Abstract
Gene silencing using small interfering RNA (siRNA) is a promising therapeutic strategy for the treatment of various diseases, in particular, cancer. Recently, our group reported on a novel gene carrier, the micelle-like nanoparticle (MNP), based on the combination of a covalent conjugate of phospholipid and polyethylenimine (PLPEI) with polyethylene glycol (PEG) and lipids. These long-circulating MNPs loaded with plasmid DNA-mediated gene expression in distal tumors after systemic administration in vivo. In the current study, we investigated the potential of MNPs for siRNA delivery. MNPs were prepared by condensing siRNA with PLPEI at a nitrogen/phosphate ratio of 10, where the binding of siRNA is complete. The addition of a PEG/lipid coating to the PLPEI complexes generated particles with sizes of ca. 200 nm and a neutral surface charge compared with positively charged PLPEI polyplexes without the additional coating. MNPs protected the loaded siRNA against enzymatic digestion and enhanced the cellular uptake of the siRNA payload. MNPs carrying green fluorescent protein (GFP)-targeted siRNA effectively downregulated the gene in cells that stably express GFP. Finally, MNPs were non-toxic at a wide range of concentrations and for different cell lines.
Keywords: Nanoparticle, Gene delivery, siRNA delivery, Polyethylenimine (PEI), Polyethylene glycol (PEG), Self-assembly, Lipid/PEG coating
Introduction
Small interfering RNA (siRNA) is currently considered a promising agent for the treatment of various diseases, including cancer (Novina and Sharp 2004; Oh and Park 2009; Siomi and Siomi 2009; Whitehead et al. 2009). However, its clinical application is limited due to its instability in the blood and poor cellular uptake following systemic administration. To overcome these drawbacks, chemically modified siRNAs and a variety of synthetic and biodegradable lipids and polymers have been developed to systemically deliver siRNA to the cytosol of target cells with variable efficacy and safety profiles (Jeong et al. 2009; Kim and Kim 2009; Schroeder et al. 2009).
Among synthetic polymers, polyethylenimine, PEI, has been used successfully for nucleic acid delivery under both in vitro and in vivo conditions (Grayson et al. 2006; Mao et al. 2006; Philipp et al. 2009; Urban-Klein et al. 2005). PEI can be synthesized in different lengths, be branched or linear, and possesses a capability of protonation of amino group at every third position. This latter feature gives PEI a high positive charge density at physiological pH and permits the condensation of negatively charged nucleotides (DNA; siRNA, ODN) into dense particles by electrostatic interactions. PEI also has an intrinsic endosomal escape mechanism known as the “proton-sponge” effect, which causes osmotic swelling and rupture of the endosome membrane that triggers the release of PEI complexes into the cytosol (Boussif et al. 1995). Unless cell-binding ligands are present, an overall positive charge is essential for the uptake of the PEI complexes by the absorptive endocytosis (Merdan et al. 2002). However, the requirement of an overall positive charge for an effective transfection is usually linked to high toxicity and represents a major problem for systemic application (Chollet et al. 2002; Kircheis et al. 1999). Positively charged complexes are prone to interact nonspecifically with a variety of components in the blood and other biological fluids or with non-target cells. These non-specific interactions cause toxic side-effects, destabilization of the complexes and their removal from circulation by the organs of the reticuloendothelial system, and thus, decrease the bioavailability of the nucleic acid molecules at the target site.
In order to improve siRNA delivery and reduce the toxicity of PEI-based complexes, several approaches have been investigated such as coupling PEI to hydrophobic moieties, including fatty acid residues (Alshamsan et al. 2009; Gusachenko et al. 2009) and degradable acrylates (Liu et al. 2009; Philipp et al. 2009) or grafting hydrophilic polyethylene glycol (PEG) to PEI (Malek et al. 2008; Mao et al. 2006; Nimesh and Chandra 2009). Additionally, noncovalent association of PEI complexes with liposomes has also been proposed. For example, the combination of PEI complexes with several liposomal formulations led to non-toxic and efficient in vitro delivery of siRNA (Schafer et al. 2010). Similarly, PEI/ODN complexes (Ko et al. 2009a) or PEI/siRNA complexes (Rothdiener et al. 2010) encapsulated in PEG-stabilized liposomes were successfully tested for targeted delivery by coupling an antibody to the surface of the liposomes.
Recently, we have reported on a novel gene carrier, the micelle-like nanoparticle (MNP), based on the combination of a covalent conjugate between phospholipid and polyethylenimine (PLPEI) with PEG and lipids. These MNPs combined the favorable properties of the low molecular weight PEI 1.8 kDa (nucleic acid condensation, low cytotoxicity, endosomal escape) with those of PEG-stabilized liposomes (in vivo stability, prolonged blood circulation) that resulted in an effective transfection of plasmid DNA in a distal tumor cell when the long-circulating MNPs were administered intravenously (Ko et al. 2009b). We hypothesized that these MNPs could also serve as a suitable carrier for siRNA delivery. In the current study, MNPs loaded with siRNA were prepared and evaluated for complex formation, nuclease stability, cytotoxicity, cellular trafficking, and gene silencing efficacy.
Materials and methods
Materials
All materials were purchased from Sigma-Aldrich unless otherwise stated. Branched PEI with a molecular weight of 1.8 or 25 kDa was purchased from Polysciences, Inc (Warrington, PA). 1-Palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine (POPC), 1,2-disrearoyl-sn-glycero-3-phosphoethanolamine-N-[methoxy (polyethyleneglycol)-2000] (PEG-PE), cholesterol, and oxidized phospholipid, 1-palmitoyl-2-azelaoyl-sn-glycero-3-phosphocholine (Az PC Ester) were purchased from Avanti Polar Lipids (Alabaster, AL). All siRNA duplexes were purchased from Dharmacon (Lafayette, CO), namely, siRNA targeting green fluorescent protein (GFP-siRNA): 5′-AUGAACUUCAGGGUCAGCUdTdT-3′ (sense) (Musacchio et al. 2010), a non-targeting control duplex, (Negative-siRNA): 5′-AGUACUGCUUACGAUACGGdTdT-3′ (sense) and 6-carboxyfluorescein (FAM)-labeled siRNA (siGLO® siRNA). RNase III was purchased from Ambion (Austin, TX). The CellTiter-Blue® Cell Viability Assay and Hoechst 33342 and Lysotracker® Red were purchased from Promega (Madison, WI). Nuclease-free water was purchased from Qiagen (Germantown, MD).
