Video abstract
Video
Keywords: peptide tag, CXCR4, intracellular targeting, self-assembling, nanoparticles, colorectal cancer
Abstract
Background
Cell-targeting peptides or proteins are appealing tools in nanomedicine and innovative medicines because they increase the local drug concentration and reduce potential side effects. CXC chemokine receptor 4 (CXCR4) is a cell surface marker associated with several severe human pathologies, including colorectal cancer, for which intracellular targeting agents are currently missing.
Results
Four different peptides that bind CXCR4 were tested for their ability to internalize a green fluorescent protein-based reporter nanoparticle into CXCR4+ cells. Among them, only the 18 mer peptide T22, an engineered segment derivative of polyphemusin II from the horseshoe crab, efficiently penetrated target cells via a rapid, receptor-specific endosomal route. This resulted in accumulation of the reporter nanoparticle in a fully fluorescent and stable form in the perinuclear region of the target cells, without toxicity either in cell culture or in an in vivo model of metastatic colorectal cancer.
Conclusion
Given the urgent demand for targeting agents in the research, diagnosis, and treatment of CXCR4-linked diseases, including colorectal cancer and human immunodeficiency virus infection, T22 appears to be a promising tag for the intracellular delivery of protein drugs, nanoparticles, and imaging agents.
Introduction
Unlike conventional therapies, nanomedicine and innovative medicines in general pursue targeted intracellular delivery of chemotherapy and imaging agents, and are expected to result in significantly lower effective therapeutic doses, production costs, and toxicity.1 Cell-penetrating peptides offer a broad potential for efficient internalization of attached cargo because of their affinity for and associated ability to cross cell membranes.2,3 However, because these activities are not dependent on specific cell surface receptors, appropriate cell targeting and biodistribution of therapeutic complexes cannot be achieved using cell-penetrating peptides. The discovery of disease-linked cell surface markers enables subsequent identification of specific ligands for receptor-mediated endocytosis. These entities should be capable of driving the uptake of large macromolecular complexes such as nanoparticles, which are useful in a therapeutic context as drug carriers and stabilizers. The CXC chemokine receptor 4 (CXCR4) plays a role in inflammation, autoimmunity, ischemia, and stem cell mobilization.4 In addition, CXCR4 is a coreceptor for the human immunodeficiency virus (HIV)5 and an important stem cell marker in several common human cancers,6,7 including metastatic colorectal cancer.8,9 Therefore, developing new therapeutic agents targeted to CXCR4 is a recognized priority in emerging medicines,4 but because CXCR4 ligands remain poorly studied and peptides suitable for CXCR4-mediated endocytosis are not available, intracellular targeting to CXCR4+ cells is still not feasible. In the present study, we describe peptide T22, a known antagonist of CXCR4, that binds to and penetrates CXCR4+ cells efficiently via CXCR4-specific endocytosis. T22 is an engineered version polyphemusin II peptide from the horseshoe crab, in which three substitutions at residues Tyr5, Lys7, and Tyr12 dramatically increase the natural affinity of this peptide for CXCR4.10,11 When fused to a self-assembling green fluorescent protein (GFP)-based building block, T22 promotes fast perinuclear accumulation of stable and highly fluorescent nanoparticles without cytotoxicity, both in cell culture and in vivo. Thus, we propose T22 as a novel cell-targeting peptide suitable for functionalization of nanoparticles and appropriate for intracellular delivery in CXCR4-associated medicines.
Materials and methods
Protein design, production, purification, and characterization
Four chimeric genes were designed inhouse and provided by Geneart (Regensburg, Germany). Using NdeI/HindIII restriction sites, these genes were introduced into pET22b (Novagen 69744-3). All the encoded proteins were produced in Escherichia coli Origami B (BL21, OmpT−, Lon−, TrxB−, Gor−, Novagen) overnight at 20°C upon addition of 0.1 mM isopropyl-β-D-thiogalactopyronaside. Bacterial cells were then centrifuged for 45 minutes (5000 g at 4°C) and resuspended in Tris buffer (Tris 20 mM, pH 8.0, NaCl 500 mM, imidazole 10 mM) in the presence of ethylenediamine tetra-acetic acid-free protease inhibitor (Complete EDTAFree, Roche, Basel, Switzerland). The cells were disrupted at 1100 psi in a French press (Thermo FA-078A) and their proteins were purified by 6 × His tag affinity chromatography using HiTrap Chelating HP 1 mL (GE Healthcare, Piscataway, NJ) columns with an AKTA purifier FPLC (GE Healthcare). Elution was achieved by a linear gradient of Tris 20 mM, pH 8.0, 500 mM NaCl, and 500 mM imidazole, and the proteins eluted were finally dialyzed against phosphate-buffered solution (140 mM NaCl, 7.5 mM Na2HPO4, 2.5 mM NaH2PO4) plus 10% glycerol, pH 7.4, against carbonate buffer (166 mM NaCO3H + 333 mM NaCl, pH 7.4) or against Tris 20 mM + NaCl 500 mM, pH 7.5. The integrity of the resulting proteins was checked by both mass spectrometry and N-terminal sequencing using the Edman degradation method, and their amounts were determined by Bradford’s assay.12 In addition, all products were analyzed by Coomassie-stained sodium dodecyl sulfate polyacrylamide gel electrophoresis and anti-His Western blot analysis. The fusion proteins were named according to N→C modular organization by the name of the CXCR4 ligand, followed by GFP and H6 (hexahistidine tail).
TEM, fluorescence determination, and dynamic light scattering
Purified proteins were diluted to 0.2 mg/mL and contrasted by evaporation of 1 nm platinum layer in carbon-coated grids. Samples were visualized in a Hitachi H-7000 transmission electron microscope. Fluorescence of the nanoparticles was determined in a Cary Eclipse fluorescence spectrophotometer (Varian Inc, Palo Alto, CA) at 510 nm using an excitation wavelength of 450 nm. The volume and size distribution of the nanoparticles was determined by dynamic light scattering at 633 nm (Zetasizer Nano ZS, Malvern Instruments Limited, Malvern, Worcestershire, UK).
Protein stability analysis
Stability of the T22-GFP-H6 (amino terminus of a His-tagged enhanced GFP) was analyzed in triplicate in human serum (S2257-5ML, Sigma, St Louis, MO) at 37°C, with agitation and at a final concentration of 0.23 μg/μL. Fluorescence was determined as described earlier, and the integrity of the T22-GFP-H6 was confirmed by sodium dodecyl sulfate polyacrylamide gel electrophoresis and further Western blotting. Nitrocellulose membranes were developed using an anti-GFP rabbit polyclonal serum.
