAD+(H) levels is what is most important when it comes to regulation of proteins and pathways responsive to perturbations in NAD+(H), such as SIRT1, SIRT3, and PARPs, and subsequent effects on cellular function and metabolism.
Shuttling of NADH into the Mitochondria
The inner mitochondrial membrane is impermeable to NAD+ and NADH (115, 143), and shuttles are required to transport NADH from the cytosol to the mitochondria (138). This is accomplished via the exchange of metabolites that are reduced in the cytosol and oxidized in mitochondria (138). In skeletal muscle these are the glycerol 3-phosphate (G3P; or α-glycerophosphate) shuttle and the malate-aspartate (M-A) shuttle (83, 138, 163–165). Considering that exercise training enhances the capacity of muscle to oxidize NADH, the activities of enzymes of the M-A shuttle are higher in trained than in untrained muscle (29, 83, 163, 165), as well as in oxidative versus glycolytic muscle (29, 163). Moreover, muscle MDH activity decreases with detraining (29). In contrast, the activity of G3P dehydrogenase, a key enzyme in the G3P shuttle, is not increased by exercise training (163, 165) but is higher in glycolytic versus oxidative muscle (83, 163). Reducing equivalents may also be transferred to the mitochondria via the lactate shuttle, which is explained in detail elsewhere (18, 60). Briefly, the lactate shuttle hypothesis posits that cytosolic pyruvate is primarily converted to lactate, which is then transported via facilitated diffusion into mitochondria, where it is converted back to pyruvate by intramitochondrial LDH (18, 19, 60). Therefore, the lactate shuttle, via the LDH reaction, would allow for transfer of NADH from the cytosol to mitochondria in a manner similar to the G3P and M-A shuttles. It should be noted that, as debated by others, there is significant controversy over the presence of LDH within pure mitochondria and the existence of a lactate shuttle in skeletal muscle mitochondria (16, 20, 59, 147, 156, 184). In recent years, the NADH/cytochrome c (Cyto c) electron transport shuttle has also been described, in which the direct transfer of electrons from cytosolic NADH to molecular oxygen inside the mitochondrial matrix is achieved at respiratory contact sites (i.e., where both mitochondrial membranes are in contact) (1, 123). The transfer capacity of the NADH/Cyto c is reported to be equivalent to the M-A shuttle (1, 123). However, whether this system is active in skeletal muscle mitochondria, or is regulated by exercise training, is unknown.
Mitochondrial Adaptations to Endurance Exercise: Role of SIRT1 and SIRT3
Sirtuins are a family of class III deacetylases that possess NAD+-dependent deacetylase and mono-ADP-ribosyltransferase activities (40, 76, 125, 174, 185, 193). Over the past decade, there has been an explosion of research on the therapeutic potential of treating various diseases via activation of sirtuins, especially SIRT1, and, more recently, SIRT3 (40, 76, 125, 174, 185, 193). In fact, a search on PubMed reveals that in just the past 12 years some 300 reviews have been published on sirtuins alone, with the majority of these focusing on SIRT1. The requirement for NAD+ for the deacetylase function of SIRT1 and SIRT3 provides a fundamental link between the activity of these proteins and perturbations in NAD+(H) status during exercise. Accordingly, our focus here is to discuss the roles of SIRT1 and SIRT3 in regulating the effects of acute and chronic exercise on mitochondrial function and biogenesis. A more general overview of sirtuin biology and function can be found elsewhere (40, 76, 125, 174, 185, 193).
SIRT1.
SIRT1 is the most studied of the mammalian sirtuins and is mainly found in the nucleus, although it also has cytosolic targets (40, 76, 174, 185, 193). Of particular importance to the focus of this review was the discovery that SIRT1 deacetylates and positively regulates the activity of PGC1α, a master regulator of mitochondrial biogenesis (5, 57, 132, 150). Thus, SIRT1 has also been put forth as a principal regulator of mitochondrial biogenesis via its ability to regulate PGC1α function. Following this, a number of studies have noted that SIRT1 gene (31, 45, 127) or protein (68, 117, 118, 121, 122, 173) levels increase in skeletal muscle in response to acute or chronic exercise, in parallel with upregulation of mitochondrial content. However, other studies have found either no effect (25, 75) or a decrease (73–75, 104) in SIRT1 protein in skeletal muscle with chronic muscle contraction (via electrical stimulation) or endurance exercise. Complimenting these latter studies, skeletal muscle SIRT1 protein content does not scale with muscle oxidative capacity or PGC1α abundance (73–75). Moreover, when SIRT1 was overexpressed in skeletal muscle, mitochondrial function and abundance [as measured by ETC and mitochondrial transcription factor A (mtTFA) protein abundance and citrate synthase activity], gene expression of mitochondrial proteins, and PGC1α gene and/or protein expression was not changed (56, 140) or even decreased (74). In C2C12 myotubes, overexpression of SIRT1 increased PGC1α gene expression and PGC1α promoter activity (5), although effects on mitochondrial biogenesis and function were not assessed. When SIRT1 protein (15, 22, 56, 57) or deacetylase activity (142) is knocked out in skeletal muscle of mice or C2C12 myotubes, there is no reduction in mitochondrial function [e.g., O2 consumption, proton conductance, activity of ETC enzymes or citrate synthase (CS)], number (as measured by mtDNA/nDNA ratio and ETC protein abundance), PGC1α gene and/or protein expression, or the gene expression of mitochondrial proteins. In contrast, PGC1α gene expression is lower in the tibialis anterior, gastrocnemius, and soleus of SIRT1-null mice, although whether this reduction impacts PGC1α protein expression, mitochondrial biogenesis, or mitochondrial function was not assessed (5). Moreover, in studies in C2C12 and mouse primary myotubes, SIRT1 knockdown downregulates mitochondrial and fatty acid oxidation gene expression, fatty acid oxidation, and CS activity (22, 57), whereas SIRT1 overexpression increases PGC1α expression, transcriptional activity, and mitochondrial genes (5, 57). Despite reductions in PGC1α gene expression, SIRT1 knockdown in C2C12 myotubes does not reduce PGC1α protein expression (56, 57).
Possible reasons for discrepancies between these different studies have recently been reviewed (70, 72). An obvious reason for many of these differences relates to differences between studying SIRT1 biology in vitro using muscle cells (particularly C2C12 muscle myotubes), versus in vivo using rodent models and adenoviral techniques. Also, the precise definition of mitochondrial biogenesis and function is different across these studies, with measurement of the gene expression of PGC1α and mitochondrial genes being a common outcome measure. While measurement of gene expression provides important information, if positive or negative effects on mitochondrial biogenesis/function are to be concluded, it will be helpful in future studies to provide a more complete assessment of mitochondrial biogenesis/function, which may include measurement of mitochondrial protein synthesis and turnover, submaximal and maximal O2 consumption, ETC enzyme activity and protein abundance, the mtDNA/nDNA ratio, or mitochondrial morphology by electron microscopy.
To resolve the incongruent findings regarding SIRT1 protein levels and mitochondrial adaptations to exercise, it has been proposed that SIRT1 activity might be the underlying mediator of these changes. Nuclear SIRT1 activity is positively correlated with oxidative capacity (i.e., CS activity, complex IV abundance) across different muscle types and is also associated with the onset of mitochondrial adaptations to acute exercise, as well as chronic changes in oxidative capacity that occur with exercise training (75). Other studies have also reported an increase in SIRT1 activity (as measured by the SIRT1 activity assay or deacetylation of PGC1α) with acute and chronic muscle contraction (22, 23, 25, 73, 75, 104, 117, 118), although no increase was found with voluntary wheel running (despite increased mitochondrial biogenesis) (25). Notably, the SIRT1 activity assay uses a peptide substrate that contains Fluor de Lys, a nonphysiological fluorescent moiety, and studies using this assay (25, 73–75, 104), may be complicated by the fact that measured SIRT1 activity is potentially an artifact of the fluorophore itself (17, 90). This assay also measures SIRT1 activity in the presence of maximal NAD+, which does not reflect the NAD+ levels in the muscle. With this in mind, measurement of the acetylation status of proposed SIRT1 targets (e.g., p53, FOXO, or PGC1α), SIRT1 binding to the promoters of known gene targets, or measurement of the gene expression of SIRT1 target genes would complement measurements of SIRT1 activity and provide a more physiological read-out of SIRT1 function.
It is important to note that SIRT1 activity can be regulated via phosphorylation (56, 69, 91, 161). Recently, Gerhart-Hines (56) demonstrated that SIRT1 was phosphorylated in its catalytic domain by protein kinase A (PKA), which is also activated by endurance exercise. In addition, activation of PKA (via forskolin) increased SIRT1 phosphorylation and activity, including induction of PGC1α expression in skeletal muscle (56). This occurred despite no increase in NAD+ (56), perhaps suggesting that SIRT1 activity (and function) could be regulated independently of NAD+ in skeletal muscle. However, the effects of exercise on SIRT1 phosphorylation in skeletal muscle are unknown.
A limitation of the aforementioned studies that investigated SIRT1 and exercise-induced mitochondrial biogenesis is that they are correlative and do not address whether SIRT1 is required for exercise-induced mitochondrial biogenesis in skeletal muscle. To address this limitation, Philp et al. (142) studied the effects of acute and chronic exercise training on muscle function, PGC1α acetylation, and mitochondrial biogenesis in mice with muscle-specific knockout of SIRT1 deacetylase activity (mKOSIRT1). In muscle from mKOSIRT1 mice, there was no compensatory upregulation in the gene expression of SIRT2–7 or the protein abundance of SIRT3 and SIRT6 (S. Schenk, A. T. White and A. Philp, unpublished observations). Similar to previous studies in mice (14), no impairment in mitochondrial function or number (e.g., abundance and/or activity of complexes I-IV of the ETC, CS activity, mtDNA/nDNA ratio) in muscle from mKOSIRT1 vs. control mice was found, nor was muscle endurance capacity impaired (142). Interestingly, mKOSIRT1 and control mice also had comparable reductions in PGC1α acetylation and induction of exercise-response genes (e.g., mitofusin 2, PDH kinase 4, cytochrome c) after acute exercise, and normal mitochondrial adaptations (e.g., abundance and/or activity of complexes I-IV of the ETC, CS activity, mtDNA/nDNA ratio) to wheel running training (142). Thus, studies in mKOSIRT1 mice reveal that SIRT1 deacetylase activity is not required for normal function of mitochondria in skeletal muscle, nor is it required for exercise-induced adaptations. Regarding PGC1α acetylation, the authors found that the acetyltransferase that regulates PGC1α transcriptional activity, general control of amino acid synthesis 5 (GCN5) (57, 116, 132), is modulated by exercise such that nuclear localization of GCN5 was reduced and less GCN5 coimmunoprecipitated with PGC1α after exercise (142). Similarly, whole body deletion of SRC-3, an upstream activator of GCN5, results in decreased PGC1α acetylation and increased mitochondrial biogenesis (36), whereas overexpression of GCN5 reduces mitochondrial gene expression and fatty acid oxidation (57). This study suggests, therefore, that the reduced acetylation of PGC1α with exercise is not due to increased deacetylation by SIRT1 but rather is a result of reduced acetylation by GCN5 (142). This is an interesting finding, and it demonstrates that PGC1α acetylation is a balance of the activities of the proteins that acetylate and deacetylate it. Currently, the mechanisms by which exercise regulates GCN5 activity, GCN5 translocation from the nucleus, and the GCN5-PGC1α interaction, are unknown.