Cell culture
The cell lines B16F10 (mouse melanoma), NIH/3T3 (mouse fibroblast), and c166-GFP cells from the c166 cell line (mouse yolk sac embryo) stably transfected with a plasmid reporter vector, pEGFP-N1, encoding for the enhanced GFP were obtained from the American Type Culture Collection (Manassas, VA). B16F10 and NIH/3T3 cells were grown at 37°C under 5% CO2 in Dulbecco’s Modified Eagle’s Medium (DMEM) supplemented with 10% fetal bovine serum (FBS) and penicillin (100 units/mL) and streptomycin (100 µg/mL). For c166-GFP cells, DMEM media was supplemented with 10% FBS and 0.2 mg/mL of Genetic in (G-418, Invitrogen, CA). Cell culture media and penicillin/streptomycin stock solutions were purchased from Cellgro (Herndon, VA). Heat-inactivated FBS was purchased from Atlanta Biologicals (Lawrenceville, GA).
Methods
Synthesis of phospholipid–polyethylenimine conjugate
The PLPEI was synthesized from PEI 1.8 kDa and oxidized phospholipid (Az PC Ester) as previously described (Ko et al. 2009b). Briefly, 12 mg of the branched PEI (7 µmol) were dissolved in 0.5 mL of chloroform and mixed with 5 mg of the oxidized PC (AzPC Ester, 7 µmol) dissolved in 1 mL of chloroform. Assuming that branched PEI has a 1:2:1 molar ratio of primary/secondary/tertiary amines, the reaction mixture corresponds to an acid-to-primary amine molar ratio of 1:10. Carbonyldiimidazole (0.5 mg, 3 µmol) was added to the above solution for the activation of acid by formation of an imidazole derivative. The mixture was incubated with 10 µL of TEA (triethylamine) at room temperature for 24 h with stirring. The chloroform was removed under nitrogen gas, and the residue was suspended with 2 mL of distilled water. The products were purified by dialysis (MWCO 2,000 Da) against dH2O and lyophilized. The PLPEI conjugate was dissolved in water to a concentration of 1 µg/µL (0.7 µg/µL as PEI).
Preparation of PLPEI/siRNA complexes
The preparation of MNPs first requires the formation of PLPEI/siRNA complexes. Complexes were prepared by mixing a fixed amount of siRNA and varying amounts of PLPEI that were separately diluted in equal volumes of PBS (pH 7.4, nuclease-free buffer). The siRNA solution was transferred to the polymer solution, mixed by vigorous pipetting and incubated for 15 min. The polymer/siRNA ratio was expressed as the nitrogen/phosphate (N/P) ratio and calculated assuming that 43 g/mol corresponds to each repeating unit of PEI containing one amine and 316 g/mol corresponds to each repeating unit of siRNA containing one phosphate.
Gel retardation studies
For gel retardation studies, complexes containing 750 ng of siRNA with varying amounts of PLPEI or PEI 1.8 kDa in PBS were electrophoresed through a 0.8% agarose gel, using the E-Gel electrophoresis system (Invitrogen Life Technologies) and evaluated under UV light.
Ethidium bromide exclusion assay
The binding of PLPEI to siRNA was examined by the fluorescence quenching method based on ethidium bromide (EtBr). The experiments were carried out by measuring the fluorescence intensity of complexes prepared from siRNA (5 µg/mL) with EtBr intercalated at a molar ratio of 2:1 (siRNA/EtBr) as increasing amounts of PLPEI or PEI 1.8 kDa were added. Fluorescence was measured using a 96-well plate reader (Multiscan MCC/340, Fisher Scientific Co.) at the excitation and emission wavelengths of 540 and 580 nm, respectively. The relative fluorescence values were determined as follows: Fr = (Fm − −Fe) × 100/(Fo − −Fe), were Fr is the relative fluorescence, Fm is the measured fluorescence, Fe is the fluorescence of EtBr in the absence of siRNA, and Fo is the initial fluorescence in the absence of the polycation.
Serum stability of complexes
The protection of siRNA within PLPEI complexes against serum degradation was evaluated by incubating PLPEI and PEI complexes at N/P ratio of 10 and naked siRNA in the presence of 50% FBS for 1 and 24 h. Samples were disrupted with heparin (50 U/µg siRNA), loaded in 0.8% agarose gel (1 µg siRNA per well) and evaluated under UV light.
Preparation of MNPs
The MNPs were assembled with PLPEI/POPC/Cholesterol/PEG-PE (4:3:3:0.3 mol/mol) and siRNA. First, 18.6 µg PLPEI (13.6 µg as PEI) and 10 µg siRNA corresponding to a N/P ratio of 10 were separately diluted in PBS and mixed to a final volume of 100 µL. A dry lipid film was prepared from the mixture of POPC, cholesterol, and PEG-PE (4.3:2.2:1.6 µg, 3:3:0.3 mol/mol). The lipid film was hydrated with preformed PLPEI complexes and incubated for 1 h at room temperature.
Stability of siRNA in MNPs against RNase digestion
Nuclease resistance of the siRNA inMNPs was determined by the treatment of the samples with 1 U of RNase III/µg siRNA for 30 min at 37°C.MNPs were disassembled by adding 0.1% Triton-X 100 and Heparin (50 U/µL) and analyzed by agarose gel electrophoresis. The integrity preservation of siRNA in MNPs was compared with that of naked siRNA.
Particle size and zeta potential measurement
The particle size and zeta potential of formulations were measured by quasi-electric light scattering using a Zeta Plus Particle Analyzer (Brookhaven Instruments Corp, Santa Barbara, CA). Scattered light was detected at 25°C at an angle of 90°. Samples (50 µL) of complexes and MNPs were diluted in 1.7 mL of nuclease-free water and measured immediately after preparation.
Cytotoxicity assays
For cytotoxicity studies, NIH/3T3, B16F10, and c166 GFP cells, were seeded in 96-well plates (104 cells/well). After 24 h, the medium was replaced with 100 µL/well of serial dilutions with equivalent concentrations of PEI for each formulation. After 4 h of incubation, the cells were washed twice with PBS and returned complete media (100 µL). After 24–48 h incubation, 20 µL of CellTiter-Blue (Promega) was added to each well and the plates re-incubated for 2 h. The fluorescence was measure at excitation and emission wavelength of 560 and 590 nm, respectively.
Study of MNP uptake
To assess the ability of MNPs to transfer siRNA into cells, carboxyfluorescein (FAM)-labeled siRNA was formulated in MNPs at the N/P ratio of 10 as described above. B16F10 melanoma cells were seeded into 6-well plates (105 cells/well) and incubated 24 h prior transfection. Cells were treated with MNPs or free siRNA at a concentration of 250 nM. After incubation for 4 h, the cells were incubated for another 24 h, washed with PBS, and trypsinized. The uptake of FAM-labeled siRNA was detected with a Becton-Dickinson FACSort™ flow cytometer (Franklin Lakes, NJ) and data analyzed with CellQuest™ software (Becton-Dickinson). Additionally, the intracellular delivery and distribution of siRNA loaded in the MNPs was studied by fluorescent microscopy. B16F10 cells were seeded in a four-chamber well slide (104 cells/well) and allowed to attach overnight. Cells were then treated for 1, 4, and 24 h with free FAM-siRNA or loaded in MNPs at a concentration of 100 nM. At the end of the incubation period, the cells were washed with PBS, incubated with Hoechst 33342 and Lysotracker® Red for 15 min and 1 h, respectively, and visualized using a Nikon Eclipse E400 fluorescent microscope.
