Abstract
The folding of the extracellular serine protease, α-lytic protease (αLP; EC 3.4.21.12) reveals a novel mechanism for stability that appears to lead to a longer functional lifetime for the protease. For αLP, stability is based not on thermodynamics, but on kinetics. Whereas this has required the coevolution of a pro region to facilitate folding, the result has been the optimization of native-state properties independent of their consequences on thermodynamic stability. Structural and mutational data lead to a model for catalysis of folding in which the pro region binds to a conserved β-hairpin in the αLP C-terminal domain, stabilizing the folding transition state and the native state. The pro region is then proteolytically degraded, leaving the active αLP trapped in a metastable conformation. This metastability appears to be a consequence of pressure to evolve properties of the native state, including a large, highly cooperative barrier to unfolding, and extreme rigidity, that reduce susceptibility to proteolytic degradation. In a test of survival under highly proteolytic conditions, homologous mammalian proteases that have not evolved kinetic stability are much more rapidly degraded than αLP. Kinetic stability as a means to longevity is likely to be a mechanism conserved among the majority of extracellular bacterial pro-proteases and may emerge as a general strategy for intracellular eukaryotic proteases subject to harsh conditions as well.
Virtually all extracellular bacterial proteases are synthesized as precursor molecules with pro regions. In every case where the function of the pro region has been investigated, it has been found to be necessary for folding and secretion (1). One of the most striking and best studied examples of pro-mediated folding is the bacterial enzyme, α-lytic protease (αLP). αLP (EC 3.4.21.12) is a 198-aa serine protease secreted by the Gram-negative soil bacterium Lysobacter enzymogenes to degrade other soil microorganisms. The overall three-dimensional fold of αLP clearly places it in the same family as the mammalian digestive serine proteases chymotrypsin, trypsin, and elastase, despite only moderate sequence homology (2). In contrast to these mammalian homologues, whose small N-terminal zymogen peptides simply prevent premature activation, αLP is synthesized with a large 166-aa N-terminal pro region (Pro) that is required for proper folding of its mature protease domain (3). In vivo, coexpression of αLP and Pro, either in cis as the natural precursor molecule or in trans as two separate polypeptide chains results in the secretion of active αLP (4), whereas expression of αLP alone leads to accumulation of the protease in the outer membrane because of apparent misfolding.
In vitro energetic studies reveal a novel means of stability for the mature protease arising from kinetics. This not only distinguishes αLP from its mammalian homologues but provides compelling support for the possibility of metastable native conformations in general. Emerging structural and energetic details of pro-mediated folding may define a theme for the folding of a wide range of homologous extracellular proteases that also contain pro regions. In addition, features of αLP’s kinetic barrier may provide insight into other proteins with metastable conformations of biological importance. Here we describe the role of kinetic stability in αLP folding, details of pro⋅αLP interactions and a possible mechanism and an evolutionary rationale for pro-mediated folding of αLP.
Folding Under Kinetic Control.
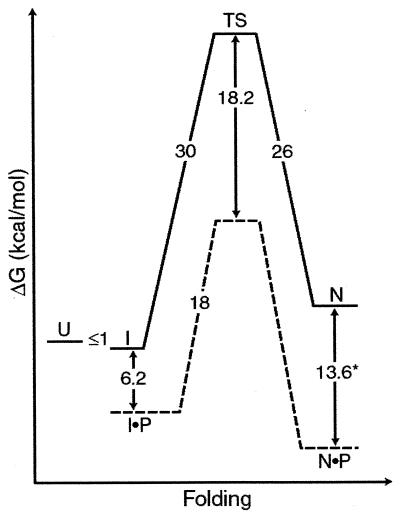
To understand the requirement of the pro region for folding, the folding free-energy landscape of αLP has been mapped in the absence of the pro region through refolding and unfolding experiments. In vitro refolding of chemically denatured αLP in the absence of Pro, by dilution from denaturant, results in an inactive molten globule-like intermediate (5). Designated the “I” state of the protease, this intermediate is monomeric and greatly expanded relative to the native state “N.” Spectroscopic data indicate that I possesses substantial secondary structure but lacks stable tertiary interactions. Urea-denaturation experiments show that I has <1 kcal/mol (1 cal = 4.18 J) stability over the unfolded state “U” (6). Under physiological conditions, the αLP I state remains stable for months without any appreciable conversion to mature enzyme. The very small fraction of I that does mature to the active N state can be measured by using a very sensitive enzymatic assay. From this assay, it has been determined that the I state refolds with an extremely slow initial rate of 1.18 × 10−11 s−1 at 4°C (t1/2 = 1,800 years), corresponding to a folding barrier height of 30 kcal/mol, as calculated by transition state theory (Fig. 1; ref. 6).
Figure 1.