How SIRT1 gene expression is regulated in response to exercise is also unknown. In liver cells, SIRT1 gene expression is regulated via opposing effects of cyclic AMP response element-binding protein (CREB) and carbohydrate response element-binding protein (ChREBP) (134), such that increased CREB binding to the SIRT1 promoter increases SIRT1 transcription, whereas ChREBP binding impairs it. CREB has also been shown to regulate PGC1α transcription (3, 4). Given that acute exercise activates CREB (49, 142), it is possible that this is responsible, at least in part, for increased SIRT1 gene transcription with exercise. The effects of exercise on ChREBP expression and activation in muscle have not been studied. It is also possible that SIRT1 gene expression is regulated by changes in NADH levels. To this end, SIRT1 gene expression is also regulated by CtBP (189), a transcriptional corepressor that has a 100-fold greater affinity for NADH than NAD+ (53, 188). While we discuss CtBP in more detail later in this review, of note here is that changes in NADH levels during or after exercise could reduce the repressive effects of CtBP on SIRT1 gene transcription in skeletal muscle.
SIRT3.
SIRT3 is considered to be a mitochondria-localized protein (8, 35, 71, 124, 136, 170, 172), although there have been some conflicting reports on its localization (166). Relevant to our discussion, in skeletal muscle, SIRT3 appears to localize solely to mitochondria (71) and scales with markers of skeletal muscle oxidative capacity (71, 137). Additionally, SIRT3 is decreased in old vs. young sedentary individuals but is higher in endurance-trained vs. sedentary individuals regardless of age (111). In line with this, exercise training or chronic electrical stimulation (71, 82, 137), but not acute exercise (71, 82), increases skeletal muscle SIRT3 protein levels and is specific to those muscles recruited during the exercise intervention. Complementing these studies, knockdown of SIRT3 in C2C12 muscle cells decreases basal and maximal O2 consumption rates and mitochondrial content and prevents PGC1α-induced activation of mitochondrial genes (86, 105). Although knockdown of SIRT3 does not reduce the total mitochondria number as measured by the abundance of complexes I, III, and V of the ETC (86), it does reduce skeletal muscle fatty acid oxidation by ∼50%, due to hyperacetylation of long-chain acyl-CoA dehydrogenase (LCAD) (81). Alternatively, overexpression of SIRT3 in C2C12 myotubes increases mitochondrial DNA content (105). Taken together, these studies suggest that SIRT3 plays an important role in regulating skeletal muscle mitochondrial biogenesis, and potentially fatty acid oxidation, in response to long-term exercise training. However, a recent paper by Yang et al. (183) in C2C12 myotubes and skeletal muscle from SIRT3-null mice counters this perspective. In their paper, the authors demonstrate that SIRT3 acts to reduce mitochondrial protein synthesis (and thus mitochondrial biogenesis) via its ability to deacetylate mitochondrial ribosomal protein L10 (MRPL10) and negatively regulate the activity of mitochondrial ribosomes. Thus, rather than increase mitochondrial protein synthesis, SIRT3 appears to have the opposite effect in skeletal muscle. The teleological implications of this will be discussed shortly.
Increased ATP utilization during exercise is matched through increased mitochondrial ATP production, which occurs via oxidation of mitochondrial NADH produced in metabolic pathways such as glycolysis, the TCA cycle and β-oxidation. Interestingly, up to one-fifth of mitochondrial proteins are acetylated, as are many of the proteins in these metabolic pathways, which has important effects on their function (95, 177, 191). Indeed, SIRT3 appears to be responsible for much of the deacetylation of mitochondrial proteins (124, 136, 170, 172). Of potential interest to ATP generation in skeletal muscle during exercise, SIRT3 deacetylates and activates the TCA cycle and ETC enzymes, including succinate dehydrogenase (SDH) (30), ubiquinol-cytochrome c reductase hinge protein (a component of complex III) (114), malate dehydrogenase (137), NDUFA9 of complex I (2), GDH (124), ATP synthase (114), and isocitrate dehydrogenase-2 (ICDH2) (168). Also, SIRT3 deacetylates and activates the β-oxidation enzyme LCAD (81). With this information in mind, we propose that a possible role of SIRT3 in skeletal muscle is the acute regulation of enzymes and pathways that generate ATP in response to ATP demand during exercise. This is supported by the fact ATP production in heart, kidney, and liver from SIRT3-null mice is reduced by more than 50% (2), although whether this is the case in skeletal muscle is unknown. In the context of the findings of Yang et al. (183) showing that SIRT3 reduces (rather than increases) mitochondrial protein synthesis, as measured by a [35S]methionine translation-based assay, this also would make teleological sense. Thus, during exercise it is necessary to generate ATP to maintain force production, so pathways that utilize energy, such as protein synthesis, would be momentarily halted. The actions of SIRT3, therefore, are akin to the effects of AMPK on enhancing energy production and inhibiting pathways that use energy for processes other than to maintain ATP production and muscle work (79, 85), albeit the effects of SIRT3 are specific to mitochondria. It will be of interest in future studies to determine whether mitochondrial biogenesis in response to exercise is impaired in SIRT3-null mice. Also, given that fatty acid oxidation increases during endurance exercise (38), it will be interesting to determine whether acute exercise alters substrate utilization in parallel with activation of SIRT3 activity and deacetylation of its downstream targets. Studies using muscle-specific SIRT3-null mice and exercise will no doubt be very informative regarding such questions.
PARPs and Mitochondrial Biogenesis in Skeletal Muscle
The PARPs are major consumers of nuclear NAD+ and therefore compete with SIRT1 for NAD+ in the nucleus (40, 76, 174, 185, 193). Considering this, a series of papers from the laboratory of Johan Auwerx recently investigated the effects of knocking down PARP1 and PARP2 on skeletal muscle mitochondrial biogenesis in C2C12 myotubes and mice. PARP1-null mice had increased levels of NAD+, reduced acetylation of SIRT1 substrates such as PGC1α and FOXO1, and increased mitochondrial biogenesis, as measured by mitochondrial gene expression, mitochondrial morphology, SDH staining, mtDNA content, and O2 consumption (7). Increased muscle SIRT1 activity may in part be due to increased protein content, although SIRT1 activity was increased in HEK 293 cells without an increase in SIRT1 protein content (7). Complementing these findings, treatment of mice with PARP1 inhibitors increased NAD+ levels and SIRT1 activity (7). The activity of other nonnuclear sirtuins including, SIRT2 and SIRT3, however, were unchanged in PARP1-null tissues (7), suggesting that the upregulation of SIRT1 in the absence of PARP1 may be due to a local change in the NAD+ pool in the nuclear compartment. Similar to PARP1, knockdown of PARP2 in C2C12 myocytes increased SIRT1 activity (6). In skeletal muscle this appeared to occur through both an increase in intracellular NAD+ levels and modulation of the SIRT1 promoter by PARP2 (6). As expected, SIRT1 activity was increased in PARP1- and PARP2-null mice, and these mice also had increases in skeletal muscle mitochondrial biogenesis (e.g., mtDNA, mitochondrial morphology and gene expression, and SDH staining), and their muscle demonstrated a more oxidative phenotype (6, 7). Moreover, PARP2-null mice had increased endurance as measured by a treadmill endurance test (6). Whether this was due to improvements in skeletal muscle per se or was a function of the changes in other tissues, such as the heart, was not determined. Collectively, these studies are very interesting and suggest that inhibition of PARPs could be used to enhance muscle mitochondrial biogenesis by increasing nuclear NAD+ levels and increasing SIRT1 activation. If exercise leads to an increase in NAD+ in the nuclear compartment, it will be interesting in the future to determine if acute exercise leads to inhibition of PARP1 and PARP2 so as to maximize NAD+ levels and SIRT1 activation, although it is notable that in vivo SIRT1 deacetylase activity is not required for the ability of exercise to enhance mitochondrial biogenesis (142). Thus, studies that cross PARP1/2-null mice with mKOSIRT1 mice, or studies with PARP inhibitors in mKOSIRT1 mice, will help to definitively determine if PARP inhibition works through SIRT1 in vivo.
Contribution of NADH to Mitochondrial Adaptations to Exercise: Possible Role of CtBP
As discussed above, CtBP is a transcriptional corepressor that is greater than 100-fold more sensitive to perturbations in cellular NADH vs. NAD+ levels (53, 188). Considering that the cytosolic/nuclear content of NAD+ in muscle is estimated to be ∼540-fold higher than NADH (42, 119), conversion of NAD+ to NADH, or vice versa, would therefore result in a greater change in NADH levels. By extension, and as reasoned by others (53, 188, 189), changes in nuclear NADH, rather than NAD+, could link perturbations in NAD+/NADH ratio to gene transcription. To this end, CtBP regulates mitochondrial morphology and function in mouse embryonic fibroblast and liver-related cells via its ability to regulate Bcl-2-associated X protein (Bax) (94). CtBP also represses the transcriptional activity of myocyte enhancer factor 2 (MEF2) (186), a key transcription factor in the regulation of mitochondrial biogenesis (39, 131) that shows increased DNA binding in response to exercise (130). The regulation of MEF2 transcriptional activity, however, is complex, as MEF2 is deacetylated by SIRT1, and deacetylation of MEF2 in vitro reduces (not increases) its transcriptional activity (126, 192). Clearly, the effects of exercise on NAD+(H), SIRT1, CtBP, and MEF2, and the subsequent transcriptional response, may represent a balance of these activating and inhibitory signals that likely involves additional levels of regulatory control, such as ubiquitination, sumoylation, and phosphorylation (67, 92, 149). Taken together, these studies point to a potentially important role of CtBP, via its sensitivity to changes in NADH, in the modulation of mitochondrial biogenesis in skeletal muscle in response to exercise.
Replenishing NAD+ Levels in Skeletal Muscle: an Important Consideration
If an increase in NAD+ during exercise leads to an increase in the activity (and thus consumption of NAD+) by SIRT1, SIRT3, PARP1, or PARP2, then it would be important for skeletal muscle to replenish NAD+ levels in the cytosolic, nuclear, and mitochondrial compartments during or after exercise. In mammals, the NAD+ biosynthetic and salvage pathways replenish NAD+, and the specifics of these pathways are reviewed elsewhere (11, 12, 84, 97, 128, 144, 190); an overview of these pathways is presented in Fig. 2. Except for research on NAM phosphoribosyltransferase [NAMPT; also known as pre-β-cell colony-enhancing factor 1 (PBEF1) or visfatin], the contribution of these pathways to replenishment of NAD+ in skeletal muscle and in response to exercise is, to date, essentially unstudied.
