Abstract
A modular microfluidic airways model system that can simulate the changes in oxygen tension in different compartments of the cystic fibrosis (CF) airways was designed, developed, and tested. The fully reconfigurable system composed of modules with different functionalities: multichannel peristaltic pumps, bubble traps, gas exchange chip, and cell culture chambers. We have successfully applied this system for studying the antibiotic therapy of Pseudomonas aeruginosa, the bacteria mainly responsible for morbidity and mortality in cystic fibrosis, in different oxygen environments. Furthermore, we have mimicked the bacterial reinoculation of the aerobic compartments (lower respiratory tract) from the anaerobic compartments (cystic fibrosis sinuses) following an antibiotic treatment. This effect is hypothesised as the one on the main reasons for recurrent lung infections in cystic fibrosis patients.
INTRODUCTION
The human airways are complex multi-compartmental habitats for infectious bacteria. In healthy humans, the majority of the airways are essentially kept sterile as a result of highly efficient clearing mechanisms.1 In cystic fibrosis (CF) patients, this clearing mechanism is severely impaired and bacterial infections inflict deteriorating health and become the major cause of mortality in these patients.2, 3
The human airways consist of at least three independent compartments, the conductive airways (the trachea, bronchi, and bronchioles), the oxygen exchange compartment (alveoli), and a third, less investigated compartment, the paranasal sinuses (maxillary sinuses, frontal sinuses, and ethmoid sinuses). In the first and the last compartment, the environment is essentially anaerobic,4 while the alveoli are highly aerated.
In healthy individuals, the conductive airways are constantly cleared by mucociliary transport of entrapped microorganisms.5 As a result, very few bacteria will ever reach the alveoli. Bacteria that do evade this clearing mechanism will rapidly be cleared by the actions of the immune system. The sinuses also have a mucociliary clearance mechanism although not as effective as the one found in the conductive airways. Large concentrations of bacteria can be found widely spread throughout the sinuses, in particular in cases of common colds, etc.6
Cystic fibrosis patients suffer from a defect in the cystic fibrosis transmembrane conductance regulator (CFTR) gene. The affected gene codes for the CFTR protein, which is a chloride channel that is present in the epithelial cell membrane. Reduced or absent function of the CFTR will lead to highly reduced secretion of chloride and accordingly water over the cell membrane.7 A direct consequence of this defect is that the mucus layer in the conductive airways becomes very viscous and the mucociliary clearance mechanisms are highly impaired.3, 8 This results in frequent and recurrent infections of the CF airways, with the risk of pneumonia. As the bacteria infect the lungs in large numbers, the immune system tries to eradicate the infection but with a reduced effect since the bacteria are embedded in mucus and more or less recalcitrant to the cellular defence.9 Instead, the lung tissue is gradually damaged by the on-going immunological exposure, eventually leading to massive pulmonary deficiency and death.10 In the clinic, the infections can be treated with cocktails of antibiotics, which can reduce or sometimes eradicate the infectious agents.11
Bacterial airway infections in patients with a normal mucosa are relatively easy to treat with antibiotics. This is unfortunately not the case for CF patients and the myriad of infections they acquire during their lifetime leave each patient with a high need for recurrent antibiotic treatments. This is a multifactorial phenomenon and there are a lot of theories that try to explain this.12, 13, 14 The most obvious reason for a treatment failure is the hindered diffusion of the antimicrobial agent through the thick and viscous mucus layer.15, 16 However, according to recent findings, the main reason may reside in limited oxygen availability in some parts of the airways.17, 18 These highly different oxygen environments are due to the human physiology of the airways and furthermore upheld by the consumption from epithelial and immune cells in the local surroundings. Immune cells (mainly polymorphonuclear neutrophils, PMNs) consume oxygen in order to produce reactive oxygen species to defeat the bacterial infection. The immense respiratory bursts by the PMNs produce anaerobic niches in the mucus layers of the CF airways.10, 19
As Pseudomonas aeruginosa (P. aeruginosa) infections are almost inescapable in CF patients, especially in older patients, this makes P. aeruginosa an important organism for studies of “oxygen” phenomena.20P. aeruginosa is a facultative anaerobic bacteria21 with reduced growth22 and metabolic activity18 at low oxygen levels. Antibiotics such as tobramycin, ciprofloxacin, and tetracycline preferentially kill the physiologically active bacteria living at high oxygen levels (aerobic environment), while colistin is more effective on the physiologically inactive bacteria growing in an anaerobic environment.10, 23
An antibiotic treatment has the potential to clear a lung infection, yet after a few months the same bacteria are very likely to reappear, possibly as a result of reinoculation from the anaerobic environment of the sinuses.24 In this context, the sinuses could very well serve as a reservoir for “sinus” bacteria, which are difficult to treat with antibiotics and can cause the reinfection of otherwise cleared patients.