Cell culture and confocal laser scanning microscopy
The cells were cultured in modified Eagle’s medium (Gibco, Rockville, MD) supplemented with 10% fetal calf serum (Gibco), and incubated at 37°C and 5% CO2 in a humidified atmosphere. Nanoparticles were added to the cell culture in the presence of Optipro medium (Gibco) 20 hours before confocal analysis, except for the time-course and internalization studies in the presence of serum (complete medium). For confocal analysis, the cells were grown on MatTek culture dishes (MatTek Corporation, Ashland, MA). The nuclei were labeled with 0.2 μg/mL Hoechst 33342 (Molecular Probes, Eugene, OR) and the plasma membranes with 2.5 μg/mL CellMaskTM Deep Red (Molecular Probes) for 10 minutes in the dark. The cells were washed in phosphate-buffered saline (Sigma-Aldrich Chemie GmbH, Steinheim, Germany). Live cells were recorded by TCS-SP5 confocal laser scanning microscopy (Leica Microsystems, Heidelberg, Germany) using a Plan Apo 63 ×/1.4 (oil HC × PL APO lambda blue) objective as described elsewhere.13 To determine particle localization inside the cell, stacks of 10–20 sections for every 0.5 μm of cell thickness were collected and three-dimensional models were generated using Imaris version 6.1.0 software (Bitplane, Zürich, Switzerland) as reported previously.14 Cell samples were analyzed after treatment with 1 mg/mL trypsin (Gibco) for 15 minutes on a FACS-Canto system (Becton Dickinson, Franklin Lakes, NJ) using a 15 mW air-cooled argon ion laser at 488 nm excitation. Fluorescence emission was measured with a D detector (530/30 nm band pass filter). Cell viability was determined by 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) assay as described elsewhere.15 An HeLa cell line was obtained from the American Type Culture Collection (reference CCL-2, Manassas, VA) and SW1417 was a generous gift from Xavier Mayol (Institut Municipal D’Investigacio Médica, Barcelona, Spain).16
Molecular modeling
Protein homology models were generated using Modeller, Phyre, and Swiss-PdbViewer as reported earlier.17 Also, different Haddock models were obtained, in which the binding residues were established using crystallographic data from multimeric forms of GFP (1GFL, 1JC0, 3GJ2, 2QLE, 1EMC) and the higher interaction energy solutions resulting from protein-protein docking calculation.
Biodistribution analysis
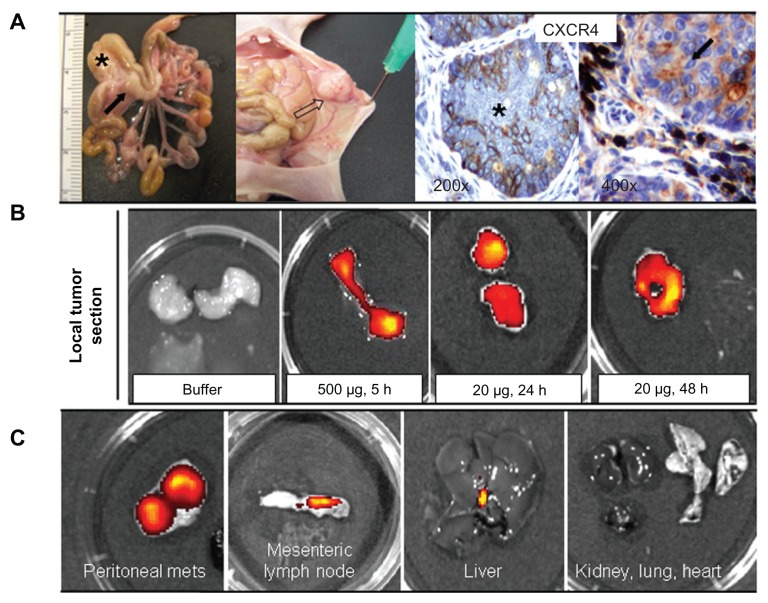
Five-week-old female Swiss nu/nu mice, weighing 18–20 g (Charles River, France) maintained in specific pathogen-free conditions, were used for the in vivo experiments. All procedures were approved by the Hospital de Sant Pau animal ethics committee. To generate a metastatic colorectal cancer model, the mice were injected with 2 million SW-1417 cells via the cecal wall, using an orthotopic cell microinjection technique.18 Two months after microinjection, when local tumor and metastases had appeared, each experimental animal received a single intravenous bolus of T22-GFP-H6 nanoparticles resuspended in a 20 mM Tris, 500 mM NaCl, pH 7.4 buffer, at a dose of 20 μg (n = 3 mice) or 500 μg (n = 3 mice). Control animals received a single bolus of empty buffer. After euthanizing the mice at 5, 24, or 48 hours post-administration, we measured ex vivo the amount of nanoparticles in normal and tumor-bearing organs from the experimental and control mice, quantifying the fluorescence emitted by each organ. To this end, primary tumors, organs bearing metastatic foci, and several samples of normal tissue (kidney, liver, lung, heart) were obtained at necropsy, cut into slices, and placed in separate wells to detect the emitted signal using IVIS® Spectrum equipment (Xenogen Biosciences, Waltham, MA). The amount of nanoparticles distributed in each tissue was calculated as the increased fluorescent (FLI) ratio. The fluorescence signal was first digitalized, displayed as a pseudocolor overlay, and expressed as radiant efficiency. Thereafter, the FLI ratio was calculated for each organ, dose, and post-treatment time, dividing the FLI signal from the nanoparticle-treated mice by the FLI signal from the control mice. Finally, all organs were collected and fixed with 4% formaldehyde in phosphate-buffered solution for 24 hours, and then embedded in paraffin for histological and immunohistochemical evaluation.
Immunohistochemistry
Sections of normal and tumor tissues 4 μm thick were stained with hematoxylin and eosin. The sections were examined histopathologically to analyze the primary tumor and to search for metastatic foci in organs with no macroscopic metastases. Paraffin-embedded tissue sections were deparaffinized, rehydrated, and washed in phosphate-buffered solution with Tween-20. Antigen retrieval was performed using citrate buffer at 120°C. After quenching peroxidase activity by incubating the slides in 3% H2O2 for 10 minutes, the slides were washed in phosphate-buffered solution with Tween-20. The slides were incubated for 30 minutes with the primary antibody against CXCR4 (1:20, Biotrend, Destin, FL) to detect expression of this receptor in normal tissue and tumor tissue. A primary anti-His antibody (1:1000; Abcam, Cambridge, UK) was used to detect nanoparticle accumulation and localization in normal and tumor ( primary or metastatic) tissue. After incubation, the samples were washed in phosphate-buffered solution with Tween-20 and incubated with the biotinylated secondary antibody for 30 minutes at room temperature. Finally, the sections were counterstained with hematoxylin and mounted using DPX mounting medium. Representative pictures were taken using Cell^B software (Olympus Soft Imaging) at 200× and 1000× magnifications.
Statistical analysis
The data were evaluated by one-way Anova analysis of variance with a confidence level of 99.9% (P < 0.001). Dose-response plots were analyzed by nonlinear regression analysis using SigmaPlot 10. The data for both the HeLa and SW1417 cells fitted into a double rectangular hyperbolic function, with a significance level of 99.9% (P < 0.001). All data were expressed as the mean ± standard error of the mean.