In vitro gene silencing
In vitro gene silencing experiments were performed in stably transfected c166-GFP cells using GFP-siRNA. A non-targeting control duplex (Negative-siRNA) was used as a non-specific control siRNA. Briefly, 24 h before transfection, cells were seeded in 12-well plates at a density of 5 × 104 per well. siRNA complexes with PLPEI or PEI 1.8 kDa polymers and MNPs were prepared at N/P of 10 as previously described. Complexes (50 µL) were added to cells to yield a final concentration of 100 nM. After 4 h of incubation, complexes were removed, fresh media was added, and the cells were further incubated for 48 h. Thereafter, cells were washed, trypsinized, and GFP downregulation was analyzed by flow cytometry.
Statistical analysis
Results are presented as mean±SD, and statistical significance of differences was evaluated by variance analysis (one-way ANOVA); p values smaller than 0.05 were considered to be significant, *p<0.05.
Results
Characterization of MNPs
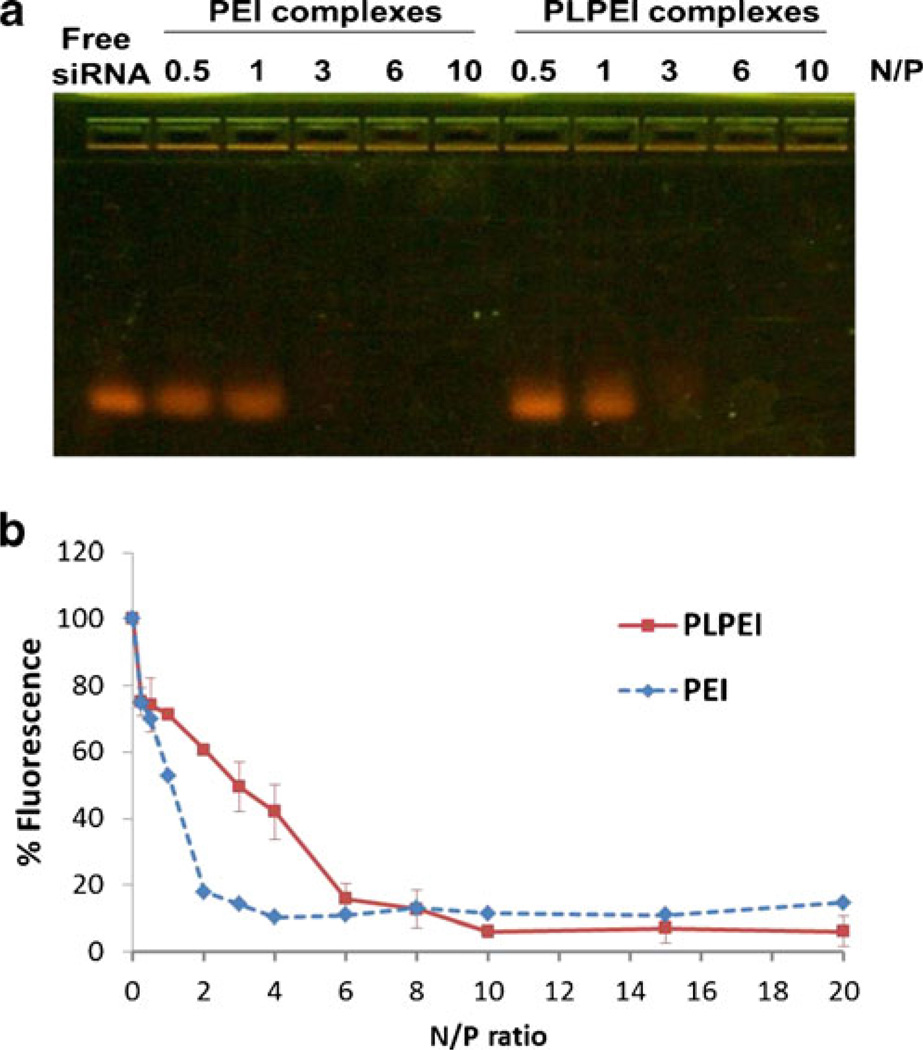
The condensation of siRNA within PLPEI complexes is a prerequisite for MNP preparation (Fig. 1). In this sense, the lipid-modified PEI is expected to possess enough positive charge to condense the siRNA and lead to formation of small particles that permit the addition of the PEG-lipid layer. Therefore, we evaluated the binding stability of PLPEI to siRNA by gel retardation and by a dye exclusion assay. PLPEI had a similar capacity to retain siRNA during gel electrophoresis compared to non-modified PEI. The N/P ratio of 3 resulted in an electrophoretic immobilization of siRNA for modified and unmodified PEIs (Fig. 2a). When the EtBr displacement assay was performed, further differences between modified and non-modified PEI were observed. As shown in Fig. 2b, as the N/P ratio of PEI complexes increased, the relative fluorescence decreased to a maximum binding degree at a N/P of 4. No differences in relative fluorescence were observed at higher ratios. The same behavior was observed in PLPEI complexes; however, the fluorescence plateau was reached at a N/P of 10. Thus, the N/P ratio of 10, when siRNA is totally condensed, was chosen for all further steps.
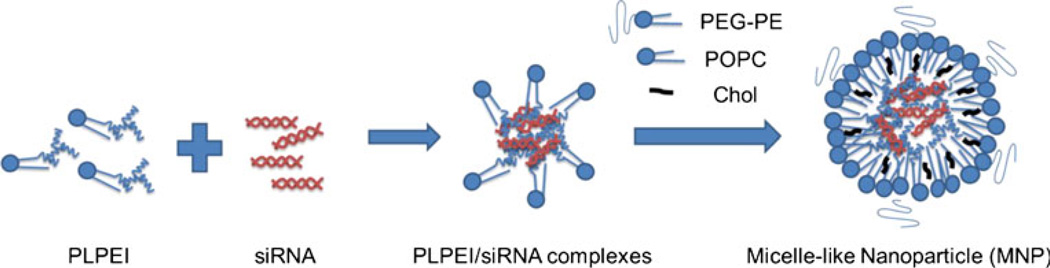
Fig. 1.
Schematic representation of the self-assembly process of micelle-like nanoparticles (MNPs)
Fig. 2.
PLPEI/siRNA complex formation. a. Gel retardation and b. relative binding affinity measured by ethidium bromide displacement assay of PLPEI and PEI complexes with siRNA at varying N/P ratios
For the preparation of MNPs, a mixture of free lipids comprising POPC, cholesterol, PEG-PE (3:3:0.3 mol/mol) was hydrated with a PLPEI complex solution (N/P 10) and kept for 1 h at RT. The hydrophobic interactions between the lipid part of PLPEI and the free lipids led to the formation of MNPs with small sizes and a neutral zeta potential. The incorporation of lipids slightly increased the size of PLPEI/siRNA complexes from 167±86 to 213±71 nm in MNPs, while the zeta potential decreased dramatically from 31±2 mV to a neutral surface charge (0±3 mV).