The now classical ways of studying CF related bacterial infections, primarily P. aeruginosa, are either to use animal models or to grow the bacteria in flow-cell systems.
A number of different animals have been tried as models of chronic infections in CF patients.25 This includes rats,26, 27, 28 guinea pigs,29 cats,30 and rhesus monkeys.31, 32 However, the most important animal model is a mouse.33, 34, 35, 36 The use of an animal model is expensive and rises ethical concerns.37, 38 Furthermore, animal models for CF related infections are still not ideal, mainly due to immunological differences between man and, e.g., mouse. Mice do not acquire spontaneous and chronic infections as seen with human CF patients.39 CFTR knock-out mice do not show the same mucus accumulation as seen in the lower airways of CF patients.40, 41
In flow-cell based systems, the bacteria are allowed to form a biofilm on a surface, as in the airways, and can then be monitored using a confocal microscope,11, 42 However, this is not a suitable model for the human airways as they are subdivided into aerobic and anaerobic compartments.
The advancement in micro- and nanofabrication and assembly, as well as better understanding of microfluidics, has made possible the development of devices for modelling different tissue organs. Thanks to these devices, the control over the environment, relations and interactions between the cells and tissues at microscale with high spatial and temporal resolution can be achieved.43, 44 The yet small but fast growing number of microdevices that can mimic different and even entire organs have been reported. These include blood vessels,45 bones,46 muscles,47 liver,48, 49, 50, 51 brain,52 guts,53 kidneys,54, 55 endothelia,56 and blood-brain barrier.57, 58
In the field of mimicking the human airways, Huh et al.59, 60 proposed a microfluidic device that can simulate injuries to the airways epithelia done by liquid plug flow. Recently, Huh et al. proposed a model of the vacuoles in the lung.61 In this work, the phagocytosis of planktonic Escherichia coli cells by neutrophils on the epithelial surface was shown.
Other works focused on the artificial lungs which could be applied to patients suffering from respiratory failure. Different solution based, e.g., on the microporous hollow fibers62, 63 or poly(dimethylsiloxane) (PDMS)64, 65, 66 has been proposed.
In our previous work,22 we have shown the possibility of using a PDMS membrane and an oxygen scavenging liquid to control the oxygen gradient within cell culture microchambers. To the best of our knowledge, the microfluidic model of different compartments of the human airways that would allow to observe the influence of the microenvironment of these compartments on the recurrence of CF related infections has not been reported previously.
Therefore, the aim of this work was to make a model system, which simulates the three compartments of the airways to better understand the interplay between them. Using this artificial airways model, we can look into the bacterial details in the three compartments, their transmitting interaction, and the states of the bacterial inhabitants before, during, and after antibiotic treatment.
MICROFLUIDIC AIRWAYS MODEL
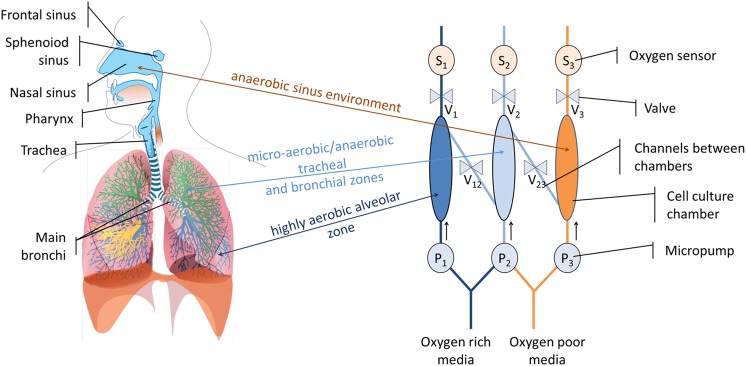
The three sections of human airways: the conductive airways (trachea, bronchi, and bronchioles), which are considered micro-aerobic, the highly aerobic gaseous exchange compartments (the alveoli), and the compartments of the paranasal sinuses, which are basically anaerobic (Fig. 1a), are reproduced in this microfluidic airways model (MAM). This is realized by constructing cell culture microchambers with different oxygen levels (Fig. 1b). A microchamber with atmospheric air oxygenated media (aerobic environment) is connected by a channel to a microchamber with culture media saturated below the atmospheric air saturation level (micro-aerobic environment). This chamber is consecutively connected to a chamber with deoxygenated media (anaerobic environment). The connections between the chambers as well as outlets can be closed and opened and the actual oxygen level in the compartments is determined by an oxygen probe. The entire system is actuated by peristaltic micropumps.