Results
Screening CXCR4 peptidic ligands for cell-targeted internalization
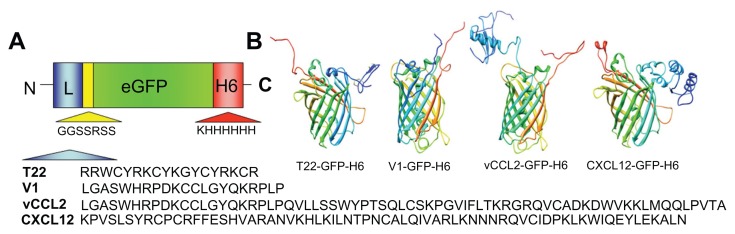
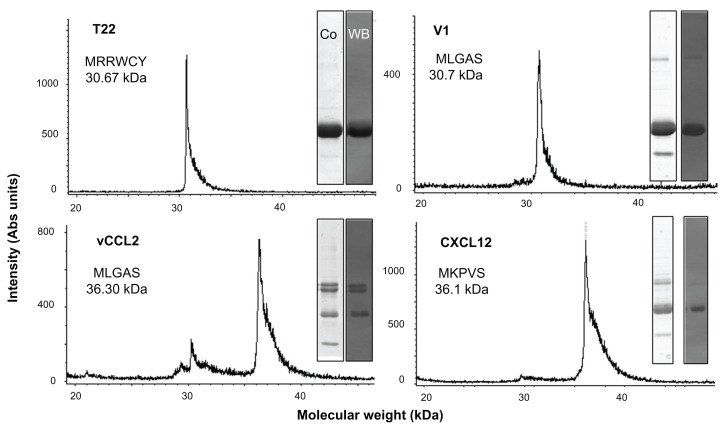
To identify peptides suitable for use as tags for CXCR4-mediated cell internalization of large macromolecular complexes, four known molecular ligands of CXCR4, namely peptide T22, the protein domains CXCL2 and vCCL2, and V1, an amino-terminal peptide of vCCL2 (Figure 1A), were tested for their ability to promote receptor-mediated delivery of attached macromolecular entities into CXCR4-expressing cells. All these protein segments were fused to GFP-H6. This fluorescent protein, when containing cationic peptides at its amino terminus, shows a tendency to self-assemble as regular-sized nanoparticles, presumably by electrostatic interaction between monomers.13 Because of their emission of fluorescence, these nanoparticles are very convenient reporters for use in internalization and trafficking studies.14 Four equivalent modular constructs differing only by the CXCR4 ligand (Figure 1B) were designed according to this strategy, produced in bacteria, and purified as full-length forms of the expected molecular mass and N-terminal amino acid sequence (Figure 2). Their reactivity in anti-His Western blot analysis indicated protein integrity also at the C-terminus. Only vCCL12 showed partial degradation at the N-terminus, but retained most of the expected molecular mass (Figure 2). This proteolytic instability was observed under all tested production conditions and in several E. coli strains tested as hosts (data not shown). In addition, the single major peak in mass spectroscopy and the unambiguous N-terminal sequence, coincident with a few dominant bands in Western blot analysis, indicated the existence of conformational isoforms of vCCL12.
Figure 1.
Features of protein constructs containing peptidic CXCR4 ligands. (A) Schematic representation of CXCR4-binding constructs indicating their modular composition. A linker (yellow box) commonly used in phage display was inserted between the protein ligand (L, blue) and eGFP (green). The amino acid sequences of the four ligands are shown. In all cases, an additional amino terminal methionine, derived from the cloning strategy adapted to Escherichia coli was expected. (B) Predicted structure of the different GFP-derived constructs. The color code of panel A is maintained here for both ligand- (blue) and H6- (red) overhanging ends.
Abbreviation: GFP, green fluorescent protein.
Figure 2.
Biochemical characterization of protein constructs upon protein purification.
Notes: Mass spectrometry of the purified constructs indicating the experimental molecular weight. The obtained N-terminal sequence is also shown, always coincident with the predicted sequence (Figure 1A). Protein integrity is also shown through Coomassie blue-stained sodium dodecyl sulfate polyacrylamide gel electrophoresis gels (Co) and by H6 immunodetection in Western blot (WB).
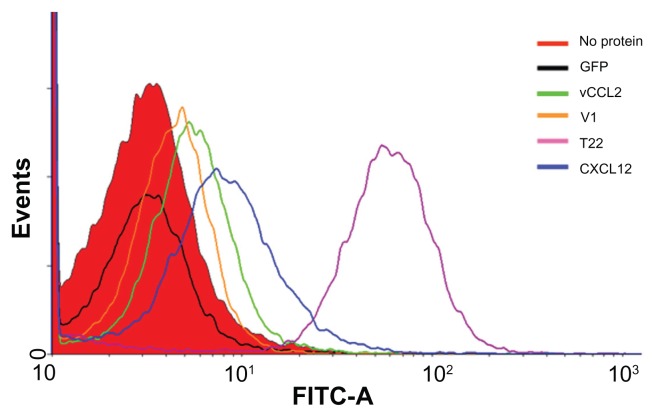
When these proteins were added to the culture medium, HeLa (CXCR4+) cells exposed to T22-GFP-H6 were tenfold more fluorescent than those exposed to V1, CXCL12, and vCCL2 fusion (Figure 3). Interestingly, cell uptake of CXCL12 and V1 has been reported previously,19,20 but the excellent performance of T22 was completely unexpected because its ability to internalize cells has not been previously suggested or described, despite its well known properties as an CXCR4 antagonist.21 As expected, the untagged parental GFP-H6 building block did not label the cells.
Figure 3.
Differential internalization of CXCR4 ligands.
Notes: Internalization of T22-GFP-H6 and alternative constructs in HeLa cells, monitored by flow cytometry 24 hours after exposure.
Abbreviation: GFP, green fluorescent protein.
CXCR4-dependent cell uptake of T22-empowered constructs
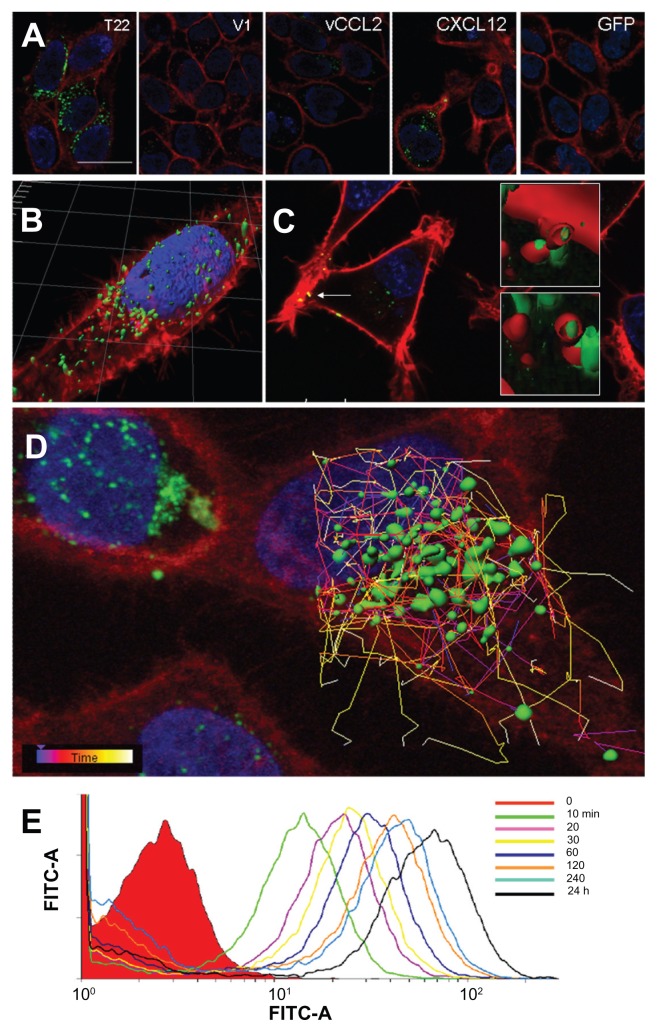
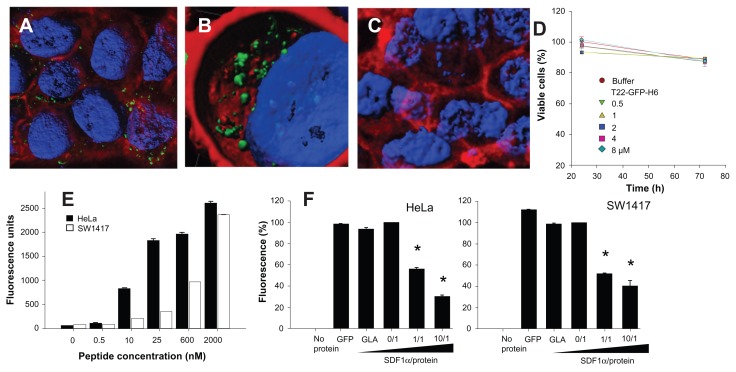
The high cell penetrability of T22-activated GFP was further confirmed by confocal microscopy of treated HeLa cell cultures (Figure 4A). In full agreement with data from Figure 3, no penetration of GFP-H6 was observed, while some uptake of CXCL12 and to a minor extent some vCCL2-GFP-H6 was seen. V1-GFP-H6 was observed inside cells as low in abundance and poorly fluorescent punctuate entities. Three- dimensional reconstructions of individual cells exposed to T22-GFP-H6 indicated perinuclear localization of fluorescence in the form of nanoparticles (Figure 4B). Discrete yellow merging signals (green particles incorporated into red membranous vesicles) were observed close to the plasma membrane (Figure 4C), indicative of uptake by endosomes. The finding of nanoparticles within the endosomes was fully confirmed by three-dimensional confocal reconstructions as yellow spots, some of them fully internalized in the cytoplasm and separate from the plasma membrane (Figure 4C, inset). However, the relative low proportion of yellow signals and rapid accumulation of nanoparticles close to the nuclear region through fast cytoplasmic trafficking (Figure 4D) were indicative of early endosomal escape. This was probably promoted by the accompanying hexahistidine tag, that has powerful endosomolytic properties,22 inducing early endosomal escape in related protein-only nanoparticles.14 Ten minutes after exposure to these T22 constructs, the uptake of nanoparticles was already evident, and the amount of intracellular fluorescence progressively increased for up to 24 hours at least (Figure 4E).