Free-energy diagram of αLP folding with and without its pro region at 4°C. In the absence its pro region (P), unfolded αLP (U) spontaneously folds to a molten globule-like intermediate (I), which proceeds at an extremely slow rate to N through a high-energy folding TS. The addition of pro region provides a catalyzed folding pathway (denoted by dashed lines) that lowers the high folding barrier and results in a thermodynamically stable inhibition complex N·P. ∗ indicates measurement at 25°C. (Modified from ref. 6.)
This remarkably high barrier prevents the intermediate and native states from being in equilibrium with each other. Therefore, to determine the relative stability of these conformations, it is necessary to compare the ratio of the folding and unfolding rates instead of using the usual equilibrium approaches. To measure the unfolding rate on a reasonable time scale, the N state must be chemically or thermally denatured. To avoid complications of autolysis when studying the rates of αLP unfolding, the active-site serine has been mutated to alanine (S195 → A; chymotrypsin homology numbering from ref. 7). During unfolding, both secondary and tertiary structure (monitored via CD and tryptophan fluorescence, respectively) are lost simultaneously in a single rate-limiting step (6). Extrapolating the data to zero denaturant gives an unfolding rate of 1.8 × 10−8 s−1 at 4°C or an unfolding barrier of 26 kcal/mol (t1/2 = 1.2 years). The ratio of this unfolding rate to the slower folding rate results in an equilibrium free energy that favors the I state by 4 kcal/mol. Over a broad range of temperatures, the I state of αLP, not the N state, is at the minimum free energy. In fact, because of the marginal stability of I, the N state is actually significantly less thermodynamically stable than either the I or U states.
To surmount the high barrier to folding and the extraordinary thermodynamic instability of the native state, αLP has coevolved the pro region, which can assist the folding of αLP when supplied in cis or in trans. Addition of Pro to I results in rapid folding to the N state (0.037 s−1) and recovery of functional protease (Fig. 1; ref. 6). Pro acts as a foldase, facilitating αLP folding by binding tightly to the folding transition state of the protease, lowering the barrier by 18.2 kcal/mol. In this manner, Pro serves as a potent catalyst, increasing the rate of αLP folding by 3 × 109. In addition, Pro is the tightest binding inhibitor known for the native protease (Ki = 3 × 10−10 M; refs. 8 and 9), making Pro a single-turnover catalyst. This tight binding serves a critical function in αLP folding by shifting the thermodynamic equilibrium in favor of folded αLP (Pro⋅N is 3.4 kcal/mol more stable than Pro⋅I; Fig. 1).
The product of the folding reaction is not active αLP but the inhibitory complex. Release of active αLP requires that the Pro region be removed by proteolysis. Once Pro is degraded, the active protease becomes kinetically trapped in the metastable N state, with the high barrier preventing unfolding to the more thermodynamically favored unfolded states. In this way, pro-mediated folding provides the only efficient means of folding αLP to its metastable native conformation.
Structures of Pro and Pro⋅αLP Complex.
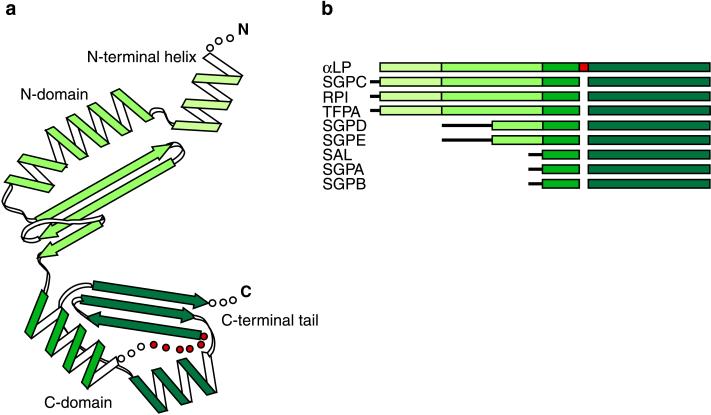
Recently determined crystal structures of Pro and the Pro⋅N complex illuminate Pro⋅αLP interactions (10). Alone, Pro adopts a novel C shaped α/β-fold, consisting of an N-terminal helix, two compact globular domains (N domain, C domain) connected by a nearly rigid hinge region, and a C-terminal tail (Fig. 2a). Each globular domain contributes a three-stranded β-sheet to the concave surface of the molecule and at least one α-helix that packs against these β-sheets to form the convex surface. The N-terminal helix appears highly flexible, changing orientations in different crystal environments. Two of the three Pro molecules in the crystallographic asymmetric unit show different conformations for the N-terminal helix, whereas the third molecule reveals the helix to be disordered. Similarly, the C-terminal tail is unseen in the Pro structure and apparently disordered in unbound Pro.
Figure 2.