Figure 1.
(a) The human airways system67 and (b) the MAM.
DESIGN
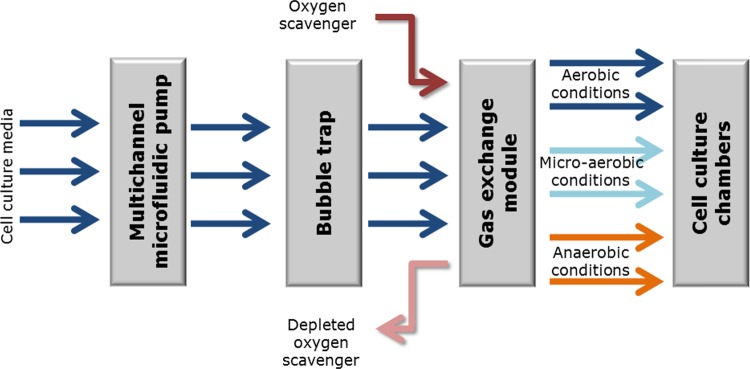
The above model was implemented, as a modular system comprised of the following distinct modules: a modified previously described multichannel microfluidic peristaltic pump,68, 69 a bubble trap, and gas exchange and cell culture chambers (Fig. 2). The system allows the simultaneously cultivation of cells in 8 chambers (3 of them facilitate aerobic environment, 2 micro-aerobic environment, and 3 anaerobic environment).
Figure 2.
Modular implementation of MAM as a multichannel setup capable of generating multiple instances of the different oxygen environments.
These modules can be attached to form a microfluidic platform. The detailed design of each of these modules and the platform is available in the supplementary material in Ref. 70.
MATERIALS AND METHODS
Microfabrication
The material of choice for most of the parts was polycarbonate (PC) (Nordisk Plast A/S, Denmark). This material exhibits low oxygen permeability (1.8 barrer71), very good mechanical and optical properties72 as well as being resistant to alcohols and oils, which is important for sterilization and microscopy. The structures in PC were obtained by micromilling (Mini-Mill/3PRO, Minitech Machinery Corp., USA) followed by solvent vapour assisted bonding. The general bonding procedure was adopted after Ogończyk et al.72 with minor changes. As a solvent we used tetrahydrofuran and the temperature and pressure was 50 °C and 1.5 MPa accordingly.
PDMS inlays were fabricated by casting the silicone mixture (Sylgard 184, Dow Corning Corp., USA) against the milled mould. The PDMS parts were bonded to PC using silicone adhesive tape (ARcare® 91005, Adhesive Research, Inc., Ireland). The tape was cut into the desired shape using laser ablation (48-5S Duo Lase carbon dioxide laser, SYNRAD Inc., USA).
The gaskets used for sealing the modules to the platform, as well as check valves, were fabricated by milling in fluoroelastomer VITON A (J-Flex Rubber Products, UK). The plunging pattern was used for fabricating the parts in fluoroelastomer.