Figure 4.
Differential internalization and intracellular trafficking of CXCR4 ligands. (A) Confocal images of HeLa cells exposed to differently tagged proteins for 24 hours. Nuclei are labeled in blue and cell membranes in red. Bar indicates 20 μm. (B) Detail of a HeLa cell exposed to T22-GFP-H6, showing the intracellular localization of nanostructured, fluorescent entities, in an isosurface representation within a three-dimensional volumetric x-y-z data field. (C) Yellow spots in the cell membrane, marked with an arrow, indicate early endosomal localization of green fluorescent particles (merging of red and green signals). In the insets, details of endosome-embedded fluorescent particles dissected by three-dimensional reconstruction. (D) Intracellular tracking of individual fluorescent particles monitored by confocal microscopy. (E) Time course monitoring of T22-GFP-H6 internalization in HeLa cells by flow cytometry.
Abbreviation: GFP, green fluorescent protein.
To exclude the possibility that T22-mediated penetration of GFP was limited to a particular cell type, we also evaluated the intracellular fluorescence in exposed CXCR4+ SW1417 cells, a human cell line used to generate a mice model of metastatic colorectal cancer for preclinical studies through orthotopic implantation.18 Again, strong penetration (Figure 5A) and perinuclear fluorescence labeling ( Figure 5B) were seen in this model, indicative of efficient penetration of the nanoparticles. No changes in cell morphology (which would have been indicative of toxicity) were observed ( Figures 4A and 5A), or when compared with cells exposed to parental GFP-H6 (Figures 4A and 5C). Furthermore, viability of T22-GFP-H6-exposed SW1417 cells remained unaffected up to 72 hours after exposure (Figure 5D), even when high protein concentrations were added. On the other hand, penetration of T22-GFP-H6 was observed to be dose-dependent (Figure 5E) in both human HeLa and SW1417 cells, and efficiently inhibited by increasing amounts of SDF1α ( Figure 5F), the natural ligand of CXCR4.23 These data confirm the specificity of intracellular delivery driven by the T22 peptide.
Figure 5.
Internalization of T22-GFP-H6 in SW1417 cells. (A) Isosurface representation of T22-GFP-H6-exposed SW1417 cells within a three-dimensional volumetric x-y-z data field. (B) The particulate nature of the protein and the perinuclear accumulation are clearly observed. (C) Isosurface representation of GFP-H6-exposed SW1417 cells showing lack of fluorescence. (D) MTT analysis of SW1417 cells exposed to different concentrations of T22-GFP-H6. As a control, we used determined viability of cells exposed to the storing buffer alone. Values are referred to cell viability of cultures not exposed to the buffer. (E) Dose-response curve of T22-GFP-H6 internalization in HeLa and SW1417 cells. Data adjusted to hyperbolic equations with r2 = 0.9620 for HeLa cells and r2 = 0.9978 for SW1417 cells (both P < 0.001). (F) Inhibition of T22-GFP-H6 internalization in HeLa and SW1417 by increasing competitor/protein ratios of the natural CXCR4 ligand SDF1α.
Notes: GFP-H6 and human GLA were included as negative controls. Asterisks indicate significant differences when comparing with any of the negative controls (P < 0.001).
Abbreviations: GFP, green fluorescent protein; GLA, α-galactosidase.
Stability and architecture of T22-empowered GFP nanoparticles
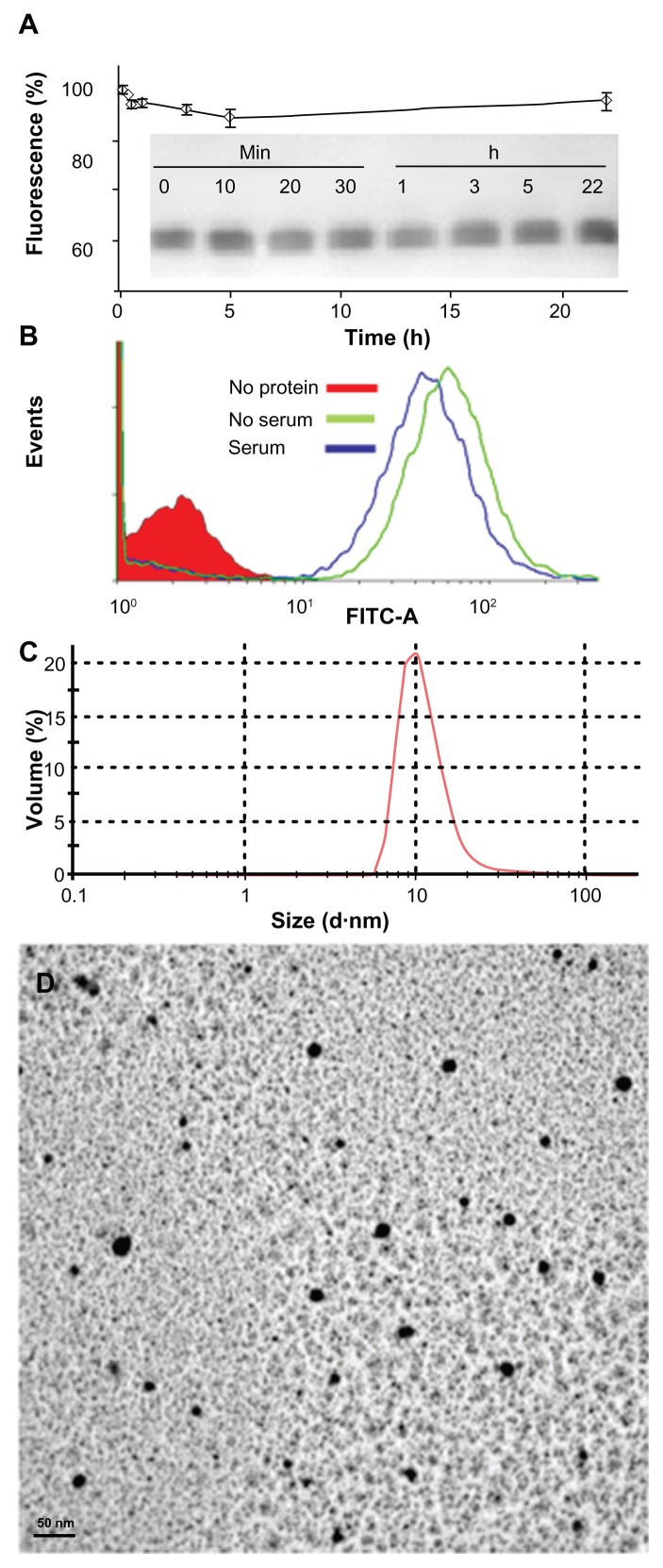
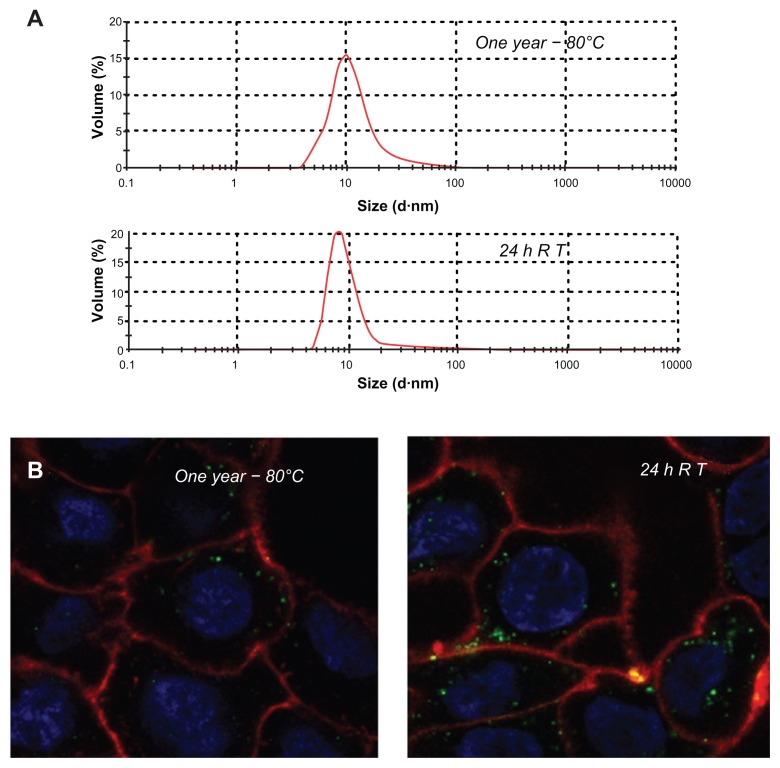