(a) Topology of Pro as described in the text. A disordered loop in the Pro C domain is shown in red. (b) Schematic of primary sequence alignments of pro regions from nine bacterial serine proteases. Alignments were determined by using the αLP Pro structure as a guide. Regions of sequence homology correspond to specific secondary structures in the Pro structure, with the Pro C-terminal domain being the most conserved region. N-terminal sequences lacking homology are depicted by thin black lines. αLP, Lysobacter enzymogenes αLP (17); SGPC, Streptomyces griseus protease C (18); RPI, Rarobacter faecitabitus protease I (19); SGPD, S. griseus protease D (20); SGPE, S. griseus protease E (21); TFPA, Themomonaspora fusca serine protease (22); SAL, Streptomyces lividans protease (23); SGPA, S. griseus protease A (24); SGPB, S. griseus protease B (24).
Sequence comparisons with homologous pro-proteases suggest that the Pro structure may be a common pro region fold. Primary sequence alignments of Pro and eight related pro regions (Fig. 2b) indicate that these homologous pro regions share common secondary-structure elements, the most conserved region being that of the Pro C-terminal domain, despite a wide range of pro region sizes. These pro regions appear compatible with the Pro structure and presumably exhibit similar mechanisms of foldase activity.
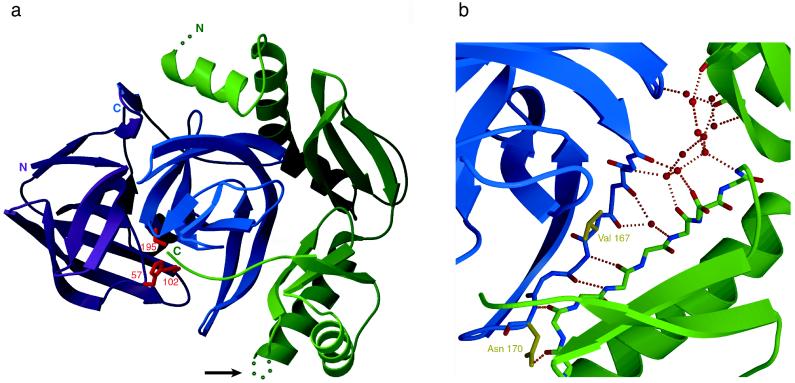
In the case of αLP, Pro⋅N complex formation does not significantly alter the Pro structure (Fig. 3a). This is surprising because the pro region by itself has quite limited stability (Tm = 28.5°, 2.3 kcal/mol; ref. 11), whereas Pro⋅N complex is greatly stabilized (13.6 kcal/mol; ref. 6). However, the only notable differences are seen in the structuring of the C-terminal tail and the positioning of the flexible N-terminal helix on protease binding. As expected for a tight-binding inhibitory complex, the Pro⋅N complex structure buries a very large surface (>4,000 Å2) in its intermolecular interface.
Figure 3.
(a) Ribbon diagram of the Pro⋅N complex structure. The αLP N and C domains are colored magenta and blue respectively, with the side chains of the catalytic triad shown in red (His-57, Asp-102 and Ser-195; chymotrypsin numbering). Illustrated in green, bound Pro inserts its C-terminal tail into the protease active site. A disordered loop in the Pro C-terminal domain, indicated by an arrow, presents a likely secondary protease cleavage site, leading to the release of active αLP from the inhibitory complex. (b) Detail of the hydrated Pro⋅N interface. A gap between Pro (green) and the αLP C domain (blue) is filled by ordered water molecules which are shown as red spheres. Some of these waters mediate hydrogen bonds (dashed orange lines) between the αLP β-hairpin and the Pro three-stranded β-sheet that form the shared five-stranded β-sheet of the Pro⋅N interface. Residues in the αLP β-hairpin that affect formation of the initial Pro⋅I Michaelis complex (Ile-167 and Asn-170) are displayed in yellow. Figures are modified from figures 2b and 3b of ref. 10.
The most striking feature of the complex structure is the fact that Pro binds almost exclusively to the αLP C domain, effectively surrounding the αLP C-terminal β-barrel. This observation raises the distinct possibility that it is the αLP C domain that cannot fold properly and is therefore the focused substrate of Pro foldase activity. In support of this, recent mutagenesis studies indicate that the structuring of the protease C domain is an integral part of the high folding barrier. Screens of libraries of chemically mutagenized αLP reveal that mutations that lower the folding transition state (as much as 3 kcal/mol) all map to the C domain of the protease (A. Derman and D.A.A., unpublished data). The most extensive and complementary interactions in the Pro⋅N interface occur between the protease and the Pro C-terminal domain. In particular, the three-stranded β-sheet in the Pro C domain pairs with an extended β-hairpin in the αLP C domain (αLP residues 166–179; chymotrypsin numbering) to form a continuous five-stranded β-sheet. Additional interactions come from the insertion of the Pro C-terminal tail into the protease active site. The Pro C tail binds in a substrate-like manner to directly occlude the protease active site, as predicted by biochemical data (25). Placement of the C tail also provides a binding pocket for the tip of the β-hairpin.