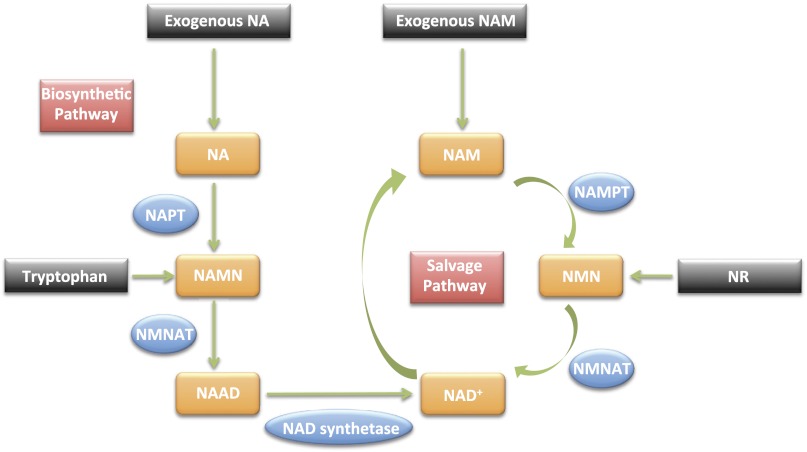
Fig. 2.
Replenishment of NAD+ through biosynthesis (de novo) and salvage pathways. Given there are many NAD+-consuming enzymes, it is essential that NAD+ be replenished to maintain compartmental NAD+ levels. This occurs through the salvage and biosynthetic pathways. NA, nicotinic acid; NAM, nicotinamide; NAMN, NA mononucleotide; NMN, NAM mononucleotide; NMNAT, NMN adenylyltransferase; NAAD, NA adenine dinucleotide; NAD+, NAM adenine dinucleotide; NAPT, NA phosphoribosyltransferase; NAMPT, NAM phosphoribosyltransferase; NR, nicotinamide riboside. Except for NAMPT, the role of these pathways in NAD+ replenishment in skeletal muscle, and in response to exercise, are essentially unknown. Molecules generated in each pathway are in orange; enzymes are in blue.
NAMPT is located in the nucleus, cytosol, and mitochondria (96, 148, 182) and is part of the NAD+ biosynthetic pathway that converts NAM to NAM mononucleotide (NMN) (40, 76, 174, 185, 193). This reaction is potentially important for maintaining the activity of SIRT1 and SIRT3, as nicotinamide (which is generated in the deacetylase reaction of sirtuins, including SIRT1 and SIRT3) is a negative regulator of SIRT1 and SIRT3 (11, 12, 84, 97, 128, 144, 169, 190). Indeed, in HEK 293 cells, NAMPT plays an important role in protecting against cell death in response to genotoxic stress by maintaining mitochondrial NAD+ levels and SIRT3 activation (182). However, in plasma from humans and mice, NAM concentrations (which range from 0.3 to 5 μM) are lower than the reported IC50 for SIRT1 inhibition but is in the range of the KM for NAMPT (24, 148, 151). Thus, whether or not NAM levels in muscle reach a level sufficient to inhibit SIRT1/SIRT3 is unknown. This aside, in rodents, endurance exercise increases NAMPT gene and/or protein expression in parallel with increased tissue NAD+ levels (23, 104). Similarly, in humans, NAMPT protein abundance is higher in trained than in untrained individuals and is increased by exercise training, although whether this increased NAD+ levels was not measured (37). Thus, in the context of increased SIRT1/SIRT3 activity during and after exercise, a coordinated increase in NAMPT activity may act to maintain SIRT1/SIRT3 activity by consuming NAM and also replenishing NAD+ (discussed below). The concentration of NAM in skeletal muscle is unknown; therefore, it will be interesting in future studies to determine whether NAMPT activity is increasing specifically in the mitochondrial, nuclear, and/or cytosolic compartments with exercise and whether this coincides with changes in NAM levels. All together, such measurements will provide important information regarding the precise contribution of NAMPT to NAD+ metabolism and the regulation of SIRT1 and SIRT3 activity in skeletal muscle in response to exercise.
To generate NAD+, NMN generated by the NAMPT reaction is converted by NMN adenylyltransferase (NMNAT) to NAD+. NMNAT can also convert nicotinic acid (NA) mononucleotide (NAMN) to NA adenine dinucleotide (NAAD), which is subsequently converted to NAD+ by NAD+ synthase. There are three isoforms of NMNAT: NMNAT1 and NMNAT2 are localized in the cytosol and nucleus, and NMNAT3 appears to be exclusively in mitochondria (113, 133). At the mRNA level, NMNAT1 is highly expressed in skeletal muscle (51, 52, 113), NMNAT2 is expressed at low levels, while NMNAT3 is very low or absent (113). The protein levels and activities of these proteins in skeletal muscle are unknown. The presence of NMNAT1 and to a lower extent NMNAT2, in skeletal muscle suggests that they may play an important role in replenishing nuclear and cytosolic NAD+ levels, and it will be interesting to see if exercise coordinately increases NAMPT and NMNAT1/2 levels in order to maintain the overall cytosolic/nuclear NAD+ pools. Regarding replenishment of mitochondrial NAD+, the inner mitochondrial membrane is impermeable to NAD+ and NADH (115, 143), which poses a potential problem for maintaining the mitochondrial NAD+ level, particularly if NAD+ consumption by SIRT3 is increased during exercise. Only recently was it demonstrated in HeLa S3 cells that NMNAT3 is the only known enzyme of NAD+ synthesis in mitochondria (133). Although NMNAT3 gene expression is very low in skeletal muscle, it will be of interest in future studies to determine if NMNAT3 activity in skeletal muscle correlates with mitochondrial density or if exercise increases the activity or abundance of NMNAT3 even independently of an increase in mitochondrial abundance. Alternatively, perhaps a different or an additional mitochondrial NAD+ salvage or biosynthetic pathway is present in skeletal muscle mitochondria.
Concluding Remarks: There Are Still Many Unanswered Questions
It has been more than 100 years since the discovery of the pyridine nucleotides NAD+ and NADH. While for much of this time NAD+(H) was considered to participate primarily in metabolic reactions that led to generation of ATP through their ability to act as substrates for enzymes or as covalent modifiers of enzyme function, these coenzymes are potentially key mediators of the adaptive response to exercise. Indeed, changes in NAD+(H) levels in concert with known NAD+(H)-sensing enzymes provide a logical link between exercise-induced metabolic stress and subsequent mitochondrial adaptations. Specifically, the effects of SIRT1, PARP1/2, and CtBP appear to manifest through their ability to directly or indirectly modulate the transcriptional response to exercise; they likely do not contribute to an immediate increase in ATP production during acute exercise (Fig. 3). Very little, however, is known about NAD+(H) dynamics in the nucleus of skeletal muscle and how this affects the transcriptionally based adaptations central to endurance exercise training. Regarding SIRT3, we propose that it acts as an acute regulator of mitochondrial ATP production via its ability to regulate the enzymatic activity of various TCA and ETC enzymes (and possibly yet to be discovered targets). An additional component of this acute regulation is proposed to include a reduction in mitochondrial protein synthesis during exercise (Fig. 3). It is possible that, during exercise, SIRT1 plays a similar role in regulating cellular protein synthesis in the cytosol via its ability to negatively regulate mammalian target of rapamycin (mTOR) and/or its interaction with tuberosclerosis complex 2 (TCS2) (58). Whether this regulation occurs in muscle or during exercise is not known. In addition, little is known regarding the coordination of NAD+ consuming and regenerating pathways in skeletal muscle and whether these two opposing events are regulated by common mechanisms. Furthermore, our understanding of the compartmentation of NAD+(H) metabolism and quantitative changes in NAD+, NADH, and the NAD+/NADH ratio in subcellular compartments in skeletal muscle at rest and in response to exercise is poor. Although technically challenging to measure, such investigation will be highly informative with respect to understanding the activation or inhibition of both NAD+(H)-responsive proteins. For example, while NAD+ can clearly activate sirtuins, NADH can act as a competitive inhibitor of SIRT1 (120). However, the relative binding affinity of NAD+ for SIRT1 is ∼1,000-fold greater than NADH, and overall, the ability of NADH to inhibit SIRT1 activity is proposed to be minimal in an in vivo setting (169). Thus, determining the precise contribution of changes in NAD+, NADH, and the NAD+/NADH ratio will be important. In the end, it is likely that a combination of changes in free NAD+ and NADH levels and the NAD+/NADH ratio within specific subcellular compartments is important. Thus, as research on NAD+(H) metabolism continues into its second century, there are still many important research questions to be resolved regarding their effects on the adaptive response to exercise in skeletal muscle. Ultimately, such research holds great promise for improving our fundamental understanding of skeletal muscle function in response to exercise, which has obvious and important implications for human health and treatment of skeletal muscle-related diseases.
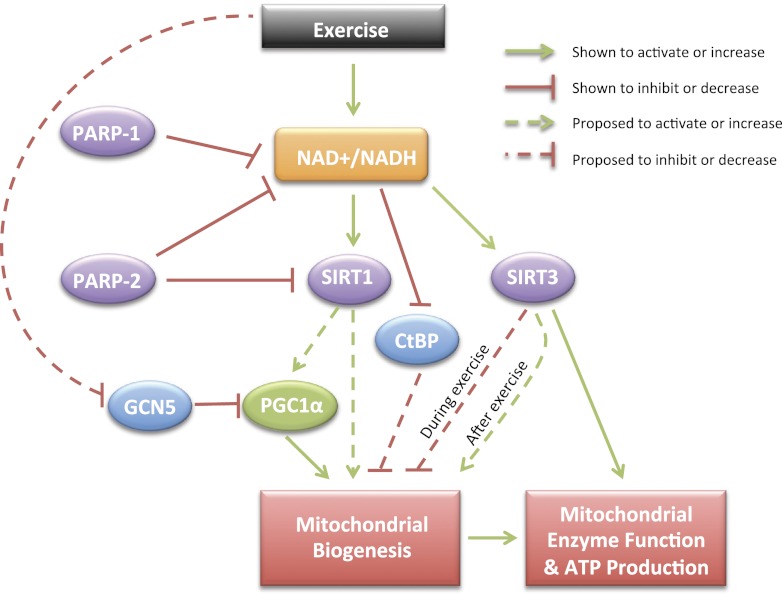
Fig. 3.