Nuclease stability of siRNA in MNPs
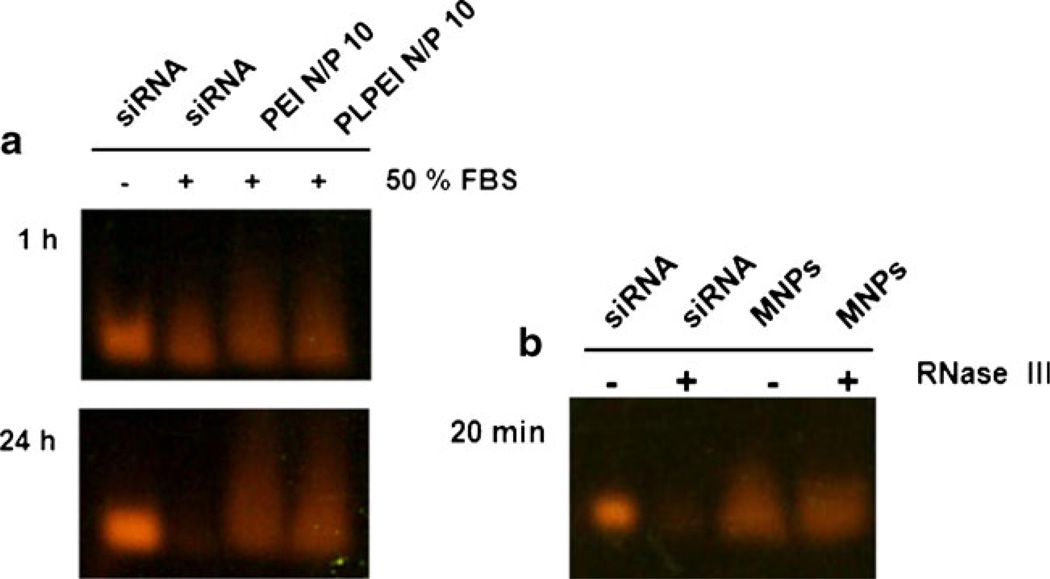
To investigate whether the MNPs can protect siRNA against nuclease digestion, we first evaluated the stability of siRNA within the PLPEI complexes. Naked siRNA and PLPEI/siRNA complexes were incubated in the presence of 50% FBS for 1 and 24 h. Figure 3a shows that the naked siRNA was completely degraded after 24 h of incubation with serum, while degradation of siRNA was prevented in PLPEI complexes at a N/P ratio of 10. Similarly, the nuclease stability of MNPs was demonstrated in the presence of RNase III. As shown in Fig. 3b, after 20 min of incubation with RNase III, naked siRNA was totally digested, and no siRNA band was detected in the agarose gel. On the contrary, siRNA bands with the intensity similar to controls were detected for MNPs incubated in the presence or in the absence of enzyme, meaning that siRNA formulated in MNPs is totally protected from enzymatic degradation.
Fig. 3.
Protection of a. siRNA within PLPEI complexes and b. MNPs against serum and RNAse III degradation
Cytotoxicity of MNPs
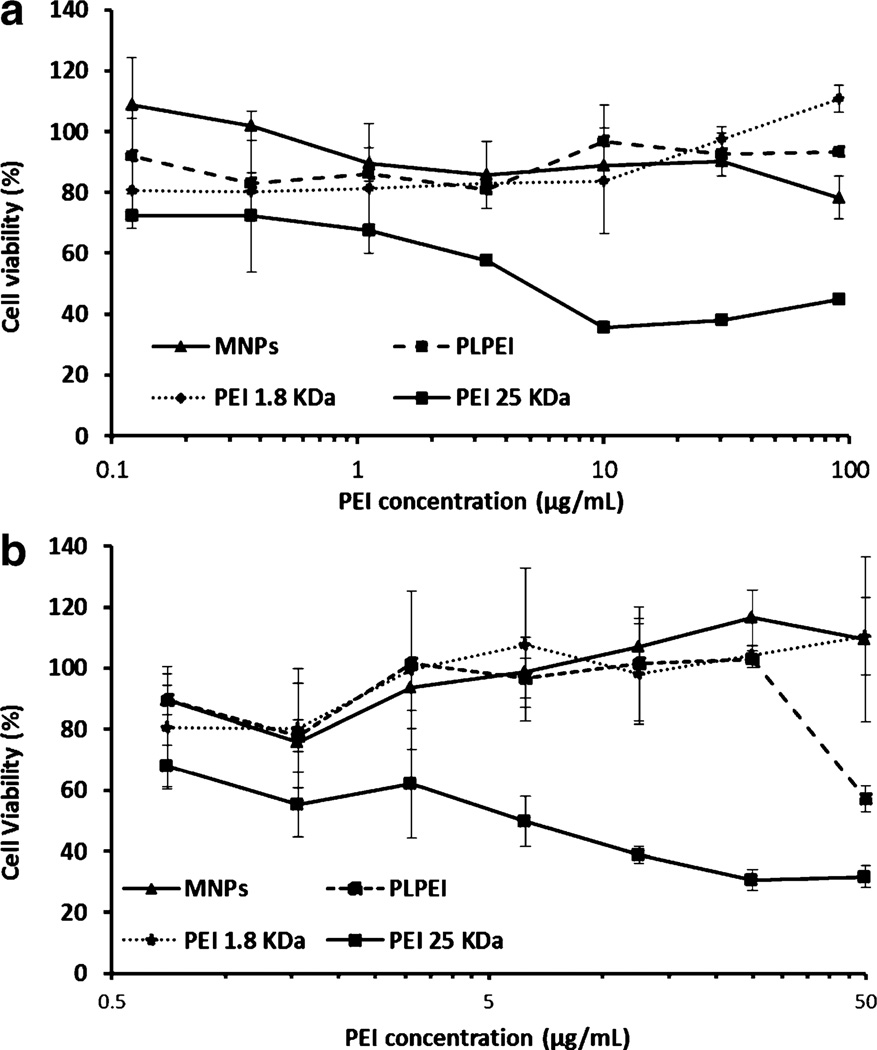
MNPs showed no toxicity towards NIH/3T3 and B16F10 cells over the concentration range of 1 to 7 µg/mL used in our in vitro experiments (Fig. 4). Moreover, the viability of cells was higher than 80% even at high concentrations (up to 50–100 µg/mL). PLPEI complexes showed a similar profile when compared to that of MNPs, although an increase in toxicity was detected at the highest concentration for B16F10 cells. In the case of non-modified PEI, PEI 1.8 kDa complexes were non-toxic over the concentration range tested, in sharp contrast to those formulated with PEI 25 kDa (positive control) that were highly toxic at a concentration of 15 µg/mL.
Fig. 4.