αLP Folding Barrier and Pro Foldase Mechanism.
The integration of prominent features of the complex structure with mutagenesis studies on both Pro and αLP provides significant insights into the origin of the folding barrier and the mechanism of Pro-catalyzed folding. Because Pro acts as a folding catalyst, it is possible to use modified Michaelis–Menten kinetics to extract functional information about the folding reaction (8). This analysis provides information on the formation of the Pro⋅I Michaelis complex (Km) and the stabilization of the folding transition state (kcat). In addition, the stability of the Pro⋅N complex can be assessed by measuring the inhibition of peptide substrate hydrolysis by Pro (Ki).
Mutations within the αLP β-hairpin loop alter both Km and kcat (8). The Km effects reveal that formation of the shared β-sheet must occur in the first step of Pro-catalyzed folding, whereas the kcat effects indicate that this extended sheet continues to play a role during folding catalysis. Unlike these hairpin mutations, removing residues from the Pro C tail (8) does not affect initial binding to the αLP I state (Km) and only marginally affects αLP N state binding (Ki), despite the Pro C tail’s high complementarity to the αLP-binding pocket. In marked contrast, these same Pro C tail truncations drastically reduce the folding rate (kcat), profoundly hindering the ability of Pro to stabilize the folding transition state (TS). Deletion of the last three residues from the Pro C tail decreases kcat by ≈300-fold, and removal of an additional fourth residue decreases folding by at least a factor of 107. The Pro C tail therefore plays a direct role in Pro foldase activity, preferentially stabilizing the folding TS over the I and N states. Preliminary data indicate that the Pro N domain also contributes to the catalytic activity of Pro. Mutations in Pro at the protease–Pro N domain interface affect TS stabilization (E.L.C., P. Chien, and D.A.A., unpublished data).
Kinetic and thermodynamic analyses suggest that the folding transition state and the native state must share many structural features. From the denaturant dependence of unfolding rates (S.S.J. and D.A.A., unpublished data), it is possible to infer that the folding transition state is significantly closer to the native state than to the folding intermediate, I. Furthermore, the extremely tight binding of the rigid Pro region to both the native state (13.6 kcal/mol) and the transition state (18.2 kcal/mol) suggests that at least the αLP C domain must be similarly structured in both states. However, they cannot be identical. Although the Pro C-terminal tail makes ideal substrate-like interactions with αLP, deletions only minimally affect the stability of Pro⋅N, while causing profound effects on the folding transition state (8). This suggests that the native Pro⋅N complex must be “strained” such that the total binding energy possible for the Pro C tail is not realized in the Pro⋅N complex. By contrast, the intrinsic binding energy of the Pro C tail does seem to be fully realized when complexed to the folding transition state, because it is stabilized by an additional 5 kcal/mol compared with Pro⋅N (Fig. 1).
Observations based on the Pro⋅N complex structure (10) suggest that this strain may be the result of poor complementarity in regions of the Pro⋅N interface, which could be improved to yield the observed additional stabilization in the Pro⋅TS complex. Most notably, there is a significant gap in the interface where the protease meets the junction of the two Pro domains. This gap contains eight ordered solvent molecules, three of which act to mediate hydrogen bonds between the αLP β-hairpin and Pro β-strand. Such highly solvated interfaces have been previously observed where two surfaces interact in two different conformational states. These “adapter” waters are seen in protein–DNA complexes(12) where waters populate the interface in nonspecific complexes yet are excluded in the specific complex. Similarly, waters are often used to adapt quaternary changes in allosteric enzymes (13, 14), with fewer waters in the higher-affinity state because of improved surface complementarity. The Pro⋅αLP TS may be similarly stabilized by excluding the bound waters, thereby reducing the entropic cost of ordering the waters and increasing the direct Pro⋅αLP interface. Because the structure of free αLP is nearly identical to that of αLP complexed with Pro, it is probable that strong αLP N state interactions prevent optimization of the Pro⋅αLP interface predicted in the TS complex. Destabilizing αLP mutations may disrupt these interactions enough to distort the Pro⋅αLP complex toward more TS-like binding.
αLP Folding Model.