Proposed mechanism for exercise-induced mitochondrial biogenesis via NAD+/NADH metabolism. PARP, poly [ADP-ribose] polymerase; SIRT, sirtuin; GCN5, general control of amino acid synthesis; PGC1α, peroxisome proliferator-activated receptor-γ coactivator 1α; CtBP, COOH-terminal binding protein. Increased ATP demand during exercise leads to an increase in the free cytosolic/nuclear and mitochondrial NAD+ level and NAD+/NADH ratio, which provides increased substrate for the NAD+-consuming enzymes (in purple), SIRT1, SIRT3, PARP1, and PARP2. Exercise also reduces the availability of NADH, the predominant covalent activator of CtBP. It is hypothesized that during exercise increased ATP production is facilitated by SIRT3-mediated deacetylation of a series of enzymes in the TCA cycle, β-oxidation, and ETC. In parallel, SIRT3 acutely reduces mitochondrial protein synthesis, which maximizes the availability of reducing equivalents for ATP production. Whether SIRT3 is required for induction of mitochondrial biogenesis after exercise remains to be determined. In response to exercise, SIRT1 is also activated by increased cytosolic/nuclear NAD+ levels, and while it likely can contribute to mitochondrial biogenesis through PGC1α-dependent and -independent mechanisms, it is not required for exercise-mediated deacetylation of PGC1α. Rather, acute exercise appears to reduce the inhibitory effect of the acetyltransferase GCN5 on PGC1α, via a mechanism that is still to be determined. PARP1 and PARP2 are able to directly or indirectly modulate SIRT1 activity (and mitochondrial biogenesis) by competing for NAD+, although the effects of exercise on the activity of these enzymes is unknown. Also, whether SIRT1 is required for the ability of PARP inhibition to induce mitochondrial biogenesis in skeletal muscle in vivo is not known. The transcriptional corepressor CtBP is activated by NADH, and it is hypothesized that during or after exercise, reductions in the cytosolic/nuclear NADH level reduces the repressive effects of CtBP on transcriptional modulators of mitochondrial biogenesis. Ultimately, increased activity of enzymes of the ETC, TCA cycle, and β-oxidation and/or increased mitochondrial number (i.e., biogenesis) leads to an enhanced capacity of mitochondria and muscle to generate ATP. Dotted lines indicate a hypothesized contribution of the pathway or that the data to date provides an incomplete perspective.
GRANTS
This research was supported in part by grants from the National Institutes of Health (R24 HD-050837, P30 AR-058878-02) including a Pilot and Feasibility Grant from the UCSD/UCLA Diabetes and Endocrinology Research Center (P30 DK-063491).
DISCLOSURES
No conflicts of interest, financial or otherwise, are declared by the author(s).
AUTHOR CONTRIBUTIONS
Author contributions: A.T.W. and S.S. conception and design of research; A.T.W. and S.S. prepared figures; A.T.W. and S.S. drafted manuscript; S.S. edited and revised manuscript; S.S. approved final version of manuscript.
ACKNOWLEDGMENTS
We are thankful to Dr. Andrew Philp for helpful discussion and input on the manuscript.
REFERENCES
- 1. Abbrescia DI, Piana GL, Lofrumento NE. Malate-aspartate shuttle and exogenous NADH/cytochrome c electron transport pathway as two independent cytosolic reducing equivalent transfer systems. Arch Biochem Biophys 518: 157– 163, 2012 [DOI] [PubMed] [Google Scholar]
- 2. Ahn BH, Kim HS, Song S, Lee IH, Liu J, Vassilopoulos A, Deng CX, Finkel T. A role for the mitochondrial deacetylase Sirt3 in regulating energy homeostasis. Proc Natl Acad Sci USA 105: 14447– 14452, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Akimoto T, Pohnert SC, Li P, Zhang M, Gumbs C, Rosenberg PB, Williams RS, Yan Z. Exercise stimulates Pgc-1alpha transcription in skeletal muscle through activation of the p38 MAPK pathway. J Biol Chem 280: 19587– 19593, 2005 [DOI] [PubMed] [Google Scholar]
- 4. Akimoto T, Sorg BS, Yan Z. Real-time imaging of peroxisome proliferator-activated receptor-γ coactivator-1α promoter activity in skeletal muscles of living mice. Am J Physiol Cell Physiol 287: C790– C796, 2004 [DOI] [PubMed] [Google Scholar]
- 5. Amat R, Planavila A, Chen SL, Iglesias R, Giralt M, Villarroya F. SIRT1 controls the transcription of the peroxisome proliferator-activated receptor-gamma co-activator-1alpha (PGC-1alpha) gene in skeletal muscle through the PGC-1alpha autoregulatory loop and interaction with MyoD. J Biol Chem 284: 21872– 21880, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Bai P, Canto C, Brunyanszki A, Huber A, Szanto M, Cen Y, Yamamoto H, Houten SM, Kiss B, Oudart H, Gergely P, Menissier-de Murcia J, Schreiber V, Sauve AA, Auwerx J. PARP-2 regulates SIRT1 expression and whole-body energy expenditure. Cell Metab 13: 450– 460, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Bai P, Canto C, Oudart H, Brunyanszki A, Cen Y, Thomas C, Yamamoto H, Huber A, Kiss B, Houtkooper RH, Schoonjans K, Schreiber V, Sauve AA, Menissier-de Murcia J, Auwerx J. PARP-1 inhibition increases mitochondrial metabolism through SIRT1 activation. Cell Metab 13: 461– 468, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Bao J, Lu Z, Joseph JJ, Carabenciov D, Dimond CC, Pang L, Samsel L, McCoy JP, Jr, Leclerc J, Nguyen P, Gius D, Sack MN. Characterization of the murine SIRT3 mitochondrial localization sequence and comparison of mitochondrial enrichment and deacetylase activity of long and short SIRT3 isoforms. J Cell Biochem 110: 238– 247, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Beard DA, Wu F, Cabrera ME, Dash RK. Modeling of cellular metabolism and microcirculatory transport. Microcirculation 15: 777– 793, 2008 [DOI] [PubMed] [Google Scholar]
- 10. Beis A, Zammit VA, Newsholme EA. Activities of 3-hydroxybutyrate dehydrogenase, 3-oxoacid CoA-transferase and acetoacetyl-CoA thiolase in relation to ketone-body utilisation in muscles from vertebrates and invertebrates. Eur J Biochem 104: 209– 215, 1980 [DOI] [PubMed] [Google Scholar]
- 11. Belenky P, Bogan KL, Brenner C. NAD+ metabolism in health and disease. Trends Biochem Sci 32: 12– 19, 2007 [DOI] [PubMed] [Google Scholar]
- 12. Berger F, Ramirez-Hernandez MH, Ziegler M. The new life of a centenarian: signalling functions of NAD(P). Trends Biochem Sci 29: 111– 118, 2004 [DOI] [PubMed] [Google Scholar]
- 13. Blinova K, Carroll S, Bose S, Smirnov AV, Harvey JJ, Knutson JR, Balaban RS. Distribution of mitochondrial NADH fluorescence lifetimes: steady-state kinetics of matrix NADH interactions. Biochemistry (Mosc) 44: 2585– 2594, 2005 [DOI] [PubMed] [Google Scholar]
- 14. Boily G, He XH, Pearce B, Jardine K, McBurney MW. SirT1-null mice develop tumors at normal rates but are poorly protected by resveratrol. Oncogene 28: 2882– 2893, 2009 [DOI] [PubMed] [Google Scholar]
- 15. Boily G, Seifert EL, Bevilacqua L, He XH, Sabourin G, Estey C, Moffat C, Crawford S, Saliba S, Jardine K, Xuan J, Evans M, Harper ME, McBurney MW. SirT1 regulates energy metabolism and response to caloric restriction in mice. PLoS ONE 3: e1759, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Bonen A, Hatta H, Holloway GP, Spriet LL, Yoshida Y. Reply to Brooks and Hashimoto, “Investigation of the lactate shuttle in skeletal muscle mitochondria”. J Physiol 584: 707– 708, 2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Borra MT, Smith BC, Denu JM. Mechanism of human SIRT1 activation by resveratrol. J Biol Chem 280: 17187– 17195, 2005 [DOI] [PubMed] [Google Scholar]
- 18. Brooks GA. Cell-cell and intracellular lactate shuttles. J Physiol 587: 5591– 5600, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Brooks GA, Dubouchaud H, Brown M, Sicurello JP, Butz CE. Role of mitochondrial lactate dehydrogenase and lactate oxidation in the intracellular lactate shuttle. Proc Natl Acad Sci USA 96: 1129– 1134, 1999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Brooks GA, Hashimoto T. Investigation of the lactate shuttle in skeletal muscle mitochondria. J Physiol 584: 705– 706; author reply 707–708, 2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Cabrera ME, Saidel GM, Kalhan SC. Modeling metabolic dynamics. From cellular processes to organ and whole body responses. Prog Biophys Mol Biol 69: 539– 557, 1998 [DOI] [PubMed] [Google Scholar]
- 22. Canto C, Gerhart-Hines Z, Feige JN, Lagouge M, Noriega L, Milne JC, Elliott PJ, Puigserver P, Auwerx J. AMPK regulates energy expenditure by modulating NAD+ metabolism and SIRT1 activity. Nature 458: 1056– 1060, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Canto C, Jiang LQ, Deshmukh AS, Mataki C, Coste A, Lagouge M, Zierath JR, Auwerx J. Interdependence of AMPK and SIRT1 for metabolic adaptation to fasting and exercise in skeletal muscle. Cell Metab 11: 213– 219, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Catz P, Shinn W, Kapetanovic IM, Kim H, Kim M, Jacobson EL, Jacobson MK, Green CE. Simultaneous determination of myristyl nicotinate, nicotinic acid, and nicotinamide in rabbit plasma by liquid chromatography-tandem mass spectrometry using methyl ethyl ketone as a deproteinization solvent. J Chromatogr B Analyt Technol Biomed Life Sci 829: 123– 135, 2005 [DOI] [PubMed] [Google Scholar]
- 25. Chabi B, Adhihetty PJ, O'Leary MF, Menzies KJ, Hood DA. Relationship between Sirt1 expression and mitochondrial proteins during conditions of chronic muscle use and disuse. J Appl Physiol 107: 1730– 1735, 2009 [DOI] [PubMed] [Google Scholar]
- 26. Chance B. Pyridine nucleotide as an indicator of the oxygen requirements for energy-linked functions of mitochondria. Circ Res 38: I31– 38, 1976 [PubMed] [Google Scholar]
