Abstract
Hepatic oxidative stress and subsequent lipid peroxidation are well-recognized consequences of sustained ethanol consumption. The covalent adduction of nucleophilic amino acid side-chains by lipid electrophiles is significantly increased in patients with alcoholic liver disease (ALD); a global assessment of in vivo protein targets and the consequences of these modifications, however, has not been conducted. In this report, we describe identification of novel protein targets for covalent adduction in a 6-week murine model for ALD. Ethanol-fed mice displayed a 2-fold increase in hepatic TBARS while immunohistochemical analysis for the reactive aldehydes 4-hydroxynonenal (4-HNE), 4-oxononenal (4-ONE), acrolein (ACR) and malondialdehyde (MDA) revealed a marked increase in the staining of modified proteins in the ethanol-treated mice. Increased protein carbonyl content was confirmed utilizing subcellular fractionation of liver homogenates followed by biotin-tagging through hydrazide chemistry, where approximately a 2-fold increase in modified proteins was observed in microsomal and cytosolic fractions. To determine targets of protein carbonylation, a secondary hydrazide method coupled to a highly sensitive 2-dimensional liquid chromatography tandem mass spectrometry (2D LC-MS/MS or MuDPIT) technique was utilized. Our results have identified 414 protein targets for modification by reactive aldehydes in ALD. The presence of novel in vivo sites of protein modification by 4-HNE (2), 4-ONE (4) and ACR (2) was also confirmed in our data set. While the precise impact of protein carbonylation in ALD remains unknown, a bioinformatic analysis of the data set has revealed key pathways associated with disease progression, including fatty acid metabolism, drug metabolism, oxidative phosphorylation and the TCA cycle. These data suggest a major role for aldehyde adduction in the pathogenesis of ALD.
Keywords: ROS, ALD, 4-HNE, 4-hydroxynonenal, 4-ONE, 4-oxononenal, acrolein, MDA, malondialdehyde, MuDPIT, 2D-LC-MS/MS, DAVID, protein adducts, protein carbonyl, TBARS, lipid peroxidation, electrophile, biotin hydrazide, in vivo adduct, GRP78, bioinformatics, steatosis, fatty liver, ethanol
Introduction
Increased reactive oxygen species (ROS) and subsequent oxidative stress has been associated with the pathogenesis of numerous disease states including Alzheimer's, atherosclerosis, diabetes, cancer and non-alcoholic- and alcoholic liver disease (ALD).1-4 ROS are known to interact with numerous biological targets including proteins, DNA and lipids,5 the last of which can result in the generation of reactive aldehydes. These highly reactive aldehydes can modify nucleophilic targets, namely the amino acid side-chains of lysine, arginine, histidine and cysteine residues, reactions generally achieved through either a Schiff-base or Michael-type reaction mechanism.5-7 Among the most studied aldehydic products of lipid peroxidation, 4-hydroxynonenal (4-HNE), 4-oxononenal (4-ONE), acrolein (ACR) and malondialdehyde (MDA) have been shown to play a role in the progression of ALD.8-10 While the reactivity of these aldehydes may differ, they all possess the ability to modify nucleophilic protein side chains.6, 11
ALD remains a prominent cause of morbidity and mortality in the United States. Among the most prevalent and predictable outcomes of sustained ethanol consumption is the increased accumulation of lipids within the liver, termed steatosis; this is mirrored by a markedly increased oxidative stress profile and provides an ideal environment for lipid peroxidation.12-14 The enhanced generation of 4-HNE, 4-ONE, ACR and MDA has been reported in rodent models for ALD, showing substantial increases in aldehyde-adducted proteins.8-11 While the impact of these modifications was previously assessed on specific proteins or pathways, a more global approach evaluating the impact of protein carbonylation on disease progression has not been reported. Given the observed increase in both lipid accumulation and ROS production, ALD remains an ideal model to investigate the biochemical consequences of protein carbonylation in a physiologically relevant disease model.
The relatively low abundance of protein carbonyls in physiological systems has greatly minimized the number of reported modifications in vivo.15, 16 Previous research in our laboratory identified protein disulfide isomerase (PDI) as a target for 4-HNE adduction in vivo, demonstrating one of only a handful of reports to detect a protein adduct in a relevant disease model.15 Enrichment strategies have recently been described, utilizing carbonyl-targeted chemical modifiers.17 In a recent publication by Codreanu et al., the application of biotin hydrazide and mass spectrometry (MS) was utilized to selectively enrich samples for 4-HNE modified proteins in a cell culture system.18 These elegant studies provided excellent avenues for research into the role of protein carbonylation in disease models of oxidative stress and lipid peroxidation.
In this report, we utilized a well-characterized murine model for ALD to investigate the role of protein carbonylation in vivo. After employing multiple enrichment strategies, we identified a total of 414 targets for aldehyde adduction with the use of 2-dimensional liquid chromatography MS/MS (2D-LC-MS/MS or MuDPIT); these studies also identified eight novel sites of in vivo protein adduction generated from 4-HNE, 4-ONE or ACR. The precise physiological consequences of these adducts remains unknown; thus, we applied bioinformatic strategies to elucidate the potential impact of protein carbonylation in our disease model. Consistent with the pathogenesis of ALD, fatty acid metabolism, oxidative phosphorylation, aldehyde dehydrogenases, drug metabolism and amino acid metabolism were all found to be significantly enriched in the collection of adducted proteins. These studies collectively highlight the potential toxicological implications of protein carbonylation in a physiologically relevant disease model and provide novel mechanistic information into the pathogenesis of ALD.
Materials and Methods
Animal Model
All animal procedures were approved by the Institutional Animal Care and Use Committee of the University of Colorado and were performed in accordance with published National Institutes of Health guidelines. Male, Wild-Type C57/BL6J mice (12 per group) were utilized for the analysis and characterization of ethanol-mediated liver damage. Briefly, mice were fed a modified Lieber-DeCarli liquid based-diet (Bio-Serv, Frenchtown, NJ) for a period of 6 weeks. The diet consisted of 45% fat-derived calories, 16% protein-derived calories, and the remaining balance comprised of either maltose-dextrin or ethanol-derived calories. Ethanol-fed mice began the study on a diet consisting of 2% (v/v) ethanol, with the ethanol-derived calories increasing on a weekly basis until sacrifice; week 6 consisted of 6.0% ethanol (v/v) or 31.8% ethanol-derived calories. Pair-fed control animals received the remaining caloric intake from carbohydrate source. Food consumption was monitored daily and body weights were measured once per week. Upon completion of the study, animals were anesthetized via intraperitoneal injection with sodium pentobarbital and euthanized by exsanguination. Livers were excised, weighed, and frozen for biochemical characterization or subjected to differential centrifugation for subcellular fractionation as previously described.10 Hepatic thiobarbituric acid reactive substances (TBARS) was measured as described.19
Immunohistochemical Staining
Following excision, livers were sectioned and placed in 10% neutral buffered formalin for 8h. Samples were processed, embedded in paraffin and mounted on slides by Colorado HistoPrep (Fort Collins, CO). One pair of slides was stained with hematoxylin and eosin (H&E) for histological characterization while the remaining slides were subjected to de-paraffinization and rehydration for immunohistochemical characterization using either custom antibodies generated in our laboratory directed against 4-HNE, 4-ONE or MDA modified proteins (Bethyl Laboratories, Montgomery, TX) or ACR modified proteins (Abcam, Cambridge, MA).