The confocal microscopy analyses shown in Figures 4 and 5 indicate that T22-GFP-H6 has a nanostructure, as has been previously demonstrated for the related construct, R9-GFP-H6.13 Although precise characterization of these assemblies is beyond the scope of this study, we were interested in confirming that T22-GFP-H6 is organized in nanoparticle form and that T22 can achieve targeted internalization of large macromolecular complexes. If so, T22 would be of broad interest for the functionalization of other categories of nanoparticles (eg, nonproteins) in emerging medicine. First, we determined that the construct was highly stable in human serum (Figure 6A) and able to internalize target cells fully in the presence of fetal bovine serum (Figure 6B). These features are suggestive of proteolytic stability, tight architecture, and regular organization of T22-GFP-H6 building blocks, in contrast with the random particle size and morphologies observed during amorphous protein aggregation, even in the form of soluble aggregates.24–27 Indeed, the mean size of T22-GFPH6, measured by dynamic light scattering, was 13.45 nm (Figure 6C), a value fully compatible with images of these nanoparticles under transmission electron microscopy (Figure 6D) showing relatively monodispersed entities. To assess further the structural and functional stability of T22-empowered nanoparticles, we determined their size distribution after storage for one year at −80°C, and also at room temperature for an additional 24 hours, followed by one additional step of freezing and thawing. As observed (Figure 7A), no important variations in nanoparticle size were observed, in agreement with their high stability in human serum (Figure 6A). After 24 hours of incubation at room temperature, we observed a slight tendency for the nanoparticles to become more compact entities, although the reduction in size was very moderate (around 1 nm on average). These nanoparticles were indistinguishable from the original material in terms of their ability to internalize cultured cells (Figure 7B). All these data confirm that the presence of T22, as it occurs with the cationic peptide R9, imparts self-organizing properties to His-tagged GFP, that cannot form multimeric complexes on its own.13
Figure 6.
Characterization of T22-empowered nanoparticles. (A) Remaining fluorescence during incubation of T22-GFP-H6 in human serum. In the inset, integrity of T22-GFP-H6 monomers monitored by Western blot. (B) Internalization of T22-GFP-H6 in HeLa cells in the presence of 10% fetal calf serum, monitored by the number of fluorescent cells. (C) Dynamic light scattering size analysis of T22-GFP-H6 nanoparticles in NaCO3H buffer. (D) Transmission electron microscopy of T22-GFP-H6 nanoparticles.
Abbreviation: GFP, green fluorescent protein.
Figure 7.
Stability of T22-GFP-H6 nanoparticles. (A) Size distribution of T22-GFP-H6 nanoparticles after one year of storage at −80°C. The same sample was further incubated at room temperature for 24 hours, submitted to one additional freezing and thawing step and analyzed. (B) These samples were tested for their ability to internalize SW1417, showing uptake images comparable with those shown in Figures 4 and 5.
Abbreviations: GFP, green fluorescent protein; RT, room temperature.
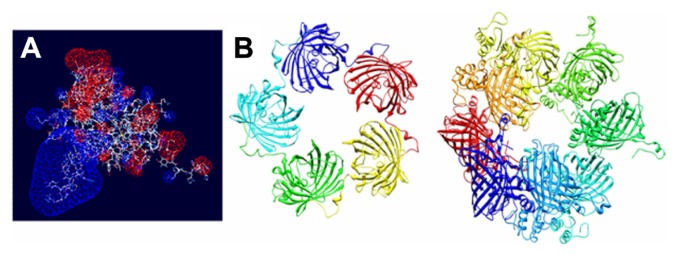
Given that T22 is also highly cationic (note its primary sequence in Figure 1A), we wondered if this peptide could promote electrostatic interaction between monomers to achieve stable nanoparticulate entities. To explore this possibility, we generated a charge map of T22-GFP-H6 (Figure 8A). The highly dipolar charge distribution of the multifunctional protein did indeed enable tight electrostatic contact between the charged sides of the GFP beta barrel and the consequent generation of regular oligomers. Two stable multimeric assemblies of T22-GFP-H6 monomers into approximately 13 nm nanoparticles are shown in Figure 8B, but alternative arrangements of the building blocks were also thermodynamically feasible (data not shown). Further structural analyses are in progress to elucidate the nature of the architectonic properties of T22, beyond those associated with its cell-targeting ability.
Figure 8.
Molecular modeling of T22-GFP-H6 monomers and nanoparticles. (A) Electrostatic field of the T22-GFP-H6 building monomer (cationic in blue and anionic in red). (B) Potential organization of T22-GFP-H6 as pentamers of 11.1 nm (left) and as octamers of 13.3 nm (right), in which the intervention of T22 assists the electrostatic self-assembling of the multimeric protein complex.
Abbreviation: GFP, green fluorescent protein.