- 27. Chance B, Connelly CM. A method for the estimation of the increase in concentration of adenosine diphosphate in muscle sarcosomes following a contraction. Nature 179: 1235– 1237, 1957 [DOI] [PubMed] [Google Scholar]
- 28. Chance B, Jobsis F. Changes in fluorescence in a frog sartorius muscle following a twitch. Nature 184: 195– 196, 1959 [Google Scholar]
- 29. Chi MM, Hintz CS, Coyle EF, Martin WH, Ivy JL, 3rd, Nemeth PM, Holloszy JO, Lowry OH. Effects of detraining on enzymes of energy metabolism in individual human muscle fibers. Am J Physiol Cell Physiol 244: C276– C287, 1983 [DOI] [PubMed] [Google Scholar]
- 30. Cimen H, Han MJ, Yang Y, Tong Q, Koc H, Koc EC. Regulation of succinate dehydrogenase activity by SIRT3 in mammalian mitochondria. Biochemistry (Mosc) 49: 304– 311, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Civitarese AE, Carling S, Heilbronn LK, Hulver MH, Ukropcova B, Deutsch WA, Smith SR, Ravussin E. Calorie restriction increases muscle mitochondrial biogenesis in healthy humans. PLoS Med 4: e76, 2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Cogswell AM, Stevens RJ, Hood DA. Properties of skeletal muscle mitochondria isolated from subsarcolemmal and intermyofibrillar regions. Am J Physiol Cell Physiol 264: C383– C389, 1993 [DOI] [PubMed] [Google Scholar]
- 33. . Connelly C. M, and Chance B Fed Proc 13: 29, 1954 [Google Scholar]
- 34. Connett RJ, Gayeski TE, Honig CR. Lactate accumulation in fully aerobic, working, dog gracilis muscle. Am J Physiol Heart Circ Physiol 246: H120– H128, 1984 [DOI] [PubMed] [Google Scholar]
- 35. Cooper HM, Spelbrink JN. The human SIRT3 protein deacetylase is exclusively mitochondrial. Biochem J 411: 279– 285, 2008 [DOI] [PubMed] [Google Scholar]
- 36. Coste A, Louet JF, Lagouge M, Lerin C, Antal MC, Meziane H, Schoonjans K, Puigserver P, O'Malley BW, Auwerx J. The genetic ablation of SRC-3 protects against obesity and improves insulin sensitivity by reducing the acetylation of PGC-1(alpha). Proc Natl Acad Sci USA 105: 17187– 17192, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Costford SR, Bajpeyi S, Pasarica M, Albarado DC, Thomas SC, Xie H, Church TS, Jubrias SA, Conley KE, Smith SR. Skeletal muscle NAMPT is induced by exercise in humans. Am J Physiol Endocrinol Metab 298: E117– E126, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Coyle EF. Substrate utilization during exercise in active people. Am J Clin Nutr 61: 968S- 979S, 1995 [DOI] [PubMed] [Google Scholar]
- 39. Czubryt MP, McAnally J, Fishman GI, Olson EN. Regulation of peroxisome proliferator-activated receptor gamma coactivator 1 alpha (PGC-1 alpha) and mitochondrial function by MEF2 and HDAC5. Proc Natl Acad Sci USA 100: 1711– 1716, 2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Dali-Youcef N, Lagouge M, Froelich S, Koehl C, Schoonjans K, Auwerx J. Sirtuins: the “magnificent seven”, function, metabolism and longevity. Ann Med 39: 335– 345, 2007 [DOI] [PubMed] [Google Scholar]
- 41. Dash RK, Dibella JA, 2nd, Cabrera ME. A computational model of skeletal muscle metabolism linking cellular adaptations induced by altered loading states to metabolic responses during exercise. Biomed Eng Online 6: 14, 2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Dash RK, Li Y, Kim J, Beard DA, Saidel GM, Cabrera ME. Metabolic dynamics in skeletal muscle during acute reduction in blood flow and oxygen supply to mitochondria: in-silico studies using a multi-scale, top-down integrated model PLoS ONE 3: e3168, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Dolle C, Niere M, Lohndal E, Ziegler M. Visualization of subcellular NAD pools and intra-organellar protein localization by poly-ADP-ribose formation. Cell Mol Life Sci 67: 433– 443, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Duboc D, Muffat-Joly M, Renault G, Degeorges M, Toussaint M, Pocidalo JJ. In situ NADH laser fluorimetry of rat fast- and slow-twitch muscles during tetanus. J Appl Physiol 64: 2692– 2695, 1988 [DOI] [PubMed] [Google Scholar]
- 45. Dumke CL, Mark Davis J, Angela Murphy E, Nieman DC, Carmichael MD, Quindry JC, Travis Triplett N, Utter AC, Gross Gowin SJ, Henson DA, McAnulty SR, McAnulty LS. Successive bouts of cycling stimulates genes associated with mitochondrial biogenesis. Eur J Appl Physiol 107: 419– 427, 2009 [DOI] [PubMed] [Google Scholar]
- 46. Easlon E, Tsang F, Skinner C, Wang C, Lin SJ. The malate-aspartate NADH shuttle components are novel metabolic longevity regulators required for calorie restriction-mediated life span extension in yeast. Genes Dev 22: 931– 944, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Edington DW. Pyridine nucleotide oxidized to reduced ratio as a regulator of muscular performance. Experientia 26: 601– 602, 1970 [DOI] [PubMed] [Google Scholar]
- 48. Edington DW, McCafferty WB. Mitochondrial size distribution analysis in the soleus muscle of trained and aged rats. Experientia 29: 692– 693, 1973 [DOI] [PubMed] [Google Scholar]
- 49. Egan B, Carson BP, Garcia-Roves PM, Chibalin AV, Sarsfield FM, Barron N, McCaffrey N, Moyna NM, Zierath JR, O'Gorman DJ. Exercise intensity-dependent regulation of peroxisome proliferator-activated receptor coactivator-1 mRNA abundance is associated with differential activation of upstream signalling kinases in human skeletal muscle. J Physiol 588: 1779– 1790, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Eisenberg BR. Quantitative ultrastructure of mammalian skeletal muscle. In: Handbook of Physiology, Section 10: Skeletal Muscle, edited by Peachey LD, Adrian RH, Geiger SR. Baltimore: Lippincott Williams and Wilkins, 1983, p. 73– 112 [Google Scholar]
- 51. Emanuelli M, Carnevali F, Saccucci F, Pierella F, Amici A, Raffaelli N, Magni G. Molecular cloning, chromosomal localization, tissue mRNA levels, bacterial expression, and enzymatic properties of human NMN adenylyltransferase. J Biol Chem 276: 406– 412, 2001 [DOI] [PubMed] [Google Scholar]
- 52. Fernando FS, Conforti L, Tosi S, Smith AD, Coleman MP. Human homologue of a gene mutated in the slow Wallerian degeneration (C57BL/Wld(s)) mouse. Gene 284: 23– 29, 2002 [DOI] [PubMed] [Google Scholar]
- 53. Fjeld CC, Birdsong WT, Goodman RH. Differential binding of NAD+ and NADH allows the transcriptional corepressor carboxyl-terminal binding protein to serve as a metabolic sensor. Proc Natl Acad Sci USA 100: 9202– 9207, 2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Frieden C. Coenzyme binding, observed by fluorescence enhancement, apparently unrelated to the enzymic activity of glutamic dehydrogenase. Biochim Biophys Acta 47: 428– 430, 1961 [DOI] [PubMed] [Google Scholar]
- 55. Friedkin M, Lehninger AL. Esterification of inorganic phosphate coupled to electron transport between dihydrodiphosphopyridine nucleotide and oxygen. J Biol Chem 178: 611– 644, 1949 [PubMed] [Google Scholar]
- 56. Gerhart-Hines Z, Dominy JE, Jr, Blattler SM, Jedrychowski MP, Banks AS, Lim JH, Chim H, Gygi SP, Puigserver P. The cAMP/PKA pathway rapidly activates SIRT1 to promote fatty acid oxidation independently of changes in NAD(+). Mol Cell 44: 851– 863, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Gerhart-Hines Z, Rodgers JT, Bare O, Lerin C, Kim SH, Mostoslavsky R, Alt FW, Wu Z, Puigserver P. Metabolic control of muscle mitochondrial function and fatty acid oxidation through SIRT1/PGC-1alpha. EMBO J 26: 1913– 1923, 2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Ghosh HS, McBurney M, Robbins PD. SIRT1 negatively regulates the mammalian target of rapamycin. PLoS ONE 5: e9199, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Gladden LB. Is there an intracellular lactate shuttle in skeletal muscle? J Physiol 582: 899, 2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Gladden LB. Lactate metabolism: a new paradigm for the third millennium. J Physiol 558: 5– 30, 2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Godfraind-de Becker A. Heat production and fluorescence changes of toad sartorius muscle during aerobic recovery after a short tetanus. J Physiol 223: 719– 734, 1972 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Graham T, Sjogaard G, Lollgen H, Saltin B. NAD in muscle of man at rest and during exercise. Pflügers Arch 376: 35– 39, 1978 [DOI] [PubMed] [Google Scholar]
- 63. Graham TE, Saltin B. Estimation of the mitochondrial redox state in human skeletal muscle during exercise. J Appl Physiol 66: 561– 566, 1989 [DOI] [PubMed] [Google Scholar]
- 64. Graham TE, Sinclair DG, Chapler CK. Metabolic intermediates and lactate diffusion in active dog skeletal muscle. Am J Physiol 231: 766– 771, 1976 [DOI] [PubMed] [Google Scholar]
- 65. Green HJ, Dusterhoft S, Dux L, Pette D. Metabolite patterns related to exhaustion, recovery and transformation of chronically stimulated rabbit fast-twitch muscle. Pflügers Arch 420: 359– 366, 1992 [DOI] [PubMed] [Google Scholar]
- 66. Green HJ, Jones S, Ball-Burnett M, Farrance B, Ranney D. Adaptations in muscle metabolism to prolonged voluntary exercise and training. J Appl Physiol 78: 138– 145, 1995 [DOI] [PubMed] [Google Scholar]
- 67. Gregoire S, Tremblay AM, Xiao L, Yang Q, Ma K, Nie J, Mao Z, Wu Z, Giguere V, Yang XJ. Control of MEF2 transcriptional activity by coordinated phosphorylation and sumoylation. J Biol Chem 281: 4423– 4433, 2006 [DOI] [PubMed] [Google Scholar]
- 68. Guerra B, Guadalupe-Grau A, Fuentes T, Ponce-Gonzalez JG, Morales-Alamo D, Olmedillas H, Guillen-Salgado J, Santana A, Calbet JA. SIRT1, AMP-activated protein kinase phosphorylation and downstream kinases in response to a single bout of sprint exercise: influence of glucose ingestion. Eur J Appl Physiol 109: 731– 743, 2010 [DOI] [PubMed] [Google Scholar]