Biotin Derivitization of Protein Carbonyls
Protein collected from subcellular fractionation was incubated in the dark with 5.0mM Biotin Hydrazide (Pierce, Rockford, IL) for 2h at room temperature while rotating. Samples were then reduced with 10.0mM sodium borohydride for 1h at room temperature. For the detection of total protein carbonyls, samples were denatured in loading buffer, separated via standard SDS-PAGE, transferred to PVDF and blotted for biotinylated proteins using an anti-biotin antibody (GeneTex, Irvine, CA). Membranes were developed using ECL-Plus Reagent (GE Healthcare, Piscataway, NJ) and chemiluminescence was visualized using a Storm 860 scanner from Molecular Dynamics (Sunnyvale, CA).
For MS analysis, subcellular fractions isolated from 6 animals were pooled. This facilitated protein identification and minimized false-positive detection. Likewise, the resulting pooled samples represented an average of the respective treatment group. Biotinylated proteins were filtered through Zeba Spin Desalting Columns (Pierce, Rockford, IL) to remove excess biotin hydrazide. Biotinylated samples were then applied to a Streptavidin HP Spintrap column (GE Healthcare, Piscataway, NJ) for 1h at room temperature. Following 5 washes with 2.0M urea in PBS, proteins were eluted in 0.1M ammonium hydroxide (5×) and samples were dried to completion in vaccuo. Proteins were resuspended in loading buffer and allowed to migrate approximately 1cm into an SDS-PAGE gel. The entire band was excised from the gel, digested with sequencing-grade trypsin (Promega, Madison, WI) in 50mM ammonium bicarbonate overnight at 37°C and the resulting peptides were prepared for analysis by 2D-LC-MS/MS as previously described.20
2-Dimensional LC-MS/MS Identification of Carbonylated Proteins
Isolated tryptic peptides were analyzed using multidimensional protein identification technology (MuDPIT) by inline strong cation exchange and reverse-phase biphasic trapping (Integra Frit column, New Objective). The LC was coupled to a nano-ESI source and Esquire HCT ion trap mass spectrometer (Bruker Daltonics, Inc., Billerica, MA). Samples were injected and eluted with a series of ammonium acetate salt plugs (25mM, 50mM, 75mM, 100mM, 125mM, 150mM, 200mM, 500mM, 1M) followed by a reverse-phase hydrophobicity ramp. Specifically, nano liquid chromatography (EASY-nLC, Proxeon) was run at a flow rate of 300 nL/min with a gradient of 5 to 40% ACN (0.2% formic acid) over 90 min on a C18 EASY-Column analytical column (100 × 0.075mm). The instrument was operated using data-dependent collision-induced dissociation (CID) MS/MS with a threshold of 10,000 total ion current (TIC.) Data analysis was performed using Mascot (v 2.1.04, www.matrixscience.com) and Esquire v5.2 data analysis package (Bruker Daltonics). Mascot was used to generate a merged file from each MuDPIT analysis (1 sample injection and 9 salt plugs), which was then imported into Scaffold 3.2 (Proteome Software, Inc., Portland OR) in order to validate, analyze, interpret and organize the complex 2D-LC-MS/MS MuDPIT data. Peptide and protein probabilities were assigned by the Protein Prophet algorithm.21 Peptide identifications were accepted if they could be established at greater than 95.0% probability as specified by the Peptide Prophet algorithm,22 and protein identifications were accepted if they could be established at greater than 95.0% probability and contained at least 1 identified peptide. Proteins containing similar peptides that could not be differentiated based on MS/MS analysis alone were grouped to satisfy the principles of parsimony. All pooled samples were run in duplicate, generating a total of 8 control samples (2 microsomal, 2 mitochondrial, 2 cytosolic and 2 nuclear) and 8 ethanol samples. All control samples were merged into one file, as well as all ethanol samples, generating a composite list of control- or ethanol-fed murine protein identifications.
Identification and Classification of Functionally Enriched Terms, Knowledge Network Analysis
The Database for Annotation, Visualization and Integrated Discovery (DAVID) v6.7 [http://david.abcc.ncifcrf.gov] was utilized to identify biologically enriched themes and terms within the protein list of interest.23 The composite list of 414 protein identifications was submitted to DAVID for analysis; background and species identified as Mus musculus.
Gene functional classification and functional annotation enrichment were all undertaken using DAVIDs default parameters and stringency measures. Functional annotations associated with Gene Ontology (GO) (biological processes, molecular function, and cellular component), protein domains (InterPro), the KEGG pathway database and functional categories (Sp Pir Keywords) were included in the analysis. Given the use of physical subcellular fractionation for our studies, all terms associated with cellular location were excluded from the analysis. Significance was determined essentially as recommended by the DAVID authors as follows: fold enrichment > 5.0, Benjamini corrected p < 0.01.23
The networks were generated using software developed in house.24, 25 Knowledge was combined from a collection of databases as described by Leach et al. 2009, except that the natural language processing component was omitted in the present study.24 The resulting network of gene IDs encodes the identified adducted proteins.24, 25 This list contained 414 gene IDs which became nodes of the combined knowledge network. For the purpose of simplicity in interpretation, only nodes contained in the KEGG database were retained. Following generation of the network, data were extracted and figures were generated utilizing Cytoscape 2.8.26
Criteria for Protein Identification and Validation of in vivo Adducts
For the identification of in vivo adducts, the stringent restrictions stated above could not be applied due to poor protein identification probabilities, perhaps a result of poor fragmentation due to the modification. Therefore, to identify adducted peptides, thresholds were lowered to allow all identifications and adducts were investigated if y-and b-ion assignments exceeded 30% of the peptide. Mass spectra were manually validated for the presence of all aldehyde modifications.
Statistical Analysis
Statistical analysis and generation of graphs was performed using GraphPad Prism 4.02 (GraphPad Software, San Diego, CA). Differences between control and ethanol-fed mice were assessed using a paired Student's t test. Differences were considered significant if p < 0.05.
Results
Sustained Ethanol Consumption Results in Increased Immunohistochemical Staining of 4-HNE, 4-ONE, ACR and MDA-Modified Proteins
Chronic ethanol consumption is widely reported to result in sustained oxidative stress, leading to enhanced generation of lipid-derived aldehydes.9, 27 Utilizing a six week murine model for early-stage ALD, we sought to investigate the role of protein carbonylation in disease progression. As shown in Figure 1a, H&E staining reveals a marked increase in hepatic lipid accumulation throughout zones 1 and 2. Using antibodies directed against 4-HNE, 4-ONE, ACR and MDA-derived protein adducts, immunohistochemical analysis was performed. Although staining is visible in the control-fed mice, an increase in immunoreactive proteins can be clearly seen in the ethanol-fed mice following staining with each antibody. To further validate the observed increase in protein adducts presented in Figure 1a, hepatic TBARS were measured, revealing roughly a 2-fold increase (P < 0.01) in nmol MDA/mg protein (Figure 1b). Collectively, these data demonstrate a state of sustained oxidative stress following 6 weeks of ethanol consumption leading to the increased generation of lipid-derived aldehydes and subsequent hepatic protein carbonyls.