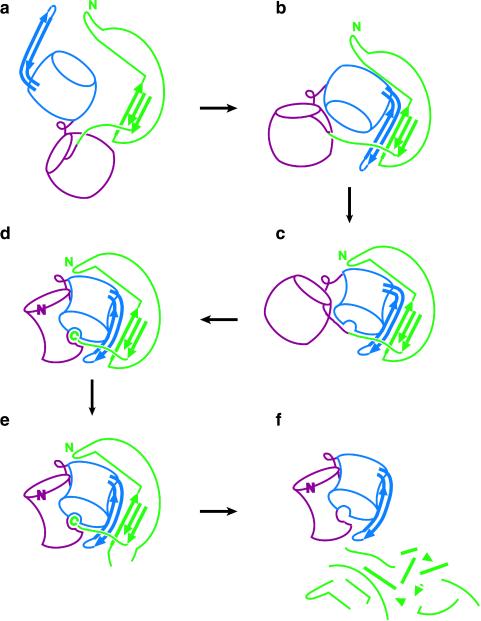
This structural and mutagenesis data can be synthesized into a model of Pro-catalyzed folding of αLP (Fig. 4; ref. 10). In this folding scheme, the N and C domains of the expanded molten globule folding intermediate are separated, and the β-hairpin is exposed to solvent. Prefolded Pro initiates protease folding by binding to the hairpin, forming a continuous five-stranded β-sheet. Efficient folding requires the Pro C tail to then bind to the nascent active site, positioning the hairpin and thereby assisting the structuring of the αLP C domain. Finally, the αLP N domain docks and folds against the C domain to complete both the catalytic triad and the packing of the N state core. Studies of the intact Pro-αLP precursor (11) support the proposed two-step folding model. Precursor refolding experiments show biphasic kinetics, with an initial fast rate equal to the rate of pro folding alone, followed by a slower rate for pro-mediated folding of αLP.
Figure 4.
Proposed model of Pro-catalyzed folding of αLP. (a) The pro domain of the Pro–αLP precursor folds, while the protease N and C domains remain separated and expanded. (b) The three-stranded β-sheet of the Pro C domain pairs with the solvent-exposed β-hairpin of the αLP C domain forming a continuous five-stranded β-sheet. (c) Substrate-like binding of the Pro–αLP junction to the nascent active site positions the β-hairpin and leads to the structuring of the αLP C domain. (d) The αLP N domain folds on docking with the αLP C domain to complete the protease active site, which can then process the Pro–αLP junction. The Pro C-terminal tail remains bound to the active site in this inhibitory complex while the new αLP N terminus repositions to its native conformation. (e) Intermolecular cleavage of secondary cleavage sites by αLP or other exogenous proteases leads to the f, eventual degradation of Pro and release of active, mature αLP. Color scheme as in Fig. 3a. Figure is modified from figure 4 of ref. 10.
During in cis folding, formation of the active site allows the protease domain of the precursor to process the Pro–αLP junction, producing the two distinct polypeptides chains of the Pro⋅N complex. After cleavage, the Pro C tail remains bound to the active site while the newly formed protease N terminus repositions to its native conformation 24 Å away. Although the Pro–αLP precursor and Pro⋅N complex show similarities in secondary and tertiary structure, the marginal stability of the precursor (2.2 kcal/mol; ref. 11) compared with the complex (10.6 kcal/mol) suggests that the rearrangement of the N terminus is critical to αLP N state stabilization. In addition to the primary intramolecular cleavage site, αLP also recognizes intermolecular cleavage sites within Pro, eventually leading to Pro degradation and release of active, mature protease. The disordered loop within the Pro C domain (Figs. 3 and 4E) presents a likely target for the requisite secondary cleavage event. This secondary cleavage site is sensitive to many other proteases besides αLP. In fact, there may be a functional synergism between the multiple proteases secreted simultaneously by the host, Lysobacter enzymogenes, in cleaving each other’s pro regions.
In the proposed folding scenario, Pro must bind to and correctly position the β-hairpin. The likely importance of this β-hairpin to the αLP folding barrier is reflected in its selective conservation among related proteases within the chymotrypsin superfamily. The β-hairpin, a common structural motif found in all 13 bacterial homologues synthesized with pro regions, is noticeably absent in other related bacterial, viral, and mammalian proteases that do not require pro regions for proper folding. Furthermore, the Pro β-strand that pairs with the hairpin loop is also highly conserved in homologous pro regions, suggesting that the hairpin and its interaction with the Pro C domain are important in structuring the protease. This observation is consistent with the fact that smaller related pro regions show sequence homology only to the Pro C domain, thereby maintaining the core structure necessary for binding the β-hairpin of the protease (Fig. 2b). The positioning of the hairpin and subsequent structuring of the αLP C domain may be a general mechanism for pro region-mediated folding of β-structures. In contrast, subtilisin, a pro-protease evolutionarily unrelated to αLP, seems to use a different method of pro-catalyzed folding. The subtilisin pro domain stabilizes a pair of α-helices in the protease instead of a β-hairpin (15). Although αLP and subtilisin have convergently evolved pro-dependent folding, they differ in both their mature protease structures and the method by which their respective pro regions achieve their active protease conformations.