- 69. Guo X, Williams JG, Schug TT, Li X. DYRK1A and DYRK3 promote cell survival through phosphorylation and activation of SIRT1. J Biol Chem 285: 13223– 13232, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Gurd BJ. Deacetylation of PGC-1alpha by SIRT1: importance for skeletal muscle function and exercise-induced mitochondrial biogenesis. Appl Physiol Nutr Metab 36: 589– 597, 2011 [DOI] [PubMed] [Google Scholar]
- 71. Gurd BJ, Holloway GP, Yoshida Y, Bonen A. In mammalian muscle, SIRT3 is present in mitochondria and not in the nucleus; and SIRT3 is upregulated by chronic muscle contraction in an adenosine monophosphate-activated protein kinase-independent manner. Metabolism 61: 733– 741, 2011 [DOI] [PubMed] [Google Scholar]
- 72. Gurd BJ, Little JP, Perry CG. Does SIRT1 determine exercise-induced skeletal muscle mitochondrial biogenesis: differences between in vitro and in vivo experiments? J Appl Physiol 112: 926– 928, 2011 [DOI] [PubMed] [Google Scholar]
- 73. Gurd BJ, Perry CG, Heigenhauser GJ, Spriet LL, Bonen A. High-intensity interval training increases SIRT1 activity in human skeletal muscle. Appl Physiol Nutr Metab 35: 350– 357, 2010 [DOI] [PubMed] [Google Scholar]
- 74. Gurd BJ, Yoshida Y, Lally J, Holloway GP, Bonen A. The deacetylase enzyme SIRT1 is not associated with oxidative capacity in rat heart and skeletal muscle and its overexpression reduces mitochondrial biogenesis. J Physiol 587: 1817– 1828, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Gurd BJ, Yoshida Y, McFarlan JT, Holloway GP, Moyes CD, Heigenhauser GJ, Spriet L, Bonen A. Nuclear SIRT1 activity, but not protein content, regulates mitochondrial biogenesis in rat and human skeletal muscle. Am J Physiol Regul Integr Comp Physiol 301: R67– R75, 2011 [DOI] [PubMed] [Google Scholar]
- 76. Haigis MC, Sinclair DA. Mammalian sirtuins: biological insights and disease relevance. Annu Rev Pathol 5: 253– 295, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Hansford RG. The control of tricarboxylate-cycle oxidations in blowfly flight muscle. The oxidized and reduced nicotinamide-adenine dinucleotide content of flight muscle and isolated mitochondria, the adenosine triphosphate and adenosine diphosphate content of mitochondria, and the energy status of the mitochondria during controlled respiration. Biochem J 146: 537– 547, 1975 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Harden A, Young WJ. The alcoholic ferment of yeast-juice. Proc R Soc Lond B Biol Sci 78: 369– 375, 1906 [Google Scholar]
- 79. Hardie DG. Energy sensing by the AMP-activated protein kinase and its effects on muscle metabolism. Proc Nutr Soc 70: 92– 99, 2011 [DOI] [PubMed] [Google Scholar]
- 80. Henriksson J, Katz A, Sahlin K. Redox state changes in human skeletal muscle after isometric contraction. J Physiol 380: 441– 451, 1986 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Hirschey MD, Shimazu T, Goetzman E, Jing E, Schwer B, Lombard DB, Grueter CA, Harris C, Biddinger S, Ilkayeva OR, Stevens RD, Li Y, Saha AK, Ruderman NB, Bain JR, Newgard CB, Farese RV, Jr, Alt FW, Kahn CR, Verdin E. SIRT3 regulates mitochondrial fatty-acid oxidation by reversible enzyme deacetylation. Nature 464: 121– 125, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Hokari F, Kawasaki E, Sakai A, Koshinaka K, Sakuma K, Kawanaka K. Muscle contractile activity regulates Sirt3 protein expression in rat skeletal muscles. J Appl Physiol 109: 332– 340, 2010 [DOI] [PubMed] [Google Scholar]
- 83. Holloszy JO. Adaptation of skeletal muscle to endurance exercise. Med Sci Sports 7: 155– 164, 1975 [PubMed] [Google Scholar]
- 84. Houtkooper RH, Canto C, Wanders RJ, Auwerx J. The secret life of NAD+: an old metabolite controlling new metabolic signaling pathways. Endocr Rev 31: 194– 223, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Jensen TE, Wojtaszewski JF, Richter EA. AMP-activated protein kinase in contraction regulation of skeletal muscle metabolism: necessary and/or sufficient? Acta Physiol (Oxf) 196: 155– 174, 2009 [DOI] [PubMed] [Google Scholar]
- 86. Jing E, Emanuelli B, Hirschey MD, Boucher J, Lee KY, Lombard D, Verdin EM, Kahn CR. Sirtuin-3 (Sirt3) regulates skeletal muscle metabolism and insulin signaling via altered mitochondrial oxidation and reactive oxygen species production. Proc Natl Acad Sci USA 108: 14608– 14613, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Jobsis FF. Spectrophotometric studies on intact muscle. II. Recovery from contractile activity. J Gen Physiol 46: 929– 969, 1963 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Jobsis FF, Duffield JC. Oxidative and glycolytic recovery metabolism in muscle. J Gen Physiol 50: 1009– 1047, 1967 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Jobsis FF, Stainsby WN. Oxidation of NADH during contractions of circulated mammalian skeletal muscle. Respir Physiol 4: 292– 300, 1968 [DOI] [PubMed] [Google Scholar]
- 90. Kaeberlein M, McDonagh T, Heltweg B, Hixon J, Westman EA, Caldwell SD, Napper A, Curtis R, DiStefano PS, Fields S, Bedalov A, Kennedy BK. Substrate-specific activation of sirtuins by resveratrol. J Biol Chem 280: 17038– 17045, 2005 [DOI] [PubMed] [Google Scholar]
- 91. Kang H, Jung JW, Kim MK, Chung JH. CK2 is the regulator of SIRT1 substrate-binding affinity, deacetylase activity and cellular response to DNA-damage. PLoS ONE 4: e6611, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Kang J, Gocke CB, Yu H. Phosphorylation-facilitated sumoylation of MEF2C negatively regulates its transcriptional activity. BMC Biochem 7: 5, 2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Katz A, Sahlin K. Effect of decreased oxygen availability on NADH and lactate contents in human skeletal muscle during exercise. Acta Physiol Scand 131: 119– 127, 1987 [DOI] [PubMed] [Google Scholar]
- 94. Kim JH, Youn HD. C-terminal binding protein maintains mitochondrial activities. Cell Death Differ 16: 584– 592, 2009 [DOI] [PubMed] [Google Scholar]
- 95. Kim SC, Sprung R, Chen Y, Xu Y, Ball H, Pei J, Cheng T, Kho Y, Xiao H, Xiao L, Grishin NV, White M, Yang XJ, Zhao Y. Substrate and functional diversity of lysine acetylation revealed by a proteomics survey. Mol Cell 23: 607– 618, 2006 [DOI] [PubMed] [Google Scholar]
- 96. Kitani T, Okuno S, Fujisawa H. Growth phase-dependent changes in the subcellular localization of pre-B-cell colony-enhancing factor. FEBS Lett 544: 74– 78, 2003 [DOI] [PubMed] [Google Scholar]
- 97. Koch-Nolte F, Fischer S, Haag F, Ziegler M. Compartmentation of NAD+-dependent signalling. FEBS Lett 585: 1651– 1656, 2011 [DOI] [PubMed] [Google Scholar]
- 98. Kohen E, Kohen C, Thorell B. A comparative study of pyridine nucleotide metabolism in yeast and mammalian cells by microfluorimetry-microelectrophoresis. Histochemie 12: 95– 106, 1968 [DOI] [PubMed] [Google Scholar]
- 99. Kohen E, Kohen C, Thorell B. Kinetics of NAD reduction in the nucleus and the cytoplasm. Histochemie 16: 170– 185, 1968 [DOI] [PubMed] [Google Scholar]
- 100. Kohen E, Kohen C, Thorell B. Metabolic response of nuclear and cytoplasmic pyridine nucleotides. Histochemie 12: 107– 119, 1968 [DOI] [PubMed] [Google Scholar]
- 101. Kohen E, Siebert G, Kohen C. Metabolism of reduced pyridine nucleotides in ascites cell nuclei. Histochemie 3: 477– 483, 1964 [DOI] [PubMed] [Google Scholar]
- 102. Kohen E, Siebert G, Kohen C. Transfer of metabolites across the nuclear membrane. A microfluorometric study. Hoppe Seylers Z Physiol Chem 352: 927– 937, 1971 [DOI] [PubMed] [Google Scholar]
- 103. Kohen E, Thorell B, Kohen C, Salmon JM. Studies on metabolic events in localized compartments of the living cell by rapid microspectrofluorometry. Adv Biol Med Phys 15: 271– 297, 1974 [DOI] [PubMed] [Google Scholar]
- 104. Koltai E, Szabo Z, Atalay M, Boldogh I, Naito H, Goto S, Nyakas C, Radak Z. Exercise alters SIRT1, SIRT6, NAD and NAMPT levels in skeletal muscle of aged rats. Mech Ageing Dev 131: 21– 28, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105. Kong X, Wang R, Xue Y, Liu X, Zhang H, Chen Y, Fang F, Chang Y. Sirtuin 3, a new target of PGC-1alpha, plays an important role in the suppression of ROS and mitochondrial biogenesis. PLoS ONE 5: e11707, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106. Korzeniewski B. Computer-aided studies on the regulation of oxidative phosphorylation during work transitions. Prog Biophys Mol Biol 107: 274– 285, 2011 [DOI] [PubMed] [Google Scholar]
- 107. Krebs HA. The redox state of nicotinamide adenine dinucleotide in the cytoplasm and mitochondria of rat liver. Adv Enzyme Regul 5: 409– 434, 1967 [DOI] [PubMed] [Google Scholar]
- 108. Krieger DA, Tate CA, McMillin-Wood J, Booth FW. Populations of rat skeletal muscle mitochondria after exercise and immobilization. J Appl Physiol 48: 23– 28, 1980 [DOI] [PubMed] [Google Scholar]
- 109. Kushmerick MJ. Energetics of muscle contraction. In: Handbook of Physiology, Section 10: Skeletal Muscle, edited by Peachey LD, Adrian RH, Geiger SR. Baltimore: Lippincott Williams and Wilkins, 1983, p. 189– 236 [Google Scholar]
- 110. Kushmerick MJ. Energy balance in muscle activity: simulations of ATPase coupled to oxidative phosphorylation and to creatine kinase. Comp Biochem Physiol B Biochem Mol Biol 120: 109– 123, 1998 [DOI] [PubMed] [Google Scholar]
- 111. Lanza IR, Short DK, Short KR, Raghavakaimal S, Basu R, Joyner MJ, McConnell JP, Nair KS. Endurance exercise as a countermeasure for aging. Diabetes 57: 2933– 2942, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112. Lau C, Dolle C, Gossmann TI, Agledal L, Niere M, Ziegler M. Isoform-specific targeting and interaction domains in human nicotinamide mononucleotide adenylyltransferases. J Biol Chem 285: 18868– 18876, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113. Lau C, Niere M, Ziegler M. The NMN/NaMN adenylyltransferase (NMNAT) protein family. Front Biosci 14: 410– 431, 2009 [DOI] [PubMed] [Google Scholar]