Figure 1.
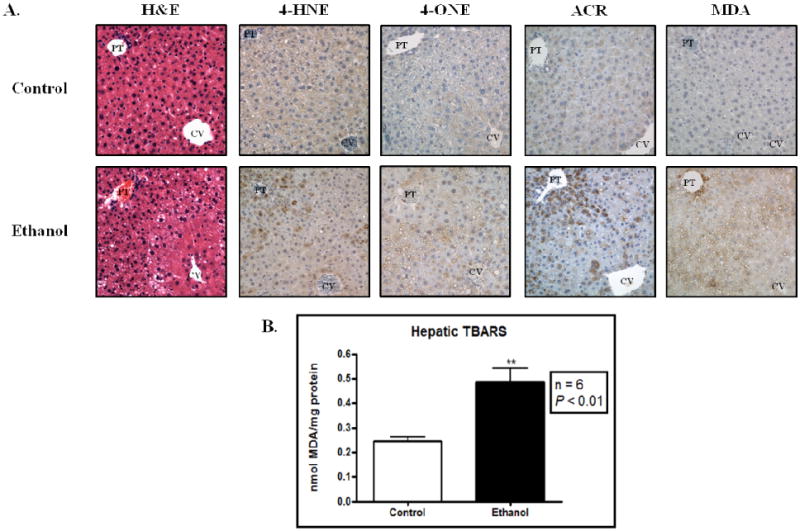
Immunohistochemical characterization of aldehyde-modified proteins in ALD. (A) Hematoxylin & Eosin (H&E) staining reveals an increased in hepatic lipid content. Staining for 4-HNE, 4-ONE, ACR and MDA-modified proteins reveals increased hepatic staining in the ethanol-fed mice (bottom). CV, central vein; PT, portal triad. (B) Hepatic TBARS content significantly increases in the ethanol-fed mice. N = 6, ** P < 0.01.
Biotin Labeling Captures Protein Carbonyls in vivo
A recent report by Codreanu et. al demonstrated the use of hydrazide chemistry and biotin labeling to selectively isolate 4-HNE modified proteins in cultured cells treated with the aldehyde.18 These studies have provided the framework for applications into physiologically relevant models of oxidative stress, such as ALD. Using subcellular fractions collected from the livers of both control and ethanol-fed mice, protein carbonyls were selectively tagged with biotin using this technique. As shown in Figure 2a, ethanol-fed mice were found to have significantly greater quantities of protein carbonyls in cytosolic and microsomal compartments. The mitochondrial and nuclear fractions were also found to have significant protein adducts, however, no statistically significant difference was observed between the control and ethanol-fed mice, an indication of the constitutive nature of protein adducts. The blot densitometry in Figure 2b, presented as percent control, confirm the increase protein adducts.
Figure 2.
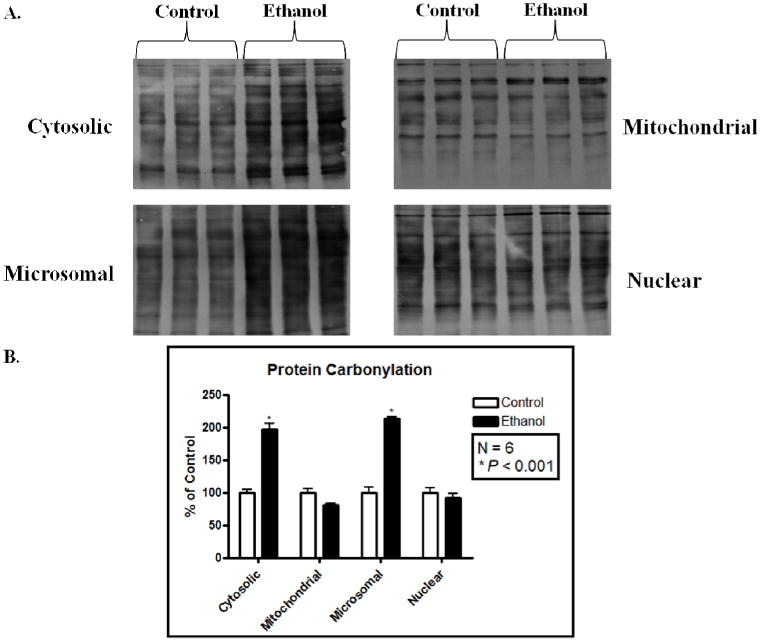
Biotin-tagging reveals an increase in protein carbonyls in microsomal and cytosolic fractions. (A) Anti-biotin western blotting on livers from three control mice (first three lanes) and three ethanol-fed mice (last three lanes), subcellular fractions are indicated. (B) Blot densitometry was quantified and is represented as a percentage of control. N = 6; * P < 0.001.
In order to isolate protein targets of aldehyde modification, we applied this biotin labeling approach in conjunction with streptavidin pulldown. To verify the specificity and efficacy of this approach, proteins were either left untreated (native) or treated with biotin hydrazide. Samples were then bound to streptavidin columns and run under SDS-PAGE as described. As shown in Supplemental Figure S1, only proteins labeled with biotin were selectively eluted. This lane was excised and prepared for MS analysis. Based on the stringent cutoffs for positive protein identification, no significant scores were found, demonstrating a high level of specificity for this method.
2D-LC-MS/MS Analysis of Subcellular Fractions for Protein Carbonyls
Previous reports from our laboratory have utilized the Lieber-DeCarli diet to identify targets of 4-HNE adduction.20, 28 These studies predominantly rely on time-consuming and labor intensive techniques such as 2-dimensional gel electrophoresis (2D-PAGE) and spot picking. While effective, the limit of detection remains inferior to enrichment strategies coupled to high-throughput mass spectrometry techniques, such as 2D-LC-MS/MS, or MuDPIT.29 Here, we have applied the latter technique to a complex mixture of enriched biotin-conjugated protein carbonyls. In an effort to maximize the number of identifications, we performed this technique on subcellular fractions collected from whole liver homogenates; these include enriched microsomal, mitochondrial, nuclear and cytosolic fractions.
Subcellular fractionation was employed to enhance the depth of proteome coverage for organelle-specific proteins and reduce sample complexity, thus optimizing detection and identification of less abundant proteins. Given the relative low abundance of protein carbonyls in physiologic systems, we performed the MS analyses on these subcellular fractions to increase the concentration of adducted proteins, concomitantly increasing selectivity and the probability of successful protein identification. As shown in Figure 3a, we identified 414 distinct protein targets for modification by reactive aldehydes in our model (50 proteins unique to the control-fed mice, 84 unique to the ethanol-fed mice and 280 common to both treatment groups). A list of the proteins identified in this study can be found in Supplemental Table S1.
Figure 3.