Physical Origins of the Folding Barrier.
Although the physical origins of the αLP folding barrier and the means by which Pro lowers this barrier remain to be determined, examination of the enthalpic and entropic contributions to the free-energy difference between the αLP I and N states provides some insights into the nature of the folding barrier. Despite the thermodynamic instability of the dLP N state, titration calorimetry experiments reveal that it is enthalpically favored over the I state by 18 kcal/mol (6). Thus, the thermodynamic stability of the I state must be entropic in origin. This means that either the I and U states are more entropically favored than in “normal” proteins, the αLP N state has lower entropy than normal, or both. One possible source of this excess entropy for I may be the high percentage of glycines found in the αLP sequence. Because glycine residues lack a side chain, they can avoid steric clashes encountered by other amino acids, thereby increasing the number of accessible conformations in the unfolded states. αLP contains 16% glycines compared with only 9% in the homologous but thermodynamically stable chymotrypsin. The 10 additional glycines found in αLP, as compared with chymotrypsin, are predicted to contribute an extra ≈7 kcal/mol of configurational entropy (16) to the unfolded state at 4°C. Removing this additional entropy would be sufficient to alter the direction of the I and N equilibrium, placing the N state at the global free-energy minimum.
The excess unfolding entropy may also be due in part to the extremely low conformational entropy of the αLP native structure. Although native states are often dynamic, αLP adopts a remarkably rigid native structure characterized by insensitivity to proteolysis, unusually low crystallographic B factors, and hydrogen-exchange protection factors on the order of >1010 for ≈40 core amides (J. Davis, J. L. Sohl, and D.A.A., unpublished data). Protection factors of this magnitude have never been observed in any other protein. Contributing to this rigidity, loops in αLP are generally shorter and therefore likely to be less flexible than those found in chymotrypsin. Many of αLP’s extra glycine residues facilitate the tight turns found in these condensed loops. With their ability to assume unusual backbone geometries, the glycines may enable tighter and more cooperative packing within the protein core. In this manner, the high glycine content can reduce the entropy of the N state while increasing the configurational entropy of the I state.
Glycine content appears to be a common feature distinguishing homologous proteases to αLP that have pro regions from those that do not (Table 1). The Streptomyces griseus proteases, along with several other pro region-containing homologues, have 16–20% glycines, whereas the mammalian digestive enzymes and other members of the trypsin serine protease family without pro regions have 6–12% glycines.
Table 1.
Glycine content of αLP relatives
| Protease | Glycines, no. | Residues, no. | Glycines, % |
|---|---|---|---|
| αLP family | |||
| L. enzymogenes αLP | 32 | 198 | 16.2 |
| R. faecitabitus protease I | 29 | 174 | 16.7 |
| S. albogriseolus protease 20 | 32 | 172 | 18.6 |
| S. fradiae protease 1 | 31 | 186 | 16.7 |
| S. griseus protease A | 32 | 182 | 17.6 |
| S. griseus protease B | 32 | 185 | 17.3 |
| S. griseus protease C | 35 | 190 | 17.9 |
| S. griseus protease D | 32 | 188 | 17.0 |
| S. griseus protease E | 32 | 183 | 17.5 |
| S. lividans protease | 35 | 171 | 20.5 |
| S. livdans protease O | 27 | 150 | 18.0 |
| T. fusca serine protease | 35 | 186 | 18.8 |
| Trypsin family | |||
| Trypsin | 24 | 224 | 10.7 |
| Chymotrypsin B | 23 | 245 | 9.4 |
| Elastase | 27 | 270 | 10.0 |
| Acrosin | 27 | 436 | 6.2 |
| Achelase 1 protease | 25 | 213 | 11.7 |
| α-Tryptase | 19 | 245 | 7.8 |
| Batroxobin | 29 | 231 | 8.7 |
| Carboxypeptidase A complex III | 25 | 240 | 10.4 |
| Coagulation factor vii | 37 | 406 | 9.1 |
| Collagenase | 22 | 230 | 9.6 |
| Complement factor B | 61 | 739 | 8.3 |
| Enteropeptidase | 77 | 1035 | 7.4 |
| Glandular kallikrein 1 | 19 | 238 | 8.0 |
| Granzyme A | 22 | 234 | 9.4 |
| Mast cell protease 7 | 22 | 244 | 9.0 |
| Natural killer cell protease 1 | 19 | 228 | 8.3 |
| Plasminogen | 59 | 790 | 7.5 |
| Prostasin precursor | 28 | 311 | 9.0 |
| Serine protease hepsin | 44 | 417 | 10.6 |
Evolution of Longevity Through Kinetic Stability.