Cultivation of P. aeruginosa strains
The P. aeruginosa laboratory strain PAO1 was used for all biofilm experiments. The PAO1 strain was originally isolated from a burn wound.73 PAO1 was fluorescently tagged at a neutral chromosomal locus with green fluorescent protein (GFP) or monomeric red fluorescent protein (mRFP1) with miniTn7 constructs as previously described.74 A P. aeruginosa medium (FAB)42, 75 supplemented with 0.3 mM glucose and 65 mM KNO3 (FAB-GN) was used for biofilm cultivation. All biofilms and batch cultures were grown at 37 °C. PAO1 was pre-cultured overnight in Luria Bertani (LB) media and prepared for inoculation in FAB-GN media. The overnight culture was diluted to an OD600 of 0.01 and subsequently 100 μl was inoculated with a Gilson P200 pipette through the designed inlets. In order to visualise the bacteria migration, the anaerobic culture chambers were inoculated with the GFP tagged strain and microaerobic and aerobic chambers were inoculated with the mRFP1 tagged strain. The device was left upside down for an hour without media flow. Following the 1 h incubation period, the media flow was started at 500 μl·h−1 per channel. After 48 h of cultivation in the growth chambers, the media was exchanged to media supplemented with 50 μg·ml−1 of the antibiotic, ciprofloxacin (Sigma-Aldrich, Denmark A/S). 24 h of antibiotic treatment was followed by staining with 1 μM SYTOX® Blue dead cell stain (Molecular Probes, Invitrogen, Denmark). The chambers were left for 48 h running on media without ciprofloxacin. This was done in order to subsequently evaluate the cells’ ability to migrate between oxygen gradients. Interconnection was made between the chambers and cells were allowed to migrate for 24 h before analysed with confocal laser scanning microscopy.
The interconnections between chambers were made with poly(vinyl chloride) (PVC) tubing attached to PEEK connector plugs that were inserted into the specific chambers with different oxygen levels.
Dissolved oxygen level control
In order to control the dissolved oxygen levels in the cell culture media, one of the PDMS gas exchange module inlays was supplied with an oxygen scavenger (10% sodium sulphite solution with 0.1 mM CoSO4 as catalyst, both from Sigma-Aldrich Denmark A/S). The second inlay was left open to atmospheric air.
Determination of the oxygen levels in the cell culture chambers was achieved using the phosphorescent oxygen-sensitive nanoprobe based on Platinum(II)-tetrakis-(pentafluorophenyl) porphine (PtTFPP) dye76 (Luxcel Biosciences, Ireland). The oxygen levels were determined by: (1) phosphorescence intensity and lifetime measurements on an Axiovert 200 wide-field microscope (Carl Zeiss, Germany) upgraded for phosphorescence lifetime imaging (LaVision Biotec, Germany), and (2) phosphorescence lifetime measurements on a multi-label plate reader (Victor2, Perkin-Elmer Life Sciences, USA). The imaging experiments were performed as described previously by Fercher et al.77 using pulsed excitation with a 390 nm light-emitting diode (LED) and emission collection at 655 ± 50 nm. Plate reader measurements were performed as described previously,76 using excitation at 340 nm and emission at 642 nm. For such measurements, the device was inoculated with non-fluorescent P. aeruginosa laboratory strain PAO1, then maintained under a flow of medium, containing 0.01 mg ml−1 of probe, for 12 h and then washed with medium. Thus, biofilms stained with the phosphorescent probe were produced in the device, which can be used to monitor oxygenation and conduct biological experiments for several days.
The correlation between the phosphorescence intensity or photoluminescence lifetime and dissolved oxygen concentration was determined by two-point calibration: at 0% and 100% of atmospheric air oxygen saturation in the culture media. The 0% atmospheric air oxygen saturation was obtained by supplementing the glucose containing culture media with glucose oxidase (Sigma-Aldrich Denmark A/S) as previously described by O’Donovan et al.78 The very good linearity of Stern-Volmer plots (τ0/τ vs [O2]) allows to use very simple two-point calibration as discussed by O’Riordan et al.79 Each cell culture chamber was calibrated separately.
Confocal microscopy and image analysis
Confocal fluorescence images were taken with a Leica TCS SP5 microscope using a 50 × /0.75 W objective. 4 random pictures were taken from each chamber. Settings for visualization of the probes were: 514 nm excitation and 613-688 nm emission for mRFP1; 488 nm excitation and 517-535 nm emission for GFP; 458 nm excitation and 475-490 nm emission for SYTOX® Blue dead stain. All images were processed by the imaris 7 software package (BITPLANE AG, Zürich, Switzerland).