T22-mediated intracellular targeting in a model of metastatic colorectal cancer
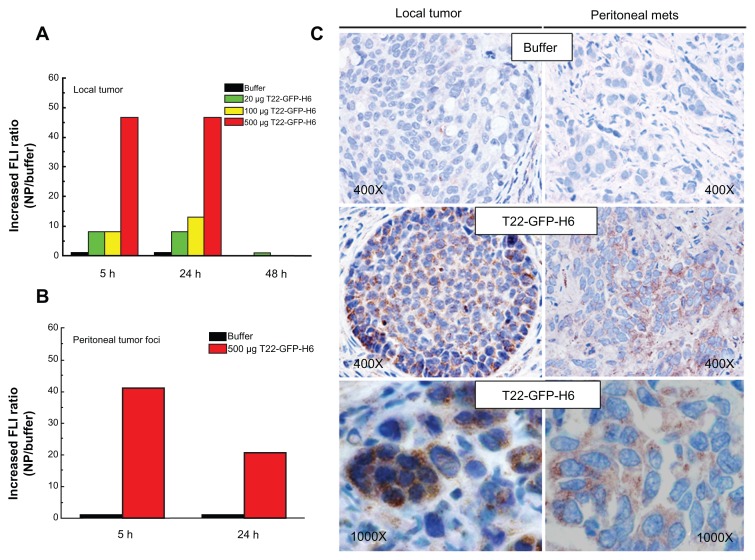
The excellent in vitro performance of T22 in receptor-specific intracellular targeting and the architectonic robustness of T22-empowered nanoparticles encouraged us to proceed further with in vivo biodistribution analyses in a CXCR4+ mouse model of metastatic colorectal cancer18 (Figure 9A). Upon tail vein administration, the green fluorescence in local tumors (Figure 9B) and metastatic foci (Figure 9C) was much more intense than the background levels observed in buffer-treated mice (Figures 9A and 10A). The fluorescent signal was dose-dependent, peaked at 5 hours, and remained relatively stable for at least 24 hours ( Figure 10A). This temporal profile is again indicative that the nanoparticles have high in vivo stability for at least one day following administration. When analyzing other tissues, we observed fluorescent metastatic foci in the peritoneum and lymph nodes but not in healthy organs (Figure 9B and C, Figure 10A and B), indicating tumor-specific targeting of the nanoparticles and accurate CXCR4-linked biodistribution. Finally, as in the cell cultures, we also observed cytosolic localization of T22-GFP-H6 by immunohistochemistry in both local tumor and metastatic foci (Figure 10C), demonstrating efficient cell internalization of the T22-functionalized constructs in vivo. No fever or other signs of toxicity were observed in any of the treated animals during the study (data not shown).
Figure 9.
Biodistribution of T22-empowered nanoparticles in an animal model of colorectal cancer. (A) Nude mouse bearing a local tumor (black asterisk), mesenteric lymph node (black arrow), and peritoneal metastases (empty arrow) after microinjecting 2 × 106 SW1417 human colorectal cancer cells into the cecal wall. The local tumor and mesenteric lymph node metastases overexpress CXCR4 in this model, as assessed by immunohistochemistry. (B) Selective biodistribution of T22-GFP-H6 in local tumor tissues 5, 24, or 48 hours after intravenous administration of 500 μg or 20 μg of nanoparticles as measured ex vivo. Fluorescence was undetectable in tumors from buffer-treated animals. (C) Accumulation of nanoparticles in peritoneal and lymph node metastases. No fluorescence was observed in any normal (liver, kidney, lung, heart) tissue, except for the biliary vesicle which showed fluorescence both in control and experimental animals.
Abbreviation: GFP, green fluorescent protein.
Figure 10.
Accumulation of T22-empowered nanoparticles in colorectal cancer metastatic foci. Enhanced green fluorescence associated with nanoparticle accumulation in local tumors (A) and peritoneal metastases (B) in experimental mice, as compared with buffer-treated controls. (C) Anti-His tag inmunostaining showing cytosolic localization of T22-GFP-H6 in local tumor tissue and peritoneal metastases in mice injected with T22-NP, which was absent in control animals injected with buffer.
Abbreviations: FLI, increased fluorescence; GFP, green fluorescent protein.
Discussion
Controlling cell targeting and penetrability of drugs and imaging agents is a major issue in emerging medicine. Successful identification of efficient intracellular targeting agents is expected to result in dramatic increases in drug stability and efficacy, as well as in significant reduction of toxicity and production costs. Given that the cell membrane is a major biological barrier for chemicals and particulate entities,28 identification of “Trojan horses” for selective intracellular delivery, ie, peptides or antibodies which selectively bind cell surface receptors and promote selective uptake of attached cargos,29 is a major demand in preclinical and clinical research, especially in cancer chemotherapy. However, very few of these peptides prove to be suitable for internalization of macromolecular complexes or nanoparticles, and lack of targeting remains a major obstacle in the design of effective drugs and full development of nanomedicine.30 In cancer chemotherapy, incorporation of monoclonal antibodies as drug targeting agents has had limited benefit for patients because of poor tumor penetration31 and unexpectedly rapid development of chemoresistance.32 Also, generic use of antitumor antibodies for targeting is controversial because only 0.001%–0.01% of these localize to the target site upon administration.33 Despite the steady identification of tumor-homing peptides,34,35 tags for receptor-dependent internalization of macromolecular complexes and nanoparticles are still unavailable.36 In this study, we identified that T22, a short amino acid segment (Figure 1A), is an unusually strong agent for intracellular targeting in CXCR4+ cells, the selectivity, stability and efficacy of penetration of which have been fully demonstrated in cell culture (Figures 3–5) and in vivo ( Figures 9 and 10). In addition, we produced T22 at high yields in a recombinant form as a domain of a highly stable modular protein ( Figure 2), demonstrating lack of toxicity of this peptide in E. coli and pointing to the feasibility of production of further (improved or adapted) engineered versions.
Interestingly, T22 had been previously identified as a CXCR4 ligand and explored in the context of antiretroviral therapies,37 because CXCR4 is a coreceptor for HIV and T22 inhibits viral attachment. However, the ability of T22 to penetrate cells was not suspected. Importantly, internalization of T22 does not depend on mere interaction with CXCR4, because other ligands tested in this study failed to promote efficient uptake (Figures 3 and 4), even showing that affinity for the receptor was higher than for T22.10 Dissociation between affinity for the receptor and endocytosis might account for the limited penetrability and poor uptake of antibody-empowered drugs.31,38 Other CXCR4 ligands previously investigated for targeted drug delivery showed very low or null penetration, even revealing themselves as agonists of CXCR4 and stimulating cell division.39 In contrast, in our hands, T22-exposed cells never showed significant proliferation compared with controls (data not shown). The efficient endosomal escape of the constructs generated might be due to the proton-sponge activity of the accompanying polyhistidines that, while useful for one-step protein purification, act also as a proton sponge, permitting endosomal disruption and delivery of the functionalized materials into the cytoplasm.22
On the other hand, when tested in an in vivo animal model of colorectal cancer, in which CXCR4+ cells were associated with aggressiveness, T22-empowered nanoparticles selectively localized not only in the primary tumor but also in metastatic foci (Figures 9C, 10B and C) confirming good stability of the protein-only nanoparticles generated in this study and suggesting that these nanoparticles might eventually be able to be attached to drugs to control tumor spread. This would be especially promising if used in the early stages of disease, because current treatment strategies for colorectal cancer are targeted to the primary tumor rather than to disseminated disease.40 Moreover, achieving higher intracellular concentrations of anticancer agents is expected to lead to a better antitumor effect, given that most of the chemotherapeutic agents used have a steep dose-response relationship.41 In the same context, precise targeting of imaging agents to metastatic foci would create additional diagnostic strategies.
Finally, T22 was able to impart self-organizing properties to GFP-H6, as has been previously shown for another cationic stretch, namely the receptor-independent cell-penetrating peptide, R9.13 The resulting building blocks, probably establishing electrostatic contact due to their dipolar nature (Figure 8), form relatively monodispersed protein-only nanoparticles about 13 nm in size (Figure 6), with proteolytic stability in human serum (Figure 6A) on systemic administration ( Figures 9 and 10) and remaining assembled and fully functional under different storage conditions (Figure 7). Interestingly, T22-empowered building blocks are highly fluorescent (22.8 fluorescence units/μg), indicative of a poor impact of the peptide on GFP. V1-GFP-H6 shows slightly lower fluorescent emission (12.4 fluorescence units/μg), being sufficiently high to monitor cell penetration, indicative of the conformational constraints imposed by the peptide on the building block.