Cytotoxicity of MNPs, PLPEI complexes, PEI 1.8 kDa complexes, and PEI 25 kDa complexes towards a. NIH/3 T3 cells and b. B16F10 cells at different PEI concentrations. Relative cell viability was expressed as a percentage of control cells
Cellular uptake of MNPs
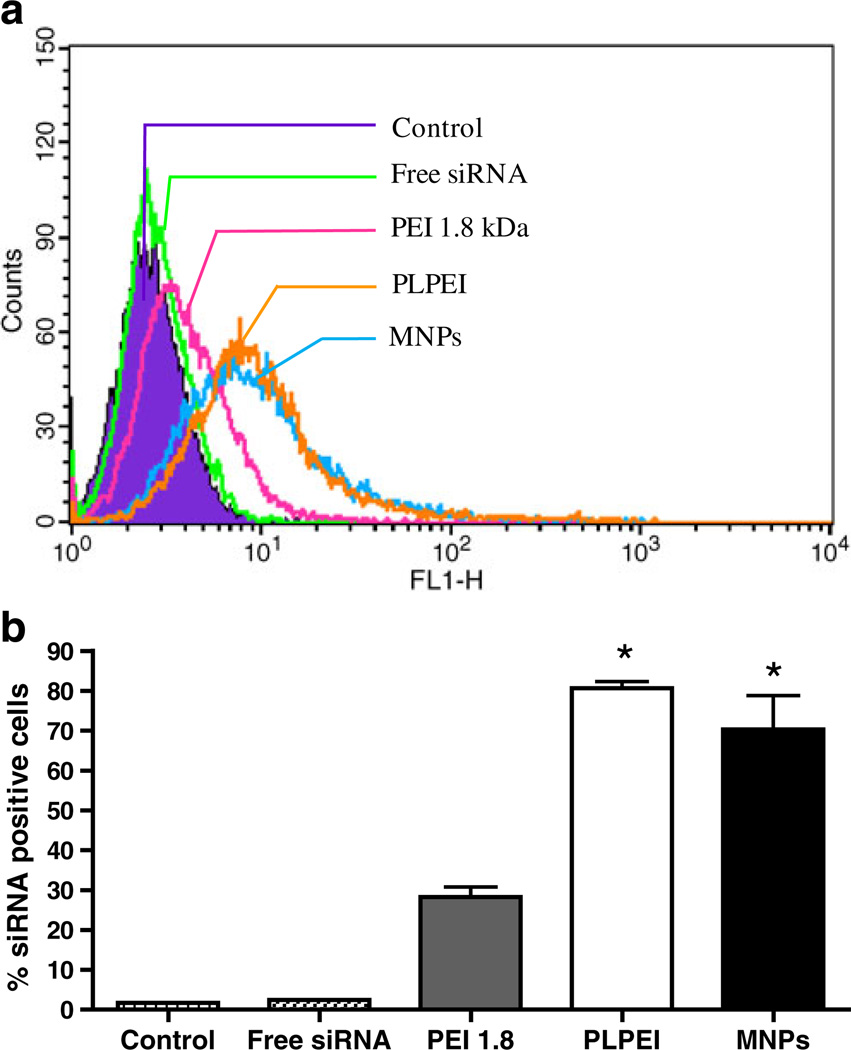
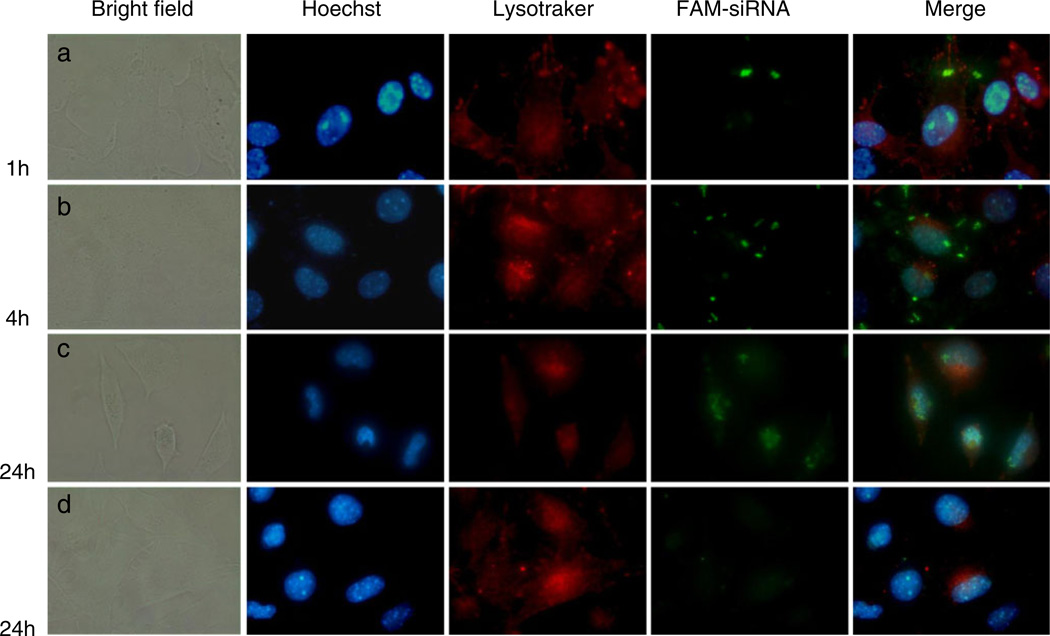
The cellular uptake of MNPs, PLPEI complexes, and PEI complexes was studied in B16F10 cells with fluorescent-labeled siRNA (FAM-siRNA). Cells were treated for 4 h with the fluorescently labeled formulations and analyzed by FACS after 24 h. The flow cytometric histogram of the different formulations is shown in Fig. 5a. All formulations generated a significant increase in the mean fluorescence of cells as compared with non-treated cells or those treated with free siRNA. As shown in Fig. 5b, more than 70% of the cells were transformed after MNP treatment. PLPEI complexes showed a 3-fold higher uptake levels than PEI complexes, whereas the increase for MNPs was 2.5-fold higher. The influence of different incubation times on the MNPs uptake is shown in Fig. 6. After 1 h of incubation of MNPs with cells, green fluorescence of siRNA was detected on the surface of the cells. After 4 h, the internalized siRNA was located in the cytoplasm. When B16F10 cells were incubated with MNPs for 1 day, the siRNA was detected in the nuclei of cells. At the same time point (24 h), naked siRNA gave almost no detectable fluorescence in cells (Fig. 6d). It is important to note that nuclear localization of the fluorescence is not due to the translocation properties of the carrier but to the siRNA itself. The FAM-siRNA used in this experiment, siGLO transfection indicator (Dharmacon, Lafayette, CO) is modified to localize finally in the nucleus after 24 h as an unmistakable signal of uptake and transfection.
Fig. 5.