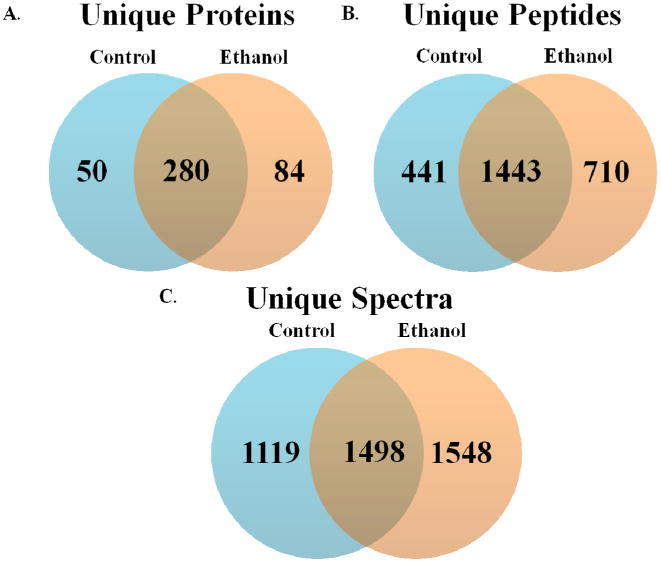
MuDPIT analysis of adducted peptides demonstrates an increase in aldehyde-modified proteins in livers of ethanol-fed mice. Pooled duplicate samples from each of the four subcellular fractions reveal an increase in (A) unique protein identifications, (B) unique peptides and (C) unique spectra.
Total protein carbonyl content can be regulated through multiple mechanisms, resulting from an increase in new protein targets, additional adducted residues per protein, and/or an increase in the number of adducted molecules of a given protein. As shown in Figure 3b, we observed 710 unique carbonylated peptides in the ethanol-fed mice, while 441 were observed in the control-fed mice. Spectral counting presented in Figure 3c revealed a similar trend, showing 1119 unique spectra in the control samples and 1548 in the ethanol-treated group. While these data remain semi-quantitative, an increase in protein identifications as well as in the number of unique peptides was observed following ethanol consumption. Our data supports current theories that protein carbonylation occurs through a “concentration-dependent trigger”, where new protein targets are adducted following saturation of low-level aldehyde targets.18, 30 In our rodent model for early-stage ALD, it is possible that these protein targets are not present under supersaturating conditions, further providing an explanation for the minimal difference in protein identifications between the two treatment groups.
Bioinformatic Analysis of Carbonylated Protein Targets in ALD
To further elucidate the impact of protein carbonylation in the alcoholic liver, the entire list of 414 proteins was converted to gene IDs and submitted to the bioinformatic tool DAVID to assess the biological processes impacted by aldehyde adduction in ALD. Using Mus musculus as the background gene list, a total of 219 molecular functions and biological processes were found to be significantly enriched (Supplemental Table S2); terms associated with subcellular location or tissue distribution were excluded from the list for clarity.
The generation of hepatic lipid droplets is considered the initiating step in the pathogenesis of ALD. Consistent with this proposition, our analyses identified the KEGG pathway associated with fatty acid metabolism (Figure 4) to have the greatest fold enrichment while achieving statistical significance (KEGG mmu00071, fold enrichment 12.20). These data suggest an ongoing cycle of altered lipid homeostasis that results in increased lipid content, increasing the pool of peroxidizable lipids. This increased lipid content mediates the further generation of reactive aldehydes, exacerbating the lipid load within the cell. Likewise, many of the other pathways and biological processes identified in these analyses have been reported to be affected during the course of ALD. In total, nine KEGG pathways involved with amino acid metabolism and degradation were found to be associated with protein carbonylation. These data agree with the proposed impact on the nutritional balance following sustained ethanol consumption.31, 32 Likewise, the peroxisome proliferator-activated receptor (PPAR) signaling pathway was significantly enriched in our dataset (KEGG mmu03320, fold enrichment: 5.03). This pathway has been a major focus for ALD research and our data may suggest an additional mechanism for its regulation.33 We also observed a significant enrichment in terms associated with the tricarboxylic acid (TCA) cycle (SP_PIR, GO 0006099 and KEGG mmu00020; fold enrichment of 25.03, 18.25 and 9.77 respectively). As indicated, the oxidative metabolism of ethanol leads to the increased production of acetyl-CoA, providing more substrate for the TCA cycle. This is thought to be the predominant effect of ethanol on the TCA cycle; our data however, support a novel mechanism whereby protein carbonylation of critical TCA enzymes may be playing an integral role in this process.
Figure 4.
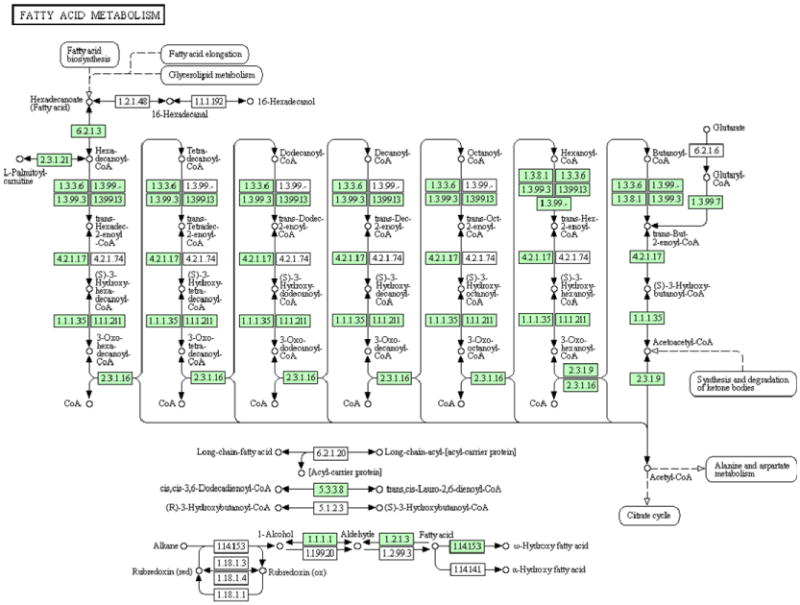
DAVID results reveal a significant enrichment in proteins involved in fatty acid metabolism. Proteins identified in our study are indicated by green shading.
DAVID provides an effective tool to analyze the cellular processes, molecular functions and pathways for a given set of proteins; however, for a large list exceeding ∼50 proteins, valid biological interpretation is arduous. To address this, we generated a network based on our data set focusing strictly on KEGG pathways to determine both the integration of the same protein in multiple pathways as well as proteins critical to numerous cellular processes. As shown in Figure 5, this analysis demonstrates the inclusion of 311 proteins identified in our list that were integrated in the network. These proteins were found to cluster based on their KEGG pathway assignments, demonstrating both heavily enriched pathways and integration of multiple proteins. The nodes (proteins) that were found in specific pathways have been highlighted using various color designations. In agreement with the DAVID analysis, fatty acid metabolism (yellow) was well-represented in the protein list. The location of this cluster demonstrates that numerous proteins in this pathway were also found to be integrated in other pathways (i.e. peroxisomes, teal). While proteins involved in fatty acid metabolism are located throughout the cell, the peroxisomes play a central role in the metabolism and oxidation of fatty acids. This analysis also demonstrated an abundance of proteins involved in amino acid metabolism (dark green), the TCA cycle (pink), PPAR signaling (purple) and drug metabolism (blue). Clustering analysis was also conducted using the 50 proteins in the “control only” group and the 84 proteins in the “ethanol only” group revealing no notable clustering or pathway enrichment (data not shown).
Figure 5.