Although the nanoscale architectonic properties imparted by T22 to the holding building block are secondary in terms of their CXCR4-dependent intracellular targeting ability, they warrant further investigation as architectonic tags for construction of protein-only nanoparticles.42,43 Self-assembling peptides, presently under intense investigation in diverse nanomedical applications,44–47 base their organizing properties on amyloid-like cross-molecular interactions, and when produced in “cell factories” such as E. coli, tend to aggregate as amorphous protein deposits.48 CXCR4 is an important cell surface marker in HIV infection,5 metastatic colorectal cancer,8,9 and other neoplasms,6 so incorporation of T22 as a targeting agent offers many opportunities for functionalization of drugs and nanoparticles for intracellular delivery, especially for disorders in which CXCR4 expression plays a pathophysiological role, including cancer, inflammation, autoimmunity, and ischemic lesions.4
Conclusion
The peptide T22, a known ligand of CXCR4, has been shown to be an unusually powerful tag for intracellular targeting in CXCR4+ cells, both in cell culture and in vivo. T22 is able to mediate the internalization of self-assembling protein-only nanoparticles 13 nm in mean diameter, keeping the stability and fluorescence emission of GFP-based building blocks. Rapid endosomal uptake and perinuclear accumulation of T22-empowered nanoparticles without cytotoxicity offer a wide spectrum of diagnostic and therapeutic opportunities for use of T22 in emerging nanomedicine to treat CXCR4-linked diseases, for which intracellular targeting agents are currently missing.
Acknowledgments
We appreciate the technical support of Fran Cortés from the Cell Culture Unit of the Servei de Cultius Cellulars, Producció d’Anticossos i Citometria, of the Servei de Microscòpia, and of the Protein Production Platform (CIBER-BBN). We also acknowledge the financial support received for the design and production of artificial viruses for gene therapy to EV, RM, and AV from FIS (PS0900165, PS0900965), MICINN (ACI2009-0919), AGAUR (2009SGR-108), and CIBER de Bioingeniería, Biomateriales y Nanomedicina, an initiative funded by the VI National R&D&i Plan 2008–2011, Iniciativa Ingenio 2010, Consolider Program, CIBER Actions and financed by the Instituto de Salud Carlos III with assistance from the European Regional Development Fund. UU and JDE have received predoctoral fellowships from ISCIII and MICINN, respectively, and AV has received an Institució Catalana de Recerca i Estudis Avançats Academia award.
Footnotes
Disclosures
UU, EV, NFM, AV, RM, IC, and MVC are cited as inventors in a patent application (EP11382005.4) covering the therapeutic use of T22. All other authors report no conflicts of interest in this work.
References
- 1.Pautler M, Brenner S. Nanomedicine: promises and challenges for the future of public health. Int J Nanomedicine. 2010;5:803–809. doi: 10.2147/IJN.S13816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ferrer-Miralles N, Vazquez E, Villaverde A. Membrane-active peptides for non-viral gene therapy: making the safest easier. Trends Biotechnol. 2008;26:267–275. doi: 10.1016/j.tibtech.2008.02.003. [DOI] [PubMed] [Google Scholar]
- 3.Milletti F. Cell-penetrating peptides: classes, origin, and current landscape. Drug Discov Today. 2012 Mar 23; doi: 10.1016/j.drudis.2012.03.002. [Epub ahead of print.] [DOI] [PubMed] [Google Scholar]
- 4.Peled A, Wald O, Burger J. Development of novel CXCR4-based therapeutics. Expert Opin Investig Drugs. 2012;21:341–353. doi: 10.1517/13543784.2012.656197. [DOI] [PubMed] [Google Scholar]
- 5.Wilen CB, Tilton JC, Doms RW. Molecular mechanisms of HIV entry. Adv Exp Med Biol. 2012;726:223–242. doi: 10.1007/978-1-4614-0980-9_10. [DOI] [PubMed] [Google Scholar]
- 6.Klonisch T, Wiechec E, Hombach-Klonisch S, et al. Cancer stem cell markers in common cancers – therapeutic implications. Trends Mol Med. 2008;14:450–460. doi: 10.1016/j.molmed.2008.08.003. [DOI] [PubMed] [Google Scholar]
- 7.Sun X, Cheng G, Hao M, et al. CXCL12/CXCR4/CXCR7 chemokine axis and cancer progression. Cancer Metastasis Rev. 2010;29:709–722. doi: 10.1007/s10555-010-9256-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kim J, Mori T, Chen SL, et al. Chemokine receptor CXCR4 expression in patients with melanoma and colorectal cancer liver metastases and the association with disease outcome. Ann Surg. 2006;244:113–120. doi: 10.1097/01.sla.0000217690.65909.9c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Liang Z, Yoon Y, Votaw J, Goodman MM, Williams L, Shim H. Silencing of CXCR4 blocks breast cancer metastasis. Cancer Res. 2005;65:967–971. [PMC free article] [PubMed] [Google Scholar]
- 10.Liang X. CXCR4, inhibitors and mechanisms of action. Chem Biol Drug Des. 2008;72:97–110. doi: 10.1111/j.1747-0285.2008.00681.x. [DOI] [PubMed] [Google Scholar]
- 11.Murakami T, Zhang TY, Koyanagi Y, et al. Inhibitory mechanism of the CXCR4 antagonist t22 against human immunodeficiency virus type 1 infection. J Virol. 1999;73:7489–7496. doi: 10.1128/jvi.73.9.7489-7496.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bradford MM. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- 13.Vazquez E, Roldan M, Díez-Gil C, et al. Protein nanodisk assembling and intracellular trafficking powered by an arginine-rich (R9) peptide. Nanomedicine (Lond) 2010;5:259–268. doi: 10.2217/nnm.09.98. [DOI] [PubMed] [Google Scholar]
- 14.Vazquez E, Cubarsi R, Unzueta U, et al. Internalization and kinetics of nuclear migration of protein-only, arginine-rich nanoparticles. Biomaterials. 2010;31:9333–9339. doi: 10.1016/j.biomaterials.2010.08.065. [DOI] [PubMed] [Google Scholar]
- 15.Seras-Franzoso J, Díez-Gil C, Vazquez E, et al. Bioadhesiveness and efficient mechanotransduction stimuli synergistically provided by bacterial inclusion bodies as scaffolds for tissue engineering. Nanomedicine (Lond) 2012;7:79–93. doi: 10.2217/nnm.11.83. [DOI] [PubMed] [Google Scholar]
- 16.Barbera VM, Martin M, Marinoso L, et al. The 18q21 region in colorectal and pancreatic cancer: independent loss of DCC and DPC4 expression. Biochim Biophys Acta. 2000;1502:283–296. doi: 10.1016/s0925-4439(00)00054-5. [DOI] [PubMed] [Google Scholar]
- 17.Baig MS, Manickam N. Homology modeling and docking studies of Comamonas testosteroni B-356 biphenyl-2,3-dioxygenase involved in degradation of polychlorinated biphenyls. Int J Biol Macromol. 2010;46:47–53. doi: 10.1016/j.ijbiomac.2009.10.014. [DOI] [PubMed] [Google Scholar]