The correlation between high glycine content and the presence of a conserved β-hairpin in the protease and the coevolution of a pro region suggests that the rigid native state and large kinetic barrier found in αLP may be conserved in other extracellular bacterial proteases. These shared properties may reflect their common function as proteases that break down microorganisms in the extracellular environment, supplying nutrients for their bacterial hosts. The utility of these proteases is compromised by their tendency to degrade themselves as well as other proteins. As such, it is presumably desirable to the host to evolve proteases that can survive as long as possible under these harsh, degradatory conditions.
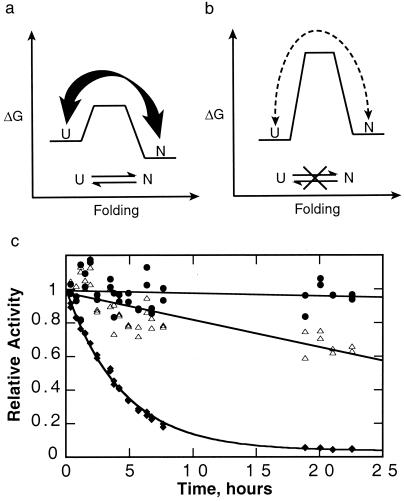
A typical protein stabilized thermodynamically without a large barrier preventing unfolding would constantly sample partially and fully unfolded states, leading to rapid destruction by exogenous proteases (Fig. 5a). By contrast, kinetic stability provides a mechanism to increase the cooperativity and raise the barrier to unfolding (Fig. 5b), thereby suppressing breathing motions and global unfolding. The result is a drastic reduction in susceptibility to proteolytic degradation.
Figure 5.
Advantages of kinetic stability. (a) A typical thermodynamically stable protein without a large barrier samples fully and partially unfolded states, making it susceptible to proteolysis. (b) A kinetically stable protein only rarely samples these unfolded states, making it much more resistant to proteolysis. In the case of αLP, the native state is less stable than the unfolded states; however, kinetic stability does not require a metastable native state. (c) αLP (●) is more resistant to proteolysis than either trypsin (▵) or chymotrypsin (♦). αLP (purified as described in ref. 25), trypsin (TPCK-treated, Worthington), and chymotrypsin (TLCK-treated, Worthington) (6.5 μM each) were mixed in 10 mM CaCl2, 50 mM Mops (pH 7.0) at 37°C. Aliquots were removed over time, and the survival of the individual proteases was measured based on their activities, which could be distinguished given their nonoverlapping specificities for different substrates (succinyl-Ala-Pro-Ala-pNA, succinyl-Ala-Ala-Pro-Arg-pNA, succinyl-Ala-Ala-Pro-Leu-pNA, used for αLP, trypsin, and chymotrypsin, respectively, all at 1 mM in 100 mM Tris, pH 8). Whereas αLP activity decreases at a rate of less than (600 hr)−1, chymotrypsin and trypsin are inactivated with rates of (4 hr)−1 and (60 hr)−1, respectively.
Preliminary experiments indicate that this has indeed been a successful strategy for extending αLP’s lifetime when compared with its thermodynamically stabilized homologues chymotrypsin and trypsin. In a survival assay where these three proteases are mixed and allowed to attack each other, αLP retains its biological activity for much longer than its mammalian counterparts (Fig. 5c). The sensitivity of trypsin and chymotrypsin to proteolysis is likely to be a necessary aspect of their regulation in vivo. Additional experiments demonstrate that the rate of αLP autolysis is comparable to the rate of its global unfolding, indicating that transient unfolding motions leading to proteolytic degradation have been suppressed. αLP has been so successfully optimized that it is vulnerable to degradation only after it completely unfolds, which occurs on an extremely slow time scale.
There is a price for kinetic stability, however. The evolution of a large barrier to unfolding and a highly rigid native state through the incorporation of glycines and other changes has, as a consequence, created an even larger barrier to folding and thermodynamically destabilized the native state of αLP. Nature’s solution has been the coevolution of a transient pro region to promote folding by both reducing the folding barrier and stabilizing the native state. Although it is expected that the general principle of longevity through kinetic stability will be shared by the majority of extracellular bacterial proteases and numerous eukaryotic proteases, the precise details of barrier height and degree of thermodynamic destabilization of the native state are likely to vary. αLP, with its large pro region and metastable native state, may be an extreme example.
Acknowledgments
We thank Dr. Nicholas Sauter for helpful discussions. S.S.J. was supported by a Howard Hughes Medical Institute Predoctoral Fellowship. D.A.A. is an Investigator of the Howard Hughes Medical Institute.