- 114. Law IK, Liu L, Xu A, Lam KS, Vanhoutte PM, Che CM, Leung PT, Wang Y. Identification and characterization of proteins interacting with SIRT1 and SIRT3: implications in the anti-aging and metabolic effects of sirtuins. Proteomics 9: 2444– 2456, 2009 [DOI] [PubMed] [Google Scholar]
- 115. Lehninger AL. Phosphorylation coupled to oxidation of dihydrodiphosphopyridine nucleotide. J Biol Chem 190: 345– 359, 1951 [PubMed] [Google Scholar]
- 116. Lerin C, Rodgers JT, Kalume DE, Kim SH, Pandey A, Puigserver P. GCN5 acetyltransferase complex controls glucose metabolism through transcriptional repression of PGC-1alpha. Cell Metab 3: 429– 438, 2006 [DOI] [PubMed] [Google Scholar]
- 117. Li L, Muhlfeld C, Niemann B, Pan R, Li R, Hilfiker-Kleiner D, Chen Y, Rohrbach S. Mitochondrial biogenesis and PGC-1alpha deacetylation by chronic treadmill exercise: differential response in cardiac and skeletal muscle. Basic Res Cardiol 106: 1221– 1234, 2011 [DOI] [PubMed] [Google Scholar]
- 118. Li L, Pan R, Li R, Niemann B, Aurich AC, Chen Y, Rohrbach S. Mitochondrial biogenesis and peroxisome proliferator-activated receptor-gamma coactivator-1alpha (PGC-1alpha) deacetylation by physical activity: intact adipocytokine signaling is required. Diabetes 60: 157– 167, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119. Li Y, Dash RK, Kim J, Saidel GM, Cabrera ME. Role of NADH/NAD+ transport activity and glycogen store on skeletal muscle energy metabolism during exercise: in silico studies. Am J Physiol Cell Physiol 296: C25– C46, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120. Lin SJ, Ford E, Haigis M, Liszt G, Guarente L. Calorie restriction extends yeast life span by lowering the level of NADH. Genes Dev 18: 12– 16, 2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121. Little JP, Safdar A, Wilkin GP, Tarnopolsky MA, Gibala MJ. A practical model of low-volume high-intensity interval training induces mitochondrial biogenesis in human skeletal muscle: potential mechanisms. J Physiol 588: 1011– 1022, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122. Ljubicic V, Joseph AM, Adhihetty PJ, Huang JH, Saleem A, Uguccioni G, Hood DA. Molecular basis for an attenuated mitochondrial adaptive plasticity in aged skeletal muscle. Aging (Albany NY) 1: 818– 830, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123. Lofrumento NE, Marzulli D, Cafagno L, La Piana G, Cipriani T. Oxidation and reduction of exogenous cytochrome c by the activity of the respiratory chain. Arch Biochem Biophys 288: 293– 301, 1991 [DOI] [PubMed] [Google Scholar]
- 124. Lombard DB, Alt FW, Cheng HL, Bunkenborg J, Streeper RS, Mostoslavsky R, Kim J, Yancopoulos G, Valenzuela D, Murphy A, Yang Y, Chen Y, Hirschey MD, Bronson RT, Haigis M, Guarente LP, Farese RV, Jr, Weissman S, Verdin E, Schwer B. Mammalian Sir2 homolog SIRT3 regulates global mitochondrial lysine acetylation. Mol Cell Biol 27: 8807– 8814, 2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125. Lombard DB, Tishkoff DX, Bao J. Mitochondrial sirtuins in the regulation of mitochondrial activity and metabolic adaptation. Handb Exp Pharmacol 206: 163– 188, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126. Ma K, Chan JK, Zhu G, Wu Z. Myocyte enhancer factor 2 acetylation by p300 enhances its DNA binding activity, transcriptional activity, and myogenic differentiation. Mol Cell Biol 25: 3575– 3582, 2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127. Marfe G, Tafani M, Pucci B, Di Stefano C, Indelicato M, Andreoli A, Russo MA, Sinibaldi-Salimei P, Manzi V. The effect of marathon on mRNA expression of anti-apoptotic and pro-apoptotic proteins and sirtuins family in male recreational long-distance runners. BMC Physiol 10: 7, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 128. Mayevsky A, Chance B. Oxidation-reduction states of NADH in vivo: from animals to clinical use. Mitochondrion 7: 330– 339, 2007 [DOI] [PubMed] [Google Scholar]
- 129. Mayevsky A, Rogatsky GG. Mitochondrial function in vivo evaluated by NADH fluorescence: from animal models to human studies. Am J Physiol Cell Physiol 292: C615– C640, 2007 [DOI] [PubMed] [Google Scholar]
- 130. McGee SL, Sparling D, Olson AL, Hargreaves M. Exercise increases MEF2- and GEF DNA-binding activity in human skeletal muscle. FASEB J 20: 348– 349, 2006 [DOI] [PubMed] [Google Scholar]
- 131. Naya FJ, Black BL, Wu H, Bassel-Duby R, Richardson JA, Hill JA, Olson EN. Mitochondrial deficiency and cardiac sudden death in mice lacking the MEF2A transcription factor. Nat Med 8: 1303– 1309, 2002 [DOI] [PubMed] [Google Scholar]
- 132. Nemoto S, Fergusson MM, Finkel T. SIRT1 functionally interacts with the metabolic regulator and transcriptional coactivator PGC-1(alpha). J Biol Chem 280: 16456– 16460, 2005 [DOI] [PubMed] [Google Scholar]
- 133. Nikiforov A, Dolle C, Niere M, Ziegler M. Pathways and subcellular compartmentation of NAD biosynthesis in human cells: from entry of extracellular precursors to mitochondrial NAD generation. J Biol Chem 286: 21767– 21778, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134. Noriega LG, Feige JN, Canto C, Yamamoto H, Yu J, Herman MA, Mataki C, Kahn BB, Auwerx J. CREB and ChREBP oppositely regulate SIRT1 expression in response to energy availability. EMBO Rep 12: 1069– 1076, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135. Olgin J, Connett RJ, Chance B. Mitochondrial redox changes during rest-work transition in dog gracilis muscle. Adv Exp Med Biol 200: 545– 554, 1986 [DOI] [PubMed] [Google Scholar]
- 136. Onyango P, Celic I, McCaffery JM, Boeke JD, Feinberg AP. SIRT3, a human SIR2 homologue, is an NAD-dependent deacetylase localized to mitochondria. Proc Natl Acad Sci USA 99: 13653– 13658, 2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137. Palacios OM, Carmona JJ, Michan S, Chen KY, Manabe Y, Ward JL, 3rd, Goodyear LJ, Tong Q. Diet and exercise signals regulate SIRT3 and activate AMPK and PGC-1alpha in skeletal muscle. Aging (Albany NY) 1: 771– 783, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138. Palmieri F. The mitochondrial transporter family (SLC25): physiological and pathological implications. Pflügers Arch 447: 689– 709, 2004 [DOI] [PubMed] [Google Scholar]
- 139. Patterson GH, Knobel SM, Arkhammar P, Thastrup O, Piston DW. Separation of the glucose-stimulated cytoplasmic and mitochondrial NAD(P)H responses in pancreatic islet beta cells. Proc Natl Acad Sci USA 97: 5203– 5207, 2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140. Pfluger PT, Herranz D, Velasco-Miguel S, Serrano M, Tschop MH. Sirt1 protects against high-fat diet-induced metabolic damage. Proc Natl Acad Sci USA 105: 9793– 9798, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141. Phillips SM, Green HJ, Tarnopolsky MA, Heigenhauser GJ, Grant SM. Progressive effect of endurance training on metabolic adaptations in working skeletal muscle. Am J Physiol Endocrinol Metab 270: E265– E272, 1996 [DOI] [PubMed] [Google Scholar]
- 142. Philp A, Chen A, Lan D, Meyer GA, Murphy AN, Knapp AE, Olfert IM, McCurdy CE, Marcotte GR, Hogan MC, Baar K, Schenk S. Sirtuin 1 (SIRT1) deacetylase activity is not required for mitochondrial biogenesis or peroxisome proliferator-activated receptor-(gamma) coactivator-1(alpha) (PGC-1alpha) deacetylation following endurance exercise. J Biol Chem 286: 30561– 30570, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143. Pittelli M, Formentini L, Faraco G, Lapucci A, Rapizzi E, Cialdai F, Romano G, Moneti G, Moroni F, Chiarugi A. Inhibition of nicotinamide phosphoribosyltransferase: cellular bioenergetics reveals a mitochondrial insensitive NAD pool. J Biol Chem 285: 34106– 34114, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144. Pollak N, Dolle C, Ziegler M. The power to reduce: pyridine nucleotides—small molecules with a multitude of functions. Biochem J 402: 205– 218, 2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145. Putman CT, Jones NL, Hultman E, Hollidge-Horvat MG, Bonen A, McConachie DR, Heigenhauser GJ. Effects of short-term submaximal training in humans on muscle metabolism in exercise. Am J Physiol Endocrinol Metab 275: E132– E139, 1998 [DOI] [PubMed] [Google Scholar]
- 146. Ragland TE, Hackett DP. Compartmentation of nicotinamide dinucleotide dehydrogenases and transhydrogenases in nonphotosynthetic plant tissues. Arch Biochem Biophys 108: 479– 489, 1964 [DOI] [PubMed] [Google Scholar]
- 147. Rasmussen HN, van Hall G, Rasmussen UF. Lactate dehydrogenase is not a mitochondrial enzyme in human and mouse vastus lateralis muscle. J Physiol 541: 575– 580, 2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148. Revollo JR, Grimm AA, Imai S. The NAD biosynthesis pathway mediated by nicotinamide phosphoribosyltransferase regulates Sir2 activity in mammalian cells. J Biol Chem 279: 50754– 50763, 2004 [DOI] [PubMed] [Google Scholar]
- 149. Riquelme C, Barthel KK, Liu X. SUMO-1 modification of MEF2A regulates its transcriptional activity. J Cell Mol Med 10: 132– 144, 2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150. Rodgers JT, Lerin C, Haas W, Gygi SP, Spiegelman BM, Puigserver P. Nutrient control of glucose homeostasis through a complex of PGC-1alpha and SIRT1. Nature 434: 113– 118, 2005 [DOI] [PubMed] [Google Scholar]
- 151. Rongvaux A, Shea RJ, Mulks MH, Gigot D, Urbain J, Leo O, Andris F. Pre-B-cell colony-enhancing factor, whose expression is up-regulated in activated lymphocytes, is a nicotinamide phosphoribosyltransferase, a cytosolic enzyme involved in NAD biosynthesis. Eur J Immunol 32: 3225– 3234, 2002 [DOI] [PubMed] [Google Scholar]