- 18.Cespedes MV, Espina C, Garcia-Cabezas MA, et al. Orthotopic microinjection of human colon cancer cells in nude mice induces tumor foci in all clinically relevant metastatic sites. Am J Pathol. 2007;170:1077–1085. doi: 10.2353/ajpath.2007.060773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Amara A, Gall SL, Schwartz O, et al. HIV coreceptor downregulation as antiviral principle: SDF-1alpha-dependent internalization of the chemokine receptor CXCR4 contributes to inhibition of HIV replication. J Exp Med. 1997;186:139–146. doi: 10.1084/jem.186.1.139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhou NM, Luo ZW, Luo JS, Hall JW, Huang ZW. A novel peptide antagonist of CXCR4 derived from the N-terminus of viral chemokine vMIP-II. Biochemistry. 2000;39:3782–3787. doi: 10.1021/bi992750v. [DOI] [PubMed] [Google Scholar]
- 21.Fujii N, Nakashima H, Tamamura H. The therapeutic potential of CXCR4 antagonists in the treatment of HIV. Expert Opin Investig Drugs. 2003;12:185–195. doi: 10.1517/13543784.12.2.185. [DOI] [PubMed] [Google Scholar]
- 22.Ferrer-Miralles N, Corchero JL, Kumar P, et al. Biological activities of histidine-rich peptides; merging biotechnology and nanomedicine. Microb Cell Fact. 2011;10:101. doi: 10.1186/1475-2859-10-101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kucia M, Jankowski K, Reca R, et al. CXCR4-SDF-1 signalling, locomotion, chemotaxis and adhesion. J Mol Histol. 2004;35:233–245. doi: 10.1023/b:hijo.0000032355.66152.b8. [DOI] [PubMed] [Google Scholar]
- 24.Toledo-Rubio V, Vazquez E, Platas G, et al. Protein aggregation and soluble aggregate formation screened by a fast microdialysis assay. J Biomol Screen. 2010;15:453–457. doi: 10.1177/1087057110363911. [DOI] [PubMed] [Google Scholar]
- 25.Domingo-Espin J, Vazquez E, Ganz J, et al. The nanoparticulate architecture of protein-based artificial viruses is supported by protein- DNA interactions. Nanomedicine (Lond) 2011;6:1047–1061. doi: 10.2217/nnm.11.28. [DOI] [PubMed] [Google Scholar]
- 26.Vazquez E, Corchero JL, Villaverde A. Post-production protein stability: trouble beyond the cell factory. Microb Cell Fact. 2011;10:60. doi: 10.1186/1475-2859-10-60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Martinez-Alonso M, Gonzalez-Montalban N, Garcia-Fruitos E, Villaverde A. The functional quality of soluble recombinant polypeptides produced in Escherichia coli is defined by a wide conformational spectrum. Appl Environ Microbiol. 2008;101:1353–1358. doi: 10.1128/AEM.01446-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Riehemann K, Schneider SW, Luger TA, Godin B, Ferrari M, Fuchs H. Nanomedicine – challenge and perspectives. Angew Chem Int Ed Engl. 2009;48:872–897. doi: 10.1002/anie.200802585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Dietz GP, Bahr M. Delivery of bioactive molecules into the cell: the Trojan horse approach. Mol Cell Neurosci. 2004;27:85–131. doi: 10.1016/j.mcn.2004.03.005. [DOI] [PubMed] [Google Scholar]
- 30.Duncan R, Gaspar R. Nanomedicine(s) under the microscope. Mol Pharm. 2011;8:2101–2141. doi: 10.1021/mp200394t. [DOI] [PubMed] [Google Scholar]
- 31.Thurber GM, Schmidt MM, Wittrup KD. Factors determining antibody distribution in tumors. Trends Pharmacol Sci. 2008;29:57–61. doi: 10.1016/j.tips.2007.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ellis LM, Hicklin DJ. Resistance to targeted therapies: refining anticancer therapy in the era of molecular oncology. Clin Cancer Res. 2009;15:7471–7478. doi: 10.1158/1078-0432.CCR-09-1070. [DOI] [PubMed] [Google Scholar]
- 33.Jain M, Venkatraman G, Batra SK. Optimization of radioimmunotherapy of solid tumors: biological impediments and their modulation. Clin Cancer Res. 2007;13:1374–1382. doi: 10.1158/1078-0432.CCR-06-2436. [DOI] [PubMed] [Google Scholar]
- 34.Laakkonen P, Vuorinen K. Homing peptides as targeted delivery vehicles. Integr Biol (Camb) 2010;2:326–337. doi: 10.1039/c0ib00013b. [DOI] [PubMed] [Google Scholar]
- 35.Enback J, Laakkonen P. Tumour-homing peptides: tools for targeting, imaging and destruction. Biochem Soc Trans. 2007;35:780–783. doi: 10.1042/BST0350780. [DOI] [PubMed] [Google Scholar]
- 36.Mocellin S, Lise M, Nitti D. Targeted therapy for colorectal cancer: mapping the way. Trends Mol Med. 2005;11:327–335. doi: 10.1016/j.molmed.2005.05.002. [DOI] [PubMed] [Google Scholar]
- 37.Rusconi S, Scozzafava A, Mastrolorenzo A, Supuran CT. An update in the development of HIV entry inhibitors. Curr Top Med Chem. 2007;7:1273–1289. doi: 10.2174/156802607781212239. [DOI] [PubMed] [Google Scholar]
- 38.Segal NH, Saltz LB. Evolving treatment of advanced colon cancer. Annu Rev Med. 2009;60:207–219. doi: 10.1146/annurev.med.60.041807.132435. [DOI] [PubMed] [Google Scholar]
- 39.Egorova A, Kiselev A, Hakli M, Ruponen M, Baranov V, Urtti A. Chemokine-derived peptides as carriers for gene delivery to CXCR4 expressing cells. J Gene Med. 2009;11:772–781. doi: 10.1002/jgm.1366. [DOI] [PubMed] [Google Scholar]
- 40.Sleeman J, Steeg PS. Cancer metastasis as a therapeutic target. Eur J Cancer. 2010;46:1177–1180. doi: 10.1016/j.ejca.2010.02.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Skipper HE, Schabel FM, Jr, Mellett LB, et al. Implications of biochemical, cytokinetic, pharmacologic, and toxicologic relationships in the design of optimal therapeutic schedules. Cancer Chemother Rep. 1970;54:431–450. [PubMed] [Google Scholar]
- 42.Villaverde A. Nanotechnology, bionanotechnology and microbial cell factories. Microb Cell Fact. 2010;9:53. doi: 10.1186/1475-2859-9-53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Vazquez E, Villaverde A. Engineering building blocks for self- assembling protein nanoparticles. Microb Cell Fact. 2010;9:101. doi: 10.1186/1475-2859-9-101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sadatmousavi P, Soltani M, Nazarian R, Jafari M, Chen P. Self-assembling peptides: potential role in tumor targeting. Curr Pharm Biotechnol. 2011;12:1089–1100. doi: 10.2174/138920111796117409. [DOI] [PubMed] [Google Scholar]
- 45.Zhao Y, Tanaka M, Kinoshita T, Higuchi M, Tan T. Self-assembling peptide nanofiber scaffolds for controlled release governed by gelator design and guest size. J Control Release. 2010;147:392–399. doi: 10.1016/j.jconrel.2010.08.002. [DOI] [PubMed] [Google Scholar]
- 46.Kyle S, Aggeli A, Ingham E, McPherson MJ. Recombinant self- assembling peptides as biomaterials for tissue engineering. Biomaterials. 2010;31:9395–9405. doi: 10.1016/j.biomaterials.2010.08.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Huang H, Sun XS. Rational design of responsive self-assembling peptides from native protein sequences (dagger) Biomacromolecules. 2010;11:3390–3394. doi: 10.1021/bm100894j. [DOI] [PubMed] [Google Scholar]
- 48.Wu W, Xing L, Zhou B, Lin Z. Active protein aggregates induced by terminally attached self-assembling peptide ELK16 in Escherichia coli. Microb Cell Fact. 2011;10:9. doi: 10.1186/1475-2859-10-9. [DOI] [PMC free article] [PubMed] [Google Scholar]