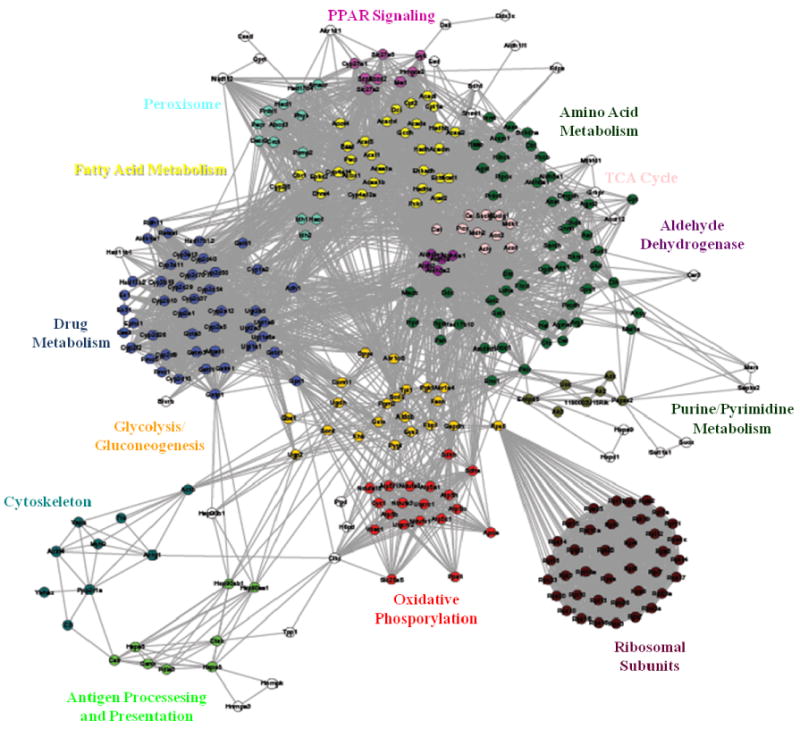
Network analysis demonstrates critical pathways impacted by protein carbonylation. The network was generated based on the KEGG assignments of the proteins identified and their co-occurrence in pathways. Colors correspond to KEGG pathways as indicated. Most proteins were found to belong to multiple pathways, demonstrating their locale in the network. Proteins with many pathway associations can be found central to the map, while peripheral proteins demonstrate segregation to a single pathway.
Identification of Novel in vivo Protein Targets for Aldehyde Adduction
The detection of in vivo protein adducts has many complicating factors including the complexity of the sample, sequence coverage, abundance of adducts, MS fragmentation technique and instrument sensitivity.17, 34 In an effort to selectively enrich our samples for aldehyde adducted proteins, we coupled subcellular fractionation, hydrazide chemistry and 2D-LC-MS/MS. The enriched fractions were then analyzed for biotin hydrazide conjugated 4-HNE, 4-ONE, ACR or MDA adducts; although additional modifications may be found, these four lipid peroxidation end-products are thought to play a major role in the pathogenesis of ALD. Figure 5 provides a schematic of an aldehyde-adducted protein (4-HNE, 4-ONE, ACR or MDA) containing the reduced hydrazone bond conjugated to biotin, yielding varying masses. Analyzing our composite list of protein identifications for these modifications, we were able to identify and validate eight new targets for aldehyde adduction in vivo. Table 1 provides a list of the proteins identified along with the peptide recovered, the modification found and the subcellular fraction containing the peptide. In total, we identified two novel in vivo protein targets for 4-HNE (nuclear factor 1 A-type and structure-specific endonuclease subunit SLX4), four for 4-ONE (CCR4-NOT transcription complex subunit 1, protein FAM135A, 78kDa glucose-regulated protein and myosin-binding protein C, fast-type) and two for ACR (neutral alpha-glucosidase AB and nesprin-1). Representative MS spectra are provided as supplemental material (Supplemental Figures S2-S9). While the impact of these adducts on the overall function of the identified proteins remains unknown, these results provide exciting avenues for future studies on protein carbonylation.
Table 1.
Identification of eight novel in vivo targets for modification by either 4-HNE, 4-ONE or MDA. The assigned UniProt ID, protein name, peptide identified, modified residue, modification detected, the fraction the peptide was recovered in, and the treatment group are indicated. ‘c’ indicates carbamidomethyl; ‘m’ indicates oxidized methionine.
| UniProt ID | Protein Name | Peptide Identified | Modified Residue | Modification | Fraction Identified | Treatment Group |
|---|---|---|---|---|---|---|
|
| ||||||
| GANAB_Mouse | Neutral alpha-glucosidase AB | TcDESSFC*K | C47 | BH-Acrolein | Microsomal | Ethanol |
| SYNE1_Mosue | Nesprin-1 | GFK*IDLNcK | K6960 | BH-Acrolein | Mitochondrial | Ethanol |
| NFIA_Mouse | Nuclear factor 1 A-type | RmSK*EEER | K71 | BH-HNE | Cytosolic | Ethanol |
| SLX4_Mouse | Structure-specific endonuclease subunit SLX4 | EQHVNRC*LDEAEK | C142 | BH-HNE | Cytosolic | Ethanol |
| CNOT1_Mouse | CCR4-NOT transcription complex subunit 1 | TAVEKAGPEmDK*R | K1520 | BH-ONE | Mitochondrial | Control |
| F135A_Mouse | Protein FAM135A | WKKSGSLLQLTC*R | C1382 | BH-ONE | Mitochondrial | Ethanol |
| GRP78_Mouse | 78 kDa glucose-regulated protein | LSSEDK*ETmEK | K592 | BH-ONE | Microsomal | Ethanol |
| MYPC2_Mouse | Myosin-binding protein C, fast-type | WYK*NGVEVRPSK | K468 | BH-ONE | Cytosolic | Ethanol |
Discussion
The covalent adduction of lipid electrophiles to nucleophilic protein side chains is proposed to play a role in the pathogenesis of numerous disease states associated with oxidative stress.1 Despite decades of research, the relatively low abundance of these aldehyde-modified proteins in physiological systems has hindered progress in the field.2, 11, 16 Previous studies evaluating protein carbonylation in disease models utilized labor intensive 2-dimensional gel electrophoresis, spot picking and western blotting.20, 28 These studies were limited not only by the abundance of aldehyde adducts, but also by the relatively low abundance of the protein itself. Reports from our laboratory, and others (i.e. see Newton et al. 2009, Roede et al. 2008, Grimsrud et al. 2007) have described redundant identifications, notably proteins in the ALDH, GST and Prdx superfamilies, while failing to identify site-specific in vivo modifications.2, 10, 35 Recent developments in mass spectrometry and enrichment strategies, however, have provided excellent new applications for research in this field.16, 18 In this report, we describe the combined use of subcellular fractionation, hydrazide chemistry and biotin-tagging with a highly sensitive MuDPIT technique to selectively enrich for low abundance protein carbonyls in an established in vivo mouse model of ALD. Our data have resulted in the identification of previously identified, highly abundant proteins as well; however this experimental approach has resulted in identification of numerous novel targets for carbonylation, including (but not limited to) cystathionine γ-lyase, apoprotein E, and the acyl-CoA dehydrogenase family of proteins (ACADS, ACADM, ACAL, ACADVL).