ABBREVIATION
- αLP
α-lytic protease
References
- 1.Baker D, Shiau A K, Agard D A. Curr Opin Cell Biol. 1993;5:966–970. doi: 10.1016/0955-0674(93)90078-5. [DOI] [PubMed] [Google Scholar]
- 2.Brayer G D, Delbaere L T J, James M N G. J Mol Biol. 1979;131:743–775. doi: 10.1016/0022-2836(79)90200-6. [DOI] [PubMed] [Google Scholar]
- 3.Silen J L, Frank D, Fujishige A, Bone R, Agard D A. J Bacteriol. 1989;171:1320–1325. doi: 10.1128/jb.171.3.1320-1325.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Silen J L, Agard D A. Nature (London) 1989;341:462–464. doi: 10.1038/341462a0. [DOI] [PubMed] [Google Scholar]
- 5.Baker D, Sohl J L, Agard D A. Nature (London) 1992;356:263–265. doi: 10.1038/356263a0. [DOI] [PubMed] [Google Scholar]
- 6.Sohl J L, Jaswal S S, Agard D A. Nature (London) 1998;395:817–819. doi: 10.1038/27470. [DOI] [PubMed] [Google Scholar]
- 7.Fujinaga M, Delbaere L T J, Brayer G D, James M N G. J Mol Biol. 1985;184:479–502. doi: 10.1016/0022-2836(85)90296-7. [DOI] [PubMed] [Google Scholar]
- 8.Peters R J, Shiau A K, Sohl J L, Anderson D E, Tang G, Silen J L, Agard D A. Biochemistry. 1998;37:12058–12067. doi: 10.1021/bi980883v. [DOI] [PubMed] [Google Scholar]
- 9.Baker D, Silen J L, Agard D A. Proteins. 1992;12:339–344. doi: 10.1002/prot.340120406. [DOI] [PubMed] [Google Scholar]
- 10.Sauter N K, Mau T, Rader S D, Agard D A. Nat Struct Biol. 1998;5:945–950. doi: 10.1038/2919. [DOI] [PubMed] [Google Scholar]
- 11.Anderson D E, Peters R J, Wilk B, Agard D. Biochemistry. 1999;38:4728–4735. doi: 10.1021/bi982165e. [DOI] [PubMed] [Google Scholar]
- 12.Gewirth D T, Sigler P B. Nat Struct Biol. 1995;2:386–394. doi: 10.1038/nsb0595-386. [DOI] [PubMed] [Google Scholar]
- 13.Schirmer T, Evans P R. Nature (London) 1990;343:140–145. doi: 10.1038/343140a0. [DOI] [PubMed] [Google Scholar]
- 14.Royer W E J, Pardanani A, Gibson Q H, Peterson E S, Friedman J M. Proc Natl Acad Sci USA. 1996;93:14526–14531. doi: 10.1073/pnas.93.25.14526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gallagher T, Gilliland G, Wang L, Bryan P. Structure (London) 1995;3:907–914. doi: 10.1016/S0969-2126(01)00225-8. [DOI] [PubMed] [Google Scholar]
- 16.D’Aquino J, Gomez J, Hilser V, Lee K, Amzel L, Freire E. Proteins. 1996;25:143–156. doi: 10.1002/(SICI)1097-0134(199606)25:2<143::AID-PROT1>3.0.CO;2-J. [DOI] [PubMed] [Google Scholar]
- 17.Silen J L, McGrath C N, Smith K R, Agard D A. Gene. 1988;69:237–244. doi: 10.1016/0378-1119(88)90434-9. [DOI] [PubMed] [Google Scholar]
- 18.Sidhu S S, Kalmar G B, Willis L G, Borgford T J. J Biol Chem. 1994;269:20167–20171. [PubMed] [Google Scholar]
- 19.Shimoi H, Iimura Y, Obata T, Tadenuma M. J Biol Chem. 1992;267:25189–25195. [PubMed] [Google Scholar]
- 20.Sidhu S S, Kalmar G B, Willis L G, Borgford T J. J Biol Chem. 1995;270:7594–7600. doi: 10.1074/jbc.270.13.7594. [DOI] [PubMed] [Google Scholar]
- 21.Sidhu S S, Kalmar G B, Borgford T J. Biochem Cell Biol. 1993;71:454–461. doi: 10.1139/o93-067. [DOI] [PubMed] [Google Scholar]
- 22.Lao G, Wilson D B. Appl Environ Microbiol. 1996;62:4256–4259. doi: 10.1128/aem.62.11.4256-4259.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Binnie C, Liao L, Walczyk E, Malek L T. Can J Microbiol. 1996;42:284–288. doi: 10.1139/m96-041. [DOI] [PubMed] [Google Scholar]
- 24.Henderson G, Krygsman P, Liu C J, Davey C C, Malek L T. J Bacteriol. 1987;169:3778–3784. doi: 10.1128/jb.169.8.3778-3784.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sohl J L, Shiau A K, Rader S D, Wilk B, Agard D A. Biochemistry. 1997;36:3894–3902. doi: 10.1021/bi962341o. [DOI] [PubMed] [Google Scholar]