Cellular uptake of FAM-labeled siRNA in various complexes and MNPs. a. Changes in FACS histograms indicative of siRNA-positive cells after 24 h of incubation following 4 h of treatment with different formulations. b. Bars represent quantitative analysis of FACS histograms in (a) to obtain the percentage of cells positive for FAM-siRNA. Data are expressed as the mean±SD (n=3; *p<0.05 vs free siRNA and PEI 1.8 complexes)
Fig. 6.
(Color online) Intracellular trafficking of a–c MNPs and d free FAM-siRNA after different incubation times with B16 cells. The nuclei (blue) were stained with Hoechst dye. The internalized FAM-siRNA appears green and the cytoplasmic boundaries (red) were marked with Lysotracker Red
Green fluorescence protein downregulation
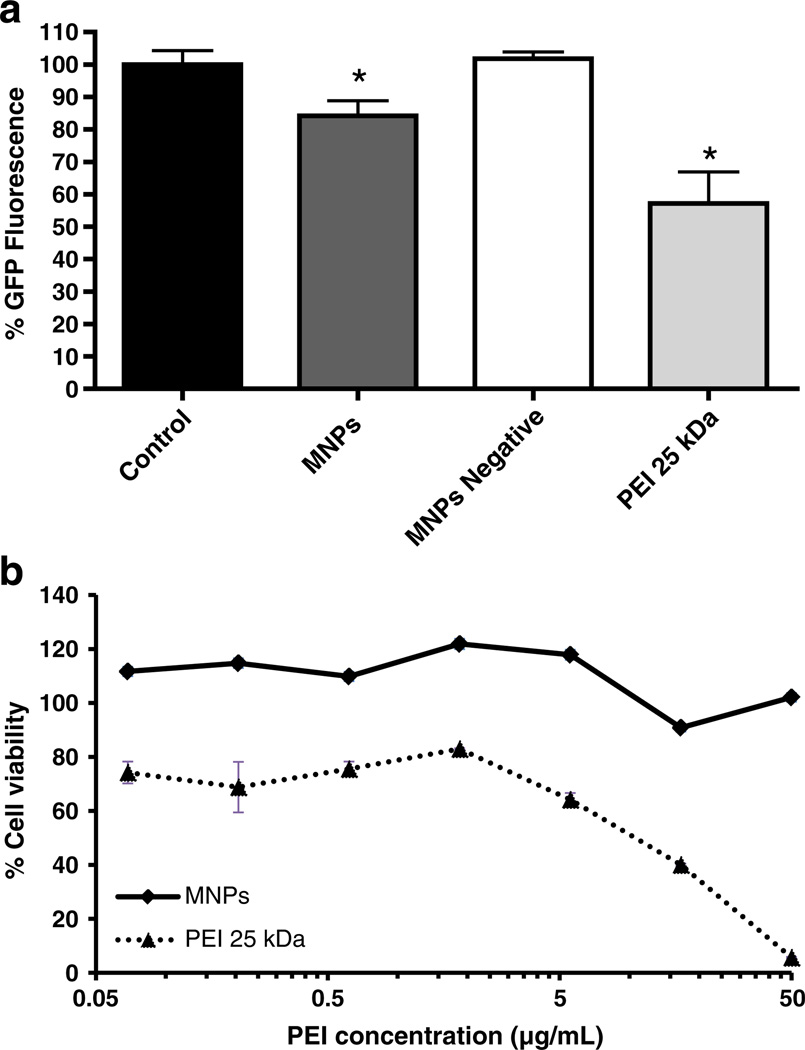
The knockdown efficacy of MNPs was assessed in stably GFP-expressing c166 cells (c166 GFP). The silencing of GFP was measured by the decrease in the mean fluorescence of cells after treatment with MNPs loaded with GFP-siRNA. As shown in Fig. 7a, cells treated with MNPs produced an almost 20% reduction of GFP fluorescence (p<0.05). This fluorescence shift was not observed when cells were treated with Negative-siRNA loaded MNPs, meaning that the decrease in the fluorescence of cells was due to the GFP knockdown and not due to the toxicity of the formulation. The absence of MNPs toxicity was also confirmed by cytotoxicity studies in GFP-c166 that were performed with the same conditions as the silencing experiments (Fig. 7b). PEI 25 kDa complexes decreased GFP fluorescence more than MNPs but also showed significantly higher toxicity to the cells.
Fig. 7.
a. siRNA-mediated downregulation of the target gene and b. toxicity of MNPs. Experiments were performed in stably transfected c166-GFP cells using GFP-siRNA. Cells were treated with MNPs for 4 h, and the GFP fluorescence was analyzed by FACS after 48 h incubation. A non-targeting control duplex (Negative-siRNA) was used as a non-specific control siRNA. PEI 25 kDa was used as positive control. Data are expressed as the mean±SD (n=3; *p<0.05 vs non-treated control cells). Cytotoxicity studies were performed under the same conditions as FACS experiments. Relative cell viability was expressed as a percentage of untreated control cells
Discussion
We previously reported a new gene delivery system consisting of micelle-like nanoparticles, MNPs, prepared by condensing plasmid DNA with lipid-modified PEI (PLPEI) and enveloping the new complexes with a PEG/lipid layer (Ko et al. 2009b). These MNPs protected the loaded DNA from enzymatic degradation, showed a reduced cytotoxicity, and demonstrated improved in vivo stability as compared to plain PEI complexes. As in the case of DNA, the in vivo application of siRNA is hampered by its rapid degradation in the plasma and fast renal clearance and inefficient uptake by tissue cells. Taking all this into account, we assumed that siRNA technology could take advantage of the good performance of MNPs as gene carriers. In this study, we prepared MNPs loaded with siRNA and characterized them for their biophysical properties, cytotoxicity, cellular uptake, and in vitro knockdown efficacy.
The first step for the preparation of MNPs is the condensation of siRNA with the lipid grafted PEI. The lipid-PEI conjugates (PLPEI) were prepared by coupling low molecular weight PEI (1.8 kDa) with the tail modified PCAz phospholipid. PEI 1.8 kDa is known to be less toxic than its high molecular weight counterparts (PEI 25 kDa, 800 kDa) due to the smaller number of primary amines present on its surface but also is less efficient in the transfection of nucleic acids to the cells (Godbey et al. 1999; Kunath et al. 2003). In addition, the incorporation of the lipid to the backbone of the polymer decreases the number of available positive groups that can interact with the siRNA. Therefore, it was important to optimize the amount of PLPEI needed for the complete binding and protection of siRNA. An N/P ratio of 10 was sufficient for the total condensation of the siRNA as small complexes (ca. 160 nm) and for its protection from serum degradation (Figs. 2 and 3a).