RESULTS
Integration of the modular microfluidic system
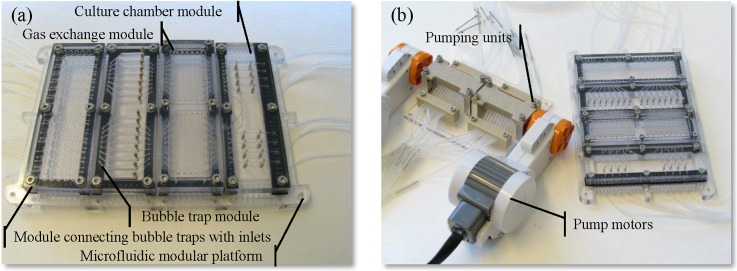
The microfluidic modules described in the supplementary material70 were successfully fabricated and assembled on a microfluidic platform (Fig. 3a). The system was integrated with 16-channel peristaltic micropump. The pump was actuated by two motors obtained from commercially available LEGO® Mindstorms® NXT 2.0 robotic kit80 (The LEGO Group, Denmark) (Fig. 3b).69
Figure 3.
(a) Microfluidic platform with modules and (b) the peristaltic micropump.
Determination of the dissolved oxygen level
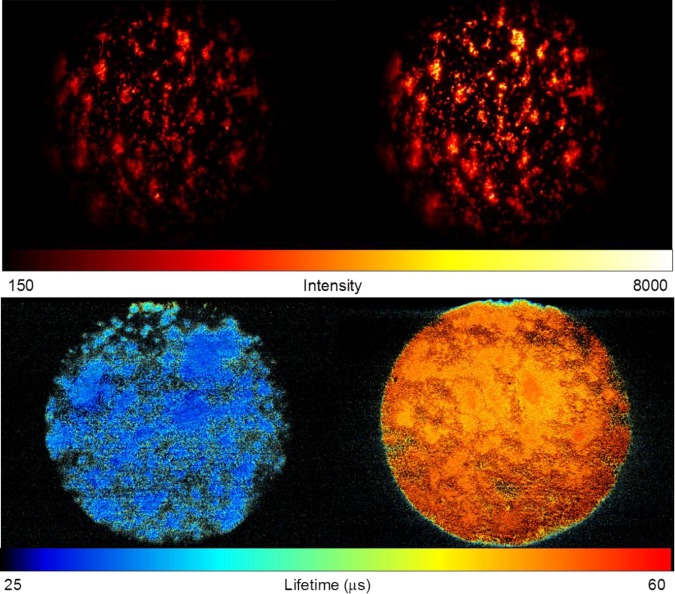
The differences between the phosphorescence intensity of the P. aerugionsa biofilm, stained with the PtTFPP nanoprobe, in the aerobic and anaerobic environments were investigated in order to determine the oxygen removal efficiency from the culture media using the gas exchange module (Fig. 4). The phosphorescence intensity increased approximately two-fold in anaerobic conditions as compared with aerobic conditions.
Figure 4.
Phosphorescence intensity (top panel) and lifetime (bottom panel) images of the P. aeruginosa biofilm stained with the PtTFPP nanoprobe in the culture chambers with high (left) and low (right) oxygen concentrations.
A two-step calibration curve was established by measuring the photoluminescence lifetime of the nanoprobe in oxygen free and atmospheric air-saturated media (see Sec. 4). The photoluminescence lifetime was determined to be 54.5 ± 1.3 μs (oxygen free media) and 29.7 ± 0.6 μs (and atmospheric air-saturated media). Oxygen concentration in atmospheric air-saturated media was determined to be 0.281 mM.81 The measurements were performed at room temperature. Assuming a reversible collisional quenching model,82 the Stern-Volmer constant was determined to be 2.97 mM−1.
The photoluminescence lifetimes of the nanoprobe in the chambers, resembling aerobic, micro-aerobic, and anaerobic environments, were 30.5 μs, 35.8 μs, and 51.1 μs, respectively, which correspond to 94.2%, 62.7%, and 7.9% of atmospheric air-saturation of the culture media according to the two-point calibration curve.