- 152. Rowan AN, Newsholme EA. Changes in the contents of adenine nucleotides and intermediates of glycolysis and the citric acid cycle in flight muscle of the locust upon flight and their relationship to the control of the cycle. Biochem J 178: 209– 216, 1979 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153. Sacktor B, Wormser-Shavit E. Regulation of metabolism in working muscle in vivo I Concentrations of some glycolytic, tricarboxylic acid cycle, and amino acid intermediates in insect flight muscle during flight. J Biol Chem 241: 624– 631, 1966 [PubMed] [Google Scholar]
- 154. Sahlin K. NADH and NADPH in human skeletal muscle at rest and during ischaemia. Clin Physiol 3: 477– 485, 1983 [DOI] [PubMed] [Google Scholar]
- 155. Sahlin K. NADH in human skeletal muscle during short-term intense exercise. Pflügers Arch 403: 193– 196, 1985 [DOI] [PubMed] [Google Scholar]
- 156. Sahlin K, Fernstrom M, Svensson M, Tonkonogi M. No evidence of an intracellular lactate shuttle in rat skeletal muscle. J Physiol 541: 569– 574, 2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157. Sahlin K, Gorski J, Edstrom L. Influence of ATP turnover and metabolite changes on IMP formation and glycolysis in rat skeletal muscle. Am J Physiol Cell Physiol 259: C409– C412, 1990 [DOI] [PubMed] [Google Scholar]
- 158. Sahlin K, Katz A. The content of NADH in rat skeletal muscle at rest and after cyanide poisoning. Biochem J 239: 245– 248, 1986 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159. Sahlin K, Katz A, Broberg S. Tricarboxylic acid cycle intermediates in human muscle during prolonged exercise. Am J Physiol Cell Physiol 259: C834– C841, 1990 [DOI] [PubMed] [Google Scholar]
- 160. Sahlin K, Katz A, Henriksson J. Redox state and lactate accumulation in human skeletal muscle during dynamic exercise. Biochem J 245: 551– 556, 1987 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161. Sasaki T, Maier B, Koclega KD, Chruszcz M, Gluba W, Stukenberg PT, Minor W, Scrable H. Phosphorylation regulates SIRT1 function. PLoS ONE 3: e4020, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162. Sauve AA, Schramm VL. Sir2 regulation by nicotinamide results from switching between base exchange and deacetylation chemistry. Biochemistry (Mosc) 42: 9249– 9256, 2003 [DOI] [PubMed] [Google Scholar]
- 163. Schantz PG, Henriksson J. Enzyme levels of the NADH shuttle systems: measurements in isolated muscle fibres from humans of differing physical activity. Acta Physiol Scand 129: 505– 515, 1987 [DOI] [PubMed] [Google Scholar]
- 164. Schantz PG, Kallman M. NADH shuttle enzymes and cytochrome b5 reductase in human skeletal muscle: effect of strength training. J Appl Physiol 67: 123– 127, 1989 [DOI] [PubMed] [Google Scholar]
- 165. Schantz PG, Sjoberg B, Svedenhag J. Malate-aspartate and alpha-glycerophosphate shuttle enzyme levels in human skeletal muscle: methodological considerations and effect of endurance training. Acta Physiol Scand 128: 397– 407, 1986 [DOI] [PubMed] [Google Scholar]
- 166. Scher MB, Vaquero A, Reinberg D. SirT3 is a nuclear NAD+-dependent histone deacetylase that translocates to the mitochondria upon cellular stress. Genes Dev 21: 920– 928, 2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167. Schiotz Thorud HM, Lunde PK, Nicolaysen G, Nicolaysen A, Helge JW, Nilsson GE, Sejersted OM. Muscle dysfunction during exercise of a single skeletal muscle in rats with congestive heart failure is not associated with reduced muscle blood supply Acta Physiol Scand 181: 173– 181, 2004 [DOI] [PubMed] [Google Scholar]
- 168. Schlicker C, Gertz M, Papatheodorou P, Kachholz B, Becker CF, Steegborn C. Substrates and regulation mechanisms for the human mitochondrial sirtuins Sirt3 and Sirt5. J Mol Biol 382: 790– 801, 2008 [DOI] [PubMed] [Google Scholar]
- 169. Schmidt MT, Smith BC, Jackson MD, Denu JM. Coenzyme specificity of Sir2 protein deacetylases: implications for physiological regulation. J Biol Chem 279: 40122– 40129, 2004 [DOI] [PubMed] [Google Scholar]
- 170. Schwer B, North BJ, Frye RA, Ott M, Verdin E. The human silent information regulator (Sir)2 homologue hSIRT3 is a mitochondrial nicotinamide adenine dinucleotide-dependent deacetylase. J Cell Biol 158: 647– 657, 2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171. Schwert GW, Takenaka Y. Lactic dehydrogenase. III. Mechanism of the reaction. J Biol Chem 223: 157– 170, 1956 [PubMed] [Google Scholar]
- 172. Shi T, Wang F, Stieren E, Tong Q. SIRT3, a mitochondrial sirtuin deacetylase, regulates mitochondrial function and thermogenesis in brown adipocytes. J Biol Chem 280: 13560– 13567, 2005 [DOI] [PubMed] [Google Scholar]
- 173. Suwa M, Nakano H, Radak Z, Kumagai S. Endurance exercise increases the SIRT1 and peroxisome proliferator-activated receptor gamma coactivator-1alpha protein expressions in rat skeletal muscle. Metabolism 57: 986– 998, 2008 [DOI] [PubMed] [Google Scholar]
- 174. Toiber D, Sebastian C, Mostoslavsky R. Characterization of nuclear sirtuins: molecular mechanisms and physiological relevance. Handb Exp Pharmacol 189– 224, 2011 [DOI] [PubMed] [Google Scholar]
- 175. Vishwasrao HD, Heikal AA, Kasischke KA, Webb WW. Conformational dependence of intracellular NADH on metabolic state revealed by associated fluorescence anisotropy. J Biol Chem 280: 25119– 25126, 2005 [DOI] [PubMed] [Google Scholar]
- 176. Wakita M, Nishimura G, Tamura M. Some characteristics of the fluorescence lifetime of reduced pyridine nucleotides in isolated mitochondria, isolated hepatocytes, and perfused rat liver in situ. J Biochem 118: 1151– 1160, 1995 [DOI] [PubMed] [Google Scholar]
- 177. Wang Q, Zhang Y, Yang C, Xiong H, Lin Y, Yao J, Li H, Xie L, Zhao W, Yao Y, Ning ZB, Zeng R, Xiong Y, Guan KL, Zhao S, Zhao GP. Acetylation of metabolic enzymes coordinates carbon source utilization and metabolic flux. Science 327: 1004– 1007, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178. Wendt IR, Chapman JB. Fluorometric studies of recovery metabolism of rat fast- and slow-twitch muscles. Am J Physiol 230: 1644– 1649, 1976 [DOI] [PubMed] [Google Scholar]
- 179. Williamson DH, Lund P, Krebs HA. The redox state of free nicotinamide-adenine dinucleotide in the cytoplasm and mitochondria of rat liver. Biochem J 103: 514– 527, 1967 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180. Winer AD, Schwert GW, Millar Lactic dehydrogenase DB. VI. Fluorimetric measurements of the complex of enzyme and reduced diphosphopyridine nucleotide. J Biol Chem 234: 1149– 1154, 1959 [PubMed] [Google Scholar]
- 181. Wolfe BR, Graham TE, Barclay JK. Hyperoxia, mitochondrial redox state, and lactate metabolism of in situ canine muscle. Am J Physiol Cell Physiol 253: C263– C268, 1987 [DOI] [PubMed] [Google Scholar]
- 182. Yang H, Yang T, Baur JA, Perez E, Matsui T, Carmona JJ, Lamming DW, Souza-Pinto NC, Bohr VA, Rosenzweig A, de Cabo R, Sauve AA, Sinclair DA. Nutrient-sensitive mitochondrial NAD+ levels dictate cell survival. Cell 130: 1095– 1107, 2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183. Yang Y, Cimen H, Han MJ, Shi T, Deng JH, Koc H, Palacios OM, Montier L, Bai Y, Tong Q, Koc EC. NAD+-dependent deacetylase SIRT3 regulates mitochondrial protein synthesis by deacetylation of the ribosomal protein MRPL10. J Biol Chem 285: 7417– 7429, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184. Yoshida Y, Holloway GP, Ljubicic V, Hatta H, Spriet LL, Hood DA, Bonen A. Negligible direct lactate oxidation in subsarcolemmal and intermyofibrillar mitochondria obtained from red and white rat skeletal muscle. J Physiol 582: 1317– 1335, 2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185. Yu J, Auwerx J. The role of sirtuins in the control of metabolic homeostasis. Ann NY Acad Sci 1173, Suppl 1: E10– E19, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186. Zhang CL, McKinsey TA, Lu JR, Olson EN. Association of COOH-terminal-binding protein (CtBP) and MEF2-interacting transcription repressor (MITR) contributes to transcriptional repression of the MEF2 transcription factor. J Biol Chem 276: 35– 39, 2001 [DOI] [PubMed] [Google Scholar]
- 187. Zhang HH, Halbleib M, Ahmad F, Manganiello VC, Greenberg AS. Tumor necrosis factor-alpha stimulates lipolysis in differentiated human adipocytes through activation of extracellular signal-related kinase and elevation of intracellular cAMP. Diabetes 51: 2929– 2935, 2002 [DOI] [PubMed] [Google Scholar]
- 188. Zhang Q, Piston DW, Goodman RH. Regulation of corepressor function by nuclear NADH. Science 295: 1895– 1897, 2002 [DOI] [PubMed] [Google Scholar]
- 189. Zhang Q, Wang SY, Fleuriel C, Leprince D, Rocheleau JV, Piston DW, Goodman RH. Metabolic regulation of SIRT1 transcription via a HIC1:CtBP corepressor complex. Proc Natl Acad Sci USA 104: 829– 833, 2007 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 190. Zhang T, Kraus WL. SIRT1-dependent regulation of chromatin and transcription: linking NAD(+) metabolism and signaling to the control of cellular functions. Biochim Biophys Acta 1804: 1666– 1675, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191. Zhao S, Xu W, Jiang W, Yu W, Lin Y, Zhang T, Yao J, Zhou L, Zeng Y, Li H, Li Y, Shi J, An W, Hancock SM, He F, Qin L, Chin J, Yang P, Chen X, Lei Q, Xiong Y, Guan KL. Regulation of cellular metabolism by protein lysine acetylation. Science 327: 1000– 1004, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192. Zhao X, Sternsdorf T, Bolger TA, Evans RM, Yao TP. Regulation of MEF2 by histone deacetylase 4- and SIRT1 deacetylase-mediated lysine modifications. Mol Cell Biol 25: 8456– 8464, 2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193. Zhong L, Mostoslavsky R. Fine tuning our cellular factories: sirtuins in mitochondrial biology. Cell Metab 13: 621– 626, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]