Lipid modification of PEI has been reported to improve the stability and uptake of the PEI complexes (Alshamsan et al. 2009; Gusachenko et al. 2009; Han et al. 2001). However, with MNPs, the lipid moiety of PLPEI plays the additional role of anchor that facilitates the envelopment of PLPEI/siRNA complexes with the lipids and PEG-PE without additional procedures (such as extrusion, sonication) usually required to prepare systems similar to MNPs (Rothdiener et al. 2010; Schafer et al. 2010). Just 1 h of incubation of PLPEI complexes with a mixture of lipids including POPC, cholesterol, and PEG-PE (3:3:0.3 mol/mol) was sufficient to permit the self-assembly of MNPs driven by the hydrophobic interactions between the exposed lipid moieties of the complexes and the free lipids. The optimal amounts of lipid and PEG-PE were chosen by taking into account our previous studies (Ko et al. 2009b). The finding of somewhat increased sizes for MNPs vs PLPEI complexes supports the notion that the lipids and PEG-lipid conjugates formed a shell around the complexes, which is also suggested by the zeta potential data. The positive surface of PLPEI complexes (31±2 mV) was decreased in MNPs to neutral (0±3 mV) because of the shielding effects of the PEG and lipids. In addition, the presence and protective effect of the MNP envelope was further demonstrated by the protection of the loaded siRNA from RNase III digestion that specifically cleaves double-stranded siRNA, a more accurate and stringent assay than the incubation in serum performed with PLPEI complexes (Fig. 3b).
Several studies carried out with plain, PEGylated, or liposomal-PEI formulations have revealed that, for a given carrier, DNA and siRNA transfection efficacy do not always correlate (Grayson et al. 2006; Malek et al. 2008; Schafer et al. 2010). Thus, the ability of MNPs to deliver siRNA into cells, already demonstrated for plasmid DNA, was evaluated in B16F10 cells with a FAM-labeled siRNA and compared with that of PEI 1.8 kDa and PLPEI (Fig. 5). On one hand, the lipid modification of PEI dramatically improves the uptake of siRNA with respect to non-modified PEI, most probably because the lipid residues provide a better interaction with the cellular membrane. Early studies demonstrated that the conjugation of cholesterol to PEI 1.8 kDa increased the DNA transfection levels compared to PEI 1.8 and 25 kDa (Han et al. 2001). The benefits of lipid-PEI conjugation were later confirmed for siRNA delivery (Alshamsan et al. 2009; Kim et al. 2007).
On the other hand, the MNPs had slightly lower cellular uptake than PLPEI complexes suggesting that the presence of PEG in MNPs may hinder the interactions with the cells (Malek et al. 2008). Still, our MNPs effectively delivered FAM-siRNA to B16F10 cells. More than 70% of the cells were siRNA-positive after the treatment with siRNA-loaded MNPs. The internalization of FAM-siRNA mediated by MNPs was confirmed by fluorescence microscopy (green dots in Fig. 6b). Once inside the cytosol, the loaded siRNA maintained its biological activity since MNPs downregulated GFP expression significantly compared with the control and with MNPs loaded with Negative-siRNA, respectively (Fig. 7a).Taken together, the uptake and silencing data indicate that the stability of MNPs is important for the final level of silencing and often an increase in the efficacy of delivery will be accompanied by an increased silencing. Studies with PEG-grafted PEI suggest that the intracellular complex stability, rather than its cellular uptake, is a major determinant of the complex bioactivity (Malek et al. 2008). In the case of MNPs, the presence of PEG and lipids does not influence their cellular uptake but provides additional stability to PLPEI complexes that might interfere with the siRNA release process in the cytoplasm. We are currently studying the effect of different phospholipids on the extracellular and intracellular stability of PLPEI complexes for improved silencing efficacy of MNPs
Finally, PEI 25 kDa, used as a positive control, showed a decrease in GFP fluorescence 1.5-fold greater than the MNPs but also had significantly higher toxicity (Fig. 7b). The lack of MNPs toxicity represents one of the major advantages over non-modified PEIs or PLPEI. MNPs were non-toxic regardless of the dose or the cell line employed for testing (Fig. 4 and 7b).
Conclusions
In this study, the suitability of MNPs for siRNA delivery and gene silencing was probed. Micelle-like nanoparticles based on a dense PLPEI/siRNA core enveloped by a lipid/PEG layer showed as good a capacity to complex and protect siRNA from serum degradation as that of PEI, but with improved biocompatibility properties including a neutral surface charge and an absence of in vitro cytotoxicity. These findings together with our previous in vivo data (long circulation time, absence of acute in vivo toxicity) suggest a promise for the application of MNPs for systemic delivery of siRNA.
Acknowledgments
This work was supported by a fellowship from the Department of Education of the Navarra Regional Government (Spain) to GN.
Contributor Information
Gemma Navarro, Center for Pharmaceutical Biotechnology and Nanomedicine, Northeastern University, 360 Huntington Ave, Boston, MA 02115, USA.
Rupa R. Sawant, Center for Pharmaceutical Biotechnology and Nanomedicine, Northeastern University, 360 Huntington Ave, Boston, MA 02115, USA
Sean Essex, Center for Pharmaceutical Biotechnology and Nanomedicine, Northeastern University, 360 Huntington Ave, Boston, MA 02115, USA.
Conchita Tros de ILarduya, Department of Pharmacy and Pharmaceutical Technology, School of Pharmacy, University of Navarra, 31080, Pamplona, Spain.
Vladimir P. Torchilin, Email: v.torchilin@neu.edu, Center for Pharmaceutical Biotechnology and Nanomedicine, Northeastern University, 360 Huntington Ave, Boston, MA 02115, USA.