The data presented here show a significant increase in total protein carbonyl content in the microsomal and cytosolic compartments of ethanol-consuming murine livers. Ethanol ingestion is well-documented to result in a substantial increase in cytochrome P450 2E1 (Cyp2E1) protein expression.36 This ER protein is thought to be the major producer of cellular ROS following ethanol consumption and correlates with the level of lipid peroxidation products in the liver.36 Thus, the data presented here provide a mechanistic explanation for the observed increase in microsomal carbonyl content. A significant increase in the mitochondrial carbonyl content was not observed but this difference may be attributed to the high abundance of protective aldehyde dehydrogenase (ALDH) enzymes in this subcellular fraction. While these discrepancies may provide additional information into the progression of ALD, our focus remained on the global impact of protein carbonylation in our model simply utilizing subcellular fractionation as an additional enrichment strategy. These enrichment strategies led to the identification of 414 targets for protein carbonylation in vivo, greatly increasing the number of identified naturally-occurring protein adducts in a physiological system. While highly abundant, previously identified proteins were found (i.e. PDI, peroxiredoxin 6, HSP90), numerous relatively low abundant proteins were also identified.28 Therefore, the present study represents one of the most comprehensive assessments of protein carbonylation in a physiologically relevant disease model that is published to date.
The level of protein carbonylation can be affected on numerous levels, varying from the number of residues adducted, to the number of molecules of any given protein. Recent hypotheses propose a “concentration-dependent trigger” model of protein carbonylation, whereby novel sites of adduction are modified only following saturation of more susceptible residues.18, 30 These studies were conducted utilizing cell culture models and supra-physiological concentrations of aldehyde. In spite of the varied approaches, the results demonstrated considerable overlap and are consistent with use of a similar mechanism. The present proteomic survey of protein carbonylation revealed only a modest increase in the number of protein identifications in the ethanol-fed mice, despite increased immunohistochemical staining of 4-HNE, 4-ONE, ACR and MDA-modified proteins and a statistically significant increase in hepatic TBARS. It is likely that these low-level targets of aldehyde modification are not saturated, thus supporting recent publications that have identified a similar trend.18, 30 Likewise, while the techniques applied in the present study have increased the concentration of these aldehyde targets in each biological sample, the limit of detection with current mass spectrometers remains a considerable problem, likely hindering the identification of countless low-level targets.
Mass spectrometry has greatly expanded this area of research and, despite decades of research, very few site-specific in vivo protein adducts have ever been detected.15, 16, 37 This can be attributed to a host of variables, including abundance of adducts, sequence coverage following enzymatic digest and stability of protein adducts during fragmentation. In a recent publication by Guo et al., the instability of these adducted residues was highlighted under CID conditions with and without the presence of a biotin tag.34 Using model peptides for His and Cys 4-HNE modifications, fragmentation was observed with and without tagging reagents such as biotin hydrazide. These studies revealed the relative instability of both non-tagged and tagged aldehyde species under CID, observing neutral loss of the aldehyde following fragmentation.34 It is likely that the instability of these adducts, in conjunction with their low abundance, has hindered the number of modifications identified in vivo. Employing subcellular fractionation, biotin-tagging and MuDPIT MS/MS techniques, we identified eight novel in vivo protein targets for aldehyde modification. We expect that additional peptides may be present following tryptic digestion, but the harsh conditions of CID contribute to a reduced number of positive identifications in our studies.34
Among the eight proteins identified with an in vivo modification, GRP78 is of particular interest. Comprised of two-domains, an N-terminal ATPase catalytic site and a C-terminal substrate binding domain, GRP78 plays a critical role in the binding of misfolded or unfolded proteins.38 The novel identification of a 4-ONE adduct on lysine 592 lies within the C-terminal substrate binding domain, potentially altering the ability of GRP78 to effectively bind and refold erred proteins. Through this activity, GRP78 plays a critical role in the initiation and regulation of the unfolded protein response (UPR).39 Previous reports have outlined a role for the UPR during the pathogenesis of ALD, while the mechanisms have remained elusive.40-43 While functional studies on adduction of GRP78 have not been completed, similar studies have been conducted regarding the impact of aldehyde-adduction on other members of the heat-shock family of proteins 11. These data provide a potential mechanism for the known UPR induction following ethanol consumption, stemming away from conventional proposed theories.41, 44
Protein carbonylation has been proposed to play a role in a host of disease states associated with sustained oxidative stress.1, 2, 9 Our murine model for early-stage ALD demonstrates a significant increase in protein carbonyl content; the precise cellular role of these protein modifications, however, remains unknown. To try to elucidate the physiological impact of protein carbonylation in ALD, we utilized multiple bioinformatic techniques. Among the biochemical pathways and biological processes associated with our list of 414 protein targets of carbonylation, fatty acid metabolism was significantly enriched. The initiating stages of disease progression are marked by lipid accumulation, and our results indicate a role for aldehyde adduction in this progression. Many of the proteins identified in this study play a major role in the both fatty acid metabolism and lipogenesis. At present, It is unknown how these modifications affect protein function; however, a loss in function of nearly all of these proteins would result in a further increase in lipid content within the cell. Among the protein targets identified, the acyl-CoA dehydrogenase enzymes (which includes ACADS, ACADM, ACAL, ACADVL) play a major role in lipid metabolism. Studies have shown that alterations in these enzymes leads to increased lipid content in the liver by inhibiting effective beta-oxidation.45 These data suggest a major role for lipid peroxidation and aldehyde adduction in mediating the additional accumulation of lipids thereby increasing the observed steatosis. Aldehyde protein modifications are likely to result in dysfunctional enzymes, leading to vicious loop of biochemical and cellular derangements. This global survey of protein carbonylation provides novel insights into the deleterious impact of these modifications in vivo and suggests compelling new avenues for research on both individual proteins as well as in other disease models.
Supplementary Material
Figure 6.
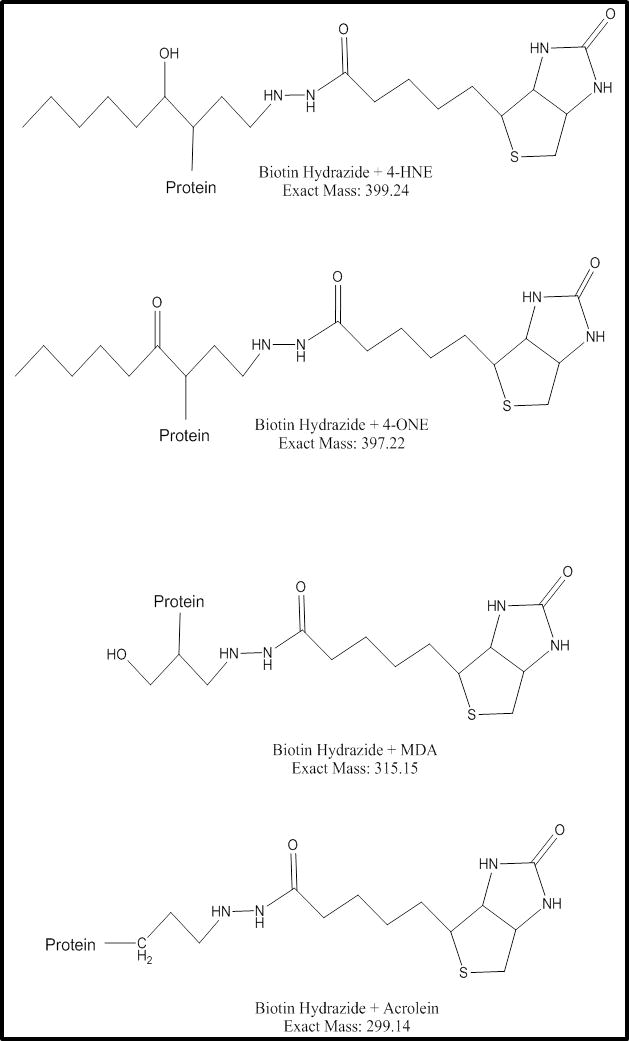
Illustration of an aldehyde adducted, biotin-tagged protein. The four aldehydes monitored in this study are shown, along with the exact mass of the tagged adduct. These modifications were monitored for validation of the in vivo adducts reported in Table 1.
Acknowledgments
We would like to acknowledge Jose D. Gomez and the mass spectrometry core at the University of Colorado Anschutz Medical Campus, School of Pharmacy, for assistance with instrumentation.
Funding Support: Studies were supported by the National Institutes of Health/National Institutes of Alcoholism and Alcohol Abuse under grant numbers R37AA09300 (DRP), R01DK074487-01 (DRP) and F31AA018606-01 (JJG).
Abbreviations
- ROS
reactive oxygen species
- ALD
alcoholic liver disease
- 4-HNE
4-hydroxynonenal
- 4-ONE
4-oxononenal
- ACR
acrolein
- MDA
malondialdehyde
- PDI
protein disulfide isomerase
- MS
mass spectrometry
- MuDPIT
multidimensional protein identification technology
- H&E
hematoxylin and eosin
- TBARS
thiobarbituric acid reactive substances
- CID
collision-induced dissociation
- DAVID
Database for Annotation, Visualization and Integrated Discovery
- GO
gene ontology
- PPAR
peroxisome proliferator-activated receptor
- TCA
tricarboxylic acid
- Cyp2E1
cytochrome P450 2E1
- ALDH
aldehyde dehydrogenase
References
- 1.Dalle-Donne I, Giustarini D, Colombo R, Rossi R, Milzani A. Protein carbonylation in human diseases. Trends Mol Med. 2003;9:169–176. doi: 10.1016/s1471-4914(03)00031-5. [DOI] [PubMed] [Google Scholar]
- 2.Grimsrud PA, Picklo MJ, Sr, Griffin TJ, Bernlohr DA. Carbonylation of adipose proteins in obesity and insulin resistance: identification of adipocyte fatty acid-binding protein as a cellular target of 4-hydroxynonenal. Mol Cell Proteomics. 2007;6:624–637. doi: 10.1074/mcp.M600120-MCP200. [DOI] [PubMed] [Google Scholar]
- 3.Albano E. Alcohol, oxidative stress and free radical damage. Proc Nutr Soc. 2006;65:278–290. doi: 10.1079/pns2006496. [DOI] [PubMed] [Google Scholar]
- 4.Mottaran E, Stewart SF, Rolla R, Vay D, Cipriani V, Moretti M, Vidali M, Sartori M, Rigamonti C, Day CP, Albano E. Lipid peroxidation contributes to immune reactions associated with alcoholic liver disease. Free Radic Biol Med. 2002;32:38–45. doi: 10.1016/s0891-5849(01)00757-2. [DOI] [PubMed] [Google Scholar]
- 5.Esterbauer H, Schaur RJ, Zollner H. Chemistry and biochemistry of 4-hydroxynonenal, malonaldehyde and related aldehydes. Free Radic Biol Med. 1991;11:81–128. doi: 10.1016/0891-5849(91)90192-6. [DOI] [PubMed] [Google Scholar]
- 6.LoPachin RM, Gavin T, Petersen DR, Barber DS. Molecular mechanisms of 4-hydroxy-2-nonenal and acrolein toxicity: nucleophilic targets and adduct formation. Chem Res Toxicol. 2009;22:1499–1508. doi: 10.1021/tx900147g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jacobs AT, Marnett LJ. Systems analysis of protein modification and cellular responses induced by electrophile stress. Acc Chem Res. 2010;43:673–683. doi: 10.1021/ar900286y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Galligan JJ, Fritz KS, Tipney H, Smathers RL, Roede JR, Shearn CT, Hunter LE, Petersen DR. Profiling impaired hepatic endoplasmic reticulum glycosylation as a consequence of ethanol ingestion. J Proteome Res. 2011;10:1837–1847. doi: 10.1021/pr101101s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sampey BP, Korourian S, Ronis MJ, Badger TM, Petersen DR. Immunohistochemical characterization of hepatic malondialdehyde and 4-hydroxynonenal modified proteins during early stages of ethanol-induced liver injury. Alcohol Clin Exp Res. 2003;27:1015–1022. doi: 10.1097/01.ALC.0000071928.16732.26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Roede JR, Stewart BJ, Petersen DR. Decreased expression of peroxiredoxin 6 in a mouse model of ethanol consumption. Free Radic Biol Med. 2008;45:1551–1558. doi: 10.1016/j.freeradbiomed.2008.08.032. [DOI] [PubMed] [Google Scholar]
- 11.Carbone DL, Doorn JA, Kiebler Z, Ickes BR, Petersen DR. Modification of heat shock protein 90 by 4-hydroxynonenal in a rat model of chronic alcoholic liver disease. J Pharmacol Exp Ther. 2005;315:8–15. doi: 10.1124/jpet.105.088088. [DOI] [PubMed] [Google Scholar]
- 12.Cederbaum AI, Lu Y, Wu D. Role of oxidative stress in alcohol-induced liver injury. Arch Toxicol. 2009;83:519–548. doi: 10.1007/s00204-009-0432-0. [DOI] [PubMed] [Google Scholar]
- 13.Seitz HK, Lieber CS, Stickel F, Salaspuro M, Schlemmer HP, Horie Y. Alcoholic liver disease: from pathophysiology to therapy. Alcohol Clin Exp Res. 2005;29:1276–1281. doi: 10.1097/01.alc.0000171896.37022.f7. [DOI] [PubMed] [Google Scholar]
- 14.Arteel G, Marsano L, Mendez C, Bentley F, McClain CJ. Advances in alcoholic liver disease. Best Pract Res Clin Gastroenterol. 2003;17:625–647. doi: 10.1016/s1521-6918(03)00053-2. [DOI] [PubMed] [Google Scholar]
- 15.Carbone DL, Doorn JA, Kiebler Z, Petersen DR. Cysteine modification by lipid peroxidation products inhibits protein disulfide isomerase. Chem Res Toxicol. 2005;18:1324–1331. doi: 10.1021/tx050078z. [DOI] [PubMed] [Google Scholar]
- 16.Chavez JD, Wu J, Bisson W, Maier CS. Site-specific proteomic analysis of lipoxidation adducts in cardiac mitochondria reveals chemical diversity of 2-alkenal adduction. J Proteomics. 2011;74:2417–2429. doi: 10.1016/j.jprot.2011.03.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fritz KS, Petersen DR. Exploring the biology of lipid peroxidation-derived protein carbonylation. Chem Res Toxicol. 2011;24:1411–1419. doi: 10.1021/tx200169n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Codreanu SG, Zhang B, Sobecki SM, Billheimer DD, Liebler DC. Global analysis of protein damage by the lipid electrophile 4-hydroxy-2-nonenal. Mol Cell Proteomics. 2009;8:670–680. doi: 10.1074/mcp.M800070-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fritz KS, Galligan JJ, Smathers RL, Roede JR, Shearn CT, Reigan P, Petersen DR. 4-Hydroxynonenal Inhibits SIRT3 via Thiol-Specific Modification. Chem Res Toxicol. 2011 doi: 10.1021/tx100355a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Roede JR, Carbone DL, Doorn JA, Kirichenko OV, Reigan P, Petersen DR. In vitro and in silico characterization of peroxiredoxin 6 modified by 4-hydroxynonenal and 4-oxononenal. Chem Res Toxicol. 2008;21:2289–2299. doi: 10.1021/tx800244u. [DOI] [PubMed] [Google Scholar]