References
- Alshamsan A, Haddadi A, Incani V, Samuel J, Lavasanifar A, Uludag H. Formulation and delivery of siRNA by oleic acid and stearic acid modified polyethylenimine. Mol Pharmaceutics. 2009;6:121–133. doi: 10.1021/mp8000815. [DOI] [PubMed] [Google Scholar]
- Boussif O, Lezoualc’h F, Zanta MA, et al. A versatile vector for gene and oligonucleotide transfer into cells in culture and in vivo: polyethylenimine. Proc Natl Acad Sci USA. 1995;92:7297–7301. doi: 10.1073/pnas.92.16.7297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chollet P, Favrot MC, Hurbin A, Coll JL. Side-effects of a systemic injection of linear polyethylenimine–DNA complexes. J Gene Med. 2002;4:84–91. doi: 10.1002/jgm.237. [DOI] [PubMed] [Google Scholar]
- Godbey WT, Wu KK, Mikos AG. Size matters: molecular weight affects the efficiency of poly(ethylenimine) as a gene delivery vehicle. J Biomed Mater Res. 1999;45:268–275. doi: 10.1002/(sici)1097-4636(19990605)45:3<268::aid-jbm15>3.0.co;2-q. [DOI] [PubMed] [Google Scholar]
- Grayson AC, Doody AM, Putnam D. Biophysical and structural characterization of polyethylenimine-mediated siRNA delivery in vitro. Pharm Res. 2006;23:1868–1876. doi: 10.1007/s11095-006-9009-2. [DOI] [PubMed] [Google Scholar]
- Gusachenko O, Kravchuk Y, Konevets D, Silnikov V, Vlassov VV, Zenkova MA. Transfection efficiency of 25-kDa PEI-cholesterol conjugates with different levels of modification. J Biomater Sci. 2009;20:1091–1110. doi: 10.1163/156856209X444448. [DOI] [PubMed] [Google Scholar]
- Han S, Mahato RI, Kim SW. Water-soluble lipopolymer for gene delivery. Bioconjug Chem. 2001;12:337–345. doi: 10.1021/bc000120w. [DOI] [PubMed] [Google Scholar]
- Jeong JH, Mok H, Oh YK, Park TG. siRNA conjugate delivery systems. Bioconjug Chem. 2009;20:5–14. doi: 10.1021/bc800278e. [DOI] [PubMed] [Google Scholar]
- Kim WJ, Kim SW. Efficient siRNA delivery with non-viral polymeric vehicles. Pharm Res. 2009;26:657–666. doi: 10.1007/s11095-008-9774-1. [DOI] [PubMed] [Google Scholar]
- Kim WJ, Chang CW, Lee M, Kim SW. Efficient siRNA delivery using water soluble lipopolymer for anti-angiogenic gene therapy. J Control Release. 2007;118:357–363. doi: 10.1016/j.jconrel.2006.12.026. [DOI] [PubMed] [Google Scholar]
- Kircheis R, Schuller S, Brunner S, et al. Polycation-based DNA complexes for tumor-targeted gene delivery in vivo. J Gene Med. 1999;1:111–120. doi: 10.1002/(SICI)1521-2254(199903/04)1:2<111::AID-JGM22>3.0.CO;2-Y. [DOI] [PubMed] [Google Scholar]
- Ko YT, Bhattacharya R, Bickel U. Liposome encapsulated polyethylenimine/ODN polyplexes for brain targeting. J Control Release. 2009a;133:230–237. doi: 10.1016/j.jconrel.2008.10.013. [DOI] [PubMed] [Google Scholar]
- Ko YT, Kale A, Hartner WC, Papahadjopoulos-Sternberg B, Torchilin VP. Self-assembling micelle-like nanoparticles based on phospholipid-polyethyleneimine conjugates for systemic gene delivery. J Control Release. 2009b;133:132–138. doi: 10.1016/j.jconrel.2008.09.079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunath K, von Harpe A, Fischer D, et al. Low-molecular-weight polyethylenimine as a non-viral vector for DNA delivery: comparison of physicochemical properties, transfection efficiency and in vivo distribution with high-molecular-weight polyethylenimine. J Control Release. 2003;89:113–125. doi: 10.1016/s0168-3659(03)00076-2. [DOI] [PubMed] [Google Scholar]
- Liu C, Zhao G, Liu J, et al. Novel biodegradable lipid nano complex for siRNA delivery significantly improving the chemosensitivity of human colon cancer stem cells to paclitaxel. J Control Release. 2009;140:277–283. doi: 10.1016/j.jconrel.2009.08.013. [DOI] [PubMed] [Google Scholar]
- Malek A, Czubayko F, Aigner A. PEG grafting of polyethylenimine (PEI) exerts different effects on DNA transfection and siRNA-induced gene targeting efficacy. J Drug Target. 2008;16:124–139. doi: 10.1080/10611860701849058. [DOI] [PubMed] [Google Scholar]
- Mao S, Neu M, Germershaus O, et al. Influence of polyethylene glycol chain length on the physicochemical and biological properties of poly(ethylene imine)-graft-poly(ethylene glycol) block copolymer/SiRNA polyplexes. Bioconjug Chem. 2006;17:1209–1218. doi: 10.1021/bc060129j. [DOI] [PubMed] [Google Scholar]
- Merdan T, Kunath K, Fischer D, Kopecek J, Kissel T. Intracellular processing of poly(ethylene imine)/ribozyme complexes can be observed in living cells by using confocal laser scanning microscopy and inhibitor experiments. Pharm Res. 2002;19:140–146. doi: 10.1023/a:1014212630566. [DOI] [PubMed] [Google Scholar]
- Musacchio T, Vaze O, D’Souza G, Torchilin VP. Effective stabilization and delivery of siRNA: reversible siRNA-phospholipid conjugate in nanosized mixed polymeric micelles. Bioconjug Chem. 2010;21:1530–1536. doi: 10.1021/bc100199c. [DOI] [PubMed] [Google Scholar]
- Nimesh S, Chandra R. Polyethylenimine nanoparticles as an efficient in vitro siRNA delivery system. Eur J Pharm Biopharm. 2009;73:43–49. doi: 10.1016/j.ejpb.2009.04.001. [DOI] [PubMed] [Google Scholar]
- Novina CD, Sharp PA. The RNAi revolution. Nature. 2004;430:161–164. doi: 10.1038/430161a. [DOI] [PubMed] [Google Scholar]
- Oh YK, Park TG. siRNA delivery systems for cancer treatment. Adv Drug Deliv Rev. 2009;61:850–862. doi: 10.1016/j.addr.2009.04.018. [DOI] [PubMed] [Google Scholar]
- Philipp A, Zhao X, Tarcha P, Wagner E, Zintchenko A. Hydrophobically modified oligoethylenimines as highly efficient transfection agents for siRNA delivery. Bioconjug Chem. 2009;20:2055–2061. doi: 10.1021/bc9001536. [DOI] [PubMed] [Google Scholar]
- Rothdiener M, Muller D, Castro PG, et al. Targeted delivery of SiRNA to CD33-positive tumor cells with liposomal carrier systems. J Control Release. 2010;144:251–258. doi: 10.1016/j.jconrel.2010.02.020. [DOI] [PubMed] [Google Scholar]
- Schafer J, Hobel S, Bakowsky U, Aigner A. Liposome–polyethylenimine complexes for enhanced DNA and siRNA delivery. Biomaterials. 2010;31:6892–6900. doi: 10.1016/j.biomaterials.2010.05.043. [DOI] [PubMed] [Google Scholar]
- Schroeder A, Levins CG, Cortez C, Langer R, Anderson DG. Lipid-based nanotherapeutics for siRNA delivery. J Intern Med. 2009;267:9–21. doi: 10.1111/j.1365-2796.2009.02189.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siomi H, Siomi MC. On the road to reading the RNA-interference code. Nature. 2009;457:396–404. doi: 10.1038/nature07754. [DOI] [PubMed] [Google Scholar]
- Urban-Klein B, Werth S, Abuharbeid S, Czubayko F, Aigner A. RNAi-mediated gene-targeting through systemic application of polyethylenimine (PEI)-complexed siRNA in vivo. Gene Ther. 2005;12:461–466. doi: 10.1038/sj.gt.3302425. [DOI] [PubMed] [Google Scholar]
- Whitehead KA, Langer R, Anderson DG. Knocking down barriers: advances in siRNA delivery. Nat Rev Drug Discov. 2009;8:129–138. doi: 10.1038/nrd2742. [DOI] [PMC free article] [PubMed] [Google Scholar]