- 21.Nesvizhskii AI, Keller A, Kolker E, Aebersold R. A statistical model for identifying proteins by tandem mass spectrometry. Anal Chem. 2003;75:4646–4658. doi: 10.1021/ac0341261. [DOI] [PubMed] [Google Scholar]
- 22.Keller A, Nesvizhskii AI, Kolker E, Aebersold R. Empirical statistical model to estimate the accuracy of peptide identifications made by MS/MS and database search. Anal Chem. 2002;74:5383–5392. doi: 10.1021/ac025747h. [DOI] [PubMed] [Google Scholar]
- 23.Huang da W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 24.Leach SM, Tipney H, Feng W, Baumgartner WA, Kasliwal P, Schuyler RP, Williams T, Spritz RA, Hunter L. Biomedical discovery acceleration, with applications to craniofacial development. PLoS Comput Biol. 2009;5:e1000215. doi: 10.1371/journal.pcbi.1000215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tipney HJ, Leach SM, Feng W, Spritz R, Williams T, Hunter L. Leveraging existing biological knowledge in the identification of candidate genes for facial dysmorphology. BMC Bioinformatics. 2009;10(2):S12. doi: 10.1186/1471-2105-10-S2-S12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Smoot ME, Ono K, Ruscheinski J, Wang PL, Ideker T. Cytoscape 2.8: new features for data integration and network visualization. Bioinformatics. 2011;27:431–432. doi: 10.1093/bioinformatics/btq675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sampey BP, Stewart BJ, Petersen DR. Ethanol-induced modulation of hepatocellular extracellular signal-regulated kinase-1/2 activity via 4-hydroxynonenal. J Biol Chem. 2007;282:1925–1937. doi: 10.1074/jbc.M610602200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Smathers RL, Galligan JJ, Stewart BJ, Petersen DR. Overview of lipid peroxidation products and hepatic protein modification in alcoholic liver disease. Chem Biol Interact. 2011;192:107–112. doi: 10.1016/j.cbi.2011.02.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wu CC, MacCoss MJ. Shotgun proteomics: tools for the analysis of complex biological systems. Curr Opin Mol Ther. 2002;4:242–250. [PubMed] [Google Scholar]
- 30.Maisonneuve E, Ducret A, Khoueiry P, Lignon S, Longhi S, Talla E, Dukan S. Rules governing selective protein carbonylation. PLoS One. 2009;4:e7269. doi: 10.1371/journal.pone.0007269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bunout D. Nutritional and metabolic effects of alcoholism: their relationship with alcoholic liver disease. Nutrition. 1999;15:583–589. doi: 10.1016/s0899-9007(99)00090-8. [DOI] [PubMed] [Google Scholar]
- 32.Lieber CS. Relationships between nutrition, alcohol use, and liver disease. Alcohol Res Health. 2003;27:220–231. [PMC free article] [PubMed] [Google Scholar]
- 33.Nagy LE. Molecular aspects of alcohol metabolism: transcription factors involved in early ethanol-induced liver injury. Annu Rev Nutr. 2004;24:55–78. doi: 10.1146/annurev.nutr.24.012003.132258. [DOI] [PubMed] [Google Scholar]
- 34.Guo J, Prokai L. To tag or not to tag: A comparative evaluation of immunoaffinity-labeling and tandem mass spectrometry for the identification and localization of posttranslational protein carbonylation by 4-hydroxy-2-nonenal, an end-product of lipid peroxidation. J Proteomics. 2011;74:2360–2369. doi: 10.1016/j.jprot.2011.07.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Newton BW, Russell WK, Russell DH, Ramaiah SK, Jayaraman A. Liver proteome analysis in a rodent model of alcoholic steatosis. J Proteome Res. 2009;8:1663–1671. doi: 10.1021/pr800905w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wu D, Cederbaum AI. Oxidative stress and alcoholic liver disease. Semin Liver Dis. 2009;29:141–154. doi: 10.1055/s-0029-1214370. [DOI] [PubMed] [Google Scholar]
- 37.Madian AG, Diaz-Maldonado N, Gao Q, Regnier FE. Oxidative stress induced carbonylation in human plasma. J Proteomics. 2011;74:2395–2416. doi: 10.1016/j.jprot.2011.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gething MJ. Role and regulation of the ER chaperone BiP. Semin Cell Dev Biol. 1999;10:465–472. doi: 10.1006/scdb.1999.0318. [DOI] [PubMed] [Google Scholar]
- 39.Malhi H, Kaufman RJ. Endoplasmic reticulum stress in liver disease. J Hepatol. 2011;54:795–809. doi: 10.1016/j.jhep.2010.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ji C, Chan C, Kaplowitz N. Predominant role of sterol response element binding proteins (SREBP) lipogenic pathways in hepatic steatosis in the murine intragastric ethanol feeding model. J Hepatol. 2006;45:717–724. doi: 10.1016/j.jhep.2006.05.009. [DOI] [PubMed] [Google Scholar]
- 41.Ji C, Kaplowitz N. Betaine decreases hyperhomocysteinemia, endoplasmic reticulum stress, and liver injury in alcohol-fed mice. Gastroenterology. 2003;124:1488–1499. doi: 10.1016/s0016-5085(03)00276-2. [DOI] [PubMed] [Google Scholar]
- 42.Ji C, Mehrian-Shai R, Chan C, Hsu YH, Kaplowitz N. Role of CHOP in hepatic apoptosis in the murine model of intragastric ethanol feeding. Alcohol Clin Exp Res. 2005;29:1496–1503. doi: 10.1097/01.alc.0000174691.03751.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kaplowitz N, Than TA, Shinohara M, Ji C. Endoplasmic reticulum stress and liver injury. Semin Liver Dis. 2007;27:367–377. doi: 10.1055/s-2007-991513. [DOI] [PubMed] [Google Scholar]
- 44.Ji C, Kaplowitz N. Hyperhomocysteinemia, endoplasmic reticulum stress, and alcoholic liver injury. World J Gastroenterol. 2004;10:1699–1708. doi: 10.3748/wjg.v10.i12.1699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zhang D, Liu ZX, Choi CS, Tian L, Kibbey R, Dong J, Cline GW, Wood PA, Shulman GI. Mitochondrial dysfunction due to long-chain Acyl-CoA dehydrogenase deficiency causes hepatic steatosis and hepatic insulin resistance. Proc Natl Acad Sci U S A. 2007;104:17075–17080. doi: 10.1073/pnas.0707060104. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