Cultivation of P. aeruginosa strains in different oxygen environments
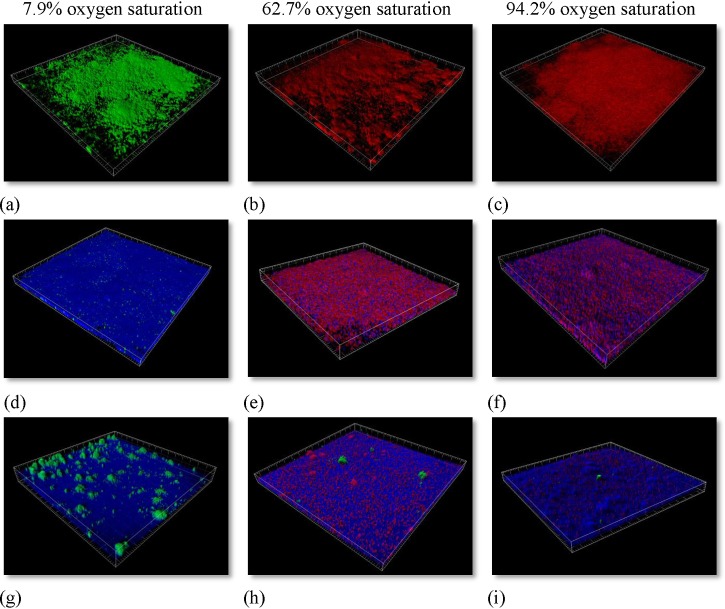
PAO1 biofilm formation was analysed 24 h after inoculation. In order to follow the trail of each specific population following each inoculation, we used different fluorescent tagged versions of PAO1. In the culture media (supplemented with nitrate as an alternative electron acceptor), highly equivalent biofilms were formed irrespective of the generated oxygen environment (Figs. 5a, 5b, 5c).
Figure 5.
3D representation of the PAO1 biofilms at different oxygen saturations in FAB-GN media (minimal media supplemented with nitrate). PAO1 expresses either the fluorescent protein GFP or mRFP1. (a)-(c) 24 h old biofilms in FAB-CN media. (d)-(f) 48 h after inoculation the cells were challenged with 50 μg·ml−1 ciprofloxacin for 24 h and then stained with dead stain SYTOX® Blue. (g)-(i) Interconnected chambers of the different oxygen saturation environments.
The green confocal image (Fig. 5a) originates from GFP tagged bacteria and was cultivated under the lowest oxygen saturation. The red biofilm derives from mRFP1 tagged bacteria and was cultivated in microaerobic (Fig. 5b) and aerobic conditions (Fig. 5c). However, under the tested conditions in a minimal media, the biomass within first 24 h of growth reached highly equivalent magnitudes of biomass regardless of oxygen tension. Under anaerobic conditions, supplemented with nitrate as electron acceptor, PAO1 has, in LB media, shown to develop 3 fold more biomass.83
In order to evaluate the efficiency of the antibiotic ciprofloxacin on PAO1 biofilms under conditions of lowered oxygen availability in a FAB-GN media, each chamber was challenged with the same ciprofloxacin concentration. After 24 h of incubation, each chamber received media supplemented with 50 μg·ml−1 of the antibiotic ciprofloxacin for a period of 24 h. The treatment would present the differences in the effectiveness of the antibiotic in a developing PAO1 biofilm. Dead cells in the biofilms were visualized by staining each chamber with Sytox® blue dead cell stain (Figs. 5d, 5e, 5f). The effect of the ciprofloxacin on the PAO1 biofilm was highly dependent on the oxygen environment. PAO1-GFP biofilm was much more susceptible to the antibiotic treatment than biofilm formed under higher oxygen concentrations. The antibiotic concentration was chosen to eradicate the majority of cells in the establishing biofilms, though low enough to allow surviving cells. PAO1 had in that sense been established enough to produce a healthy biofilm and represent a community associated environment.
Following the 24 h treatment with ciprofloxacin, the chambers were taken off the antibiotic containing media for 48 h and connections between the different oxygen environments were made (Figs. 5g, 5h, 5i) (see connection details in the supplementary material in Ref. 70). This enabled tracking of the bacteria in a novel way that has previously not been possible. We setup the system in a way to follow in which direction, if any, the surviving bacteria would move. As the ciprofloxacin treatment had been stopped, the only difference between the chambers was the differences in oxygen concentrations. The small green clusters on Figs. 5a, 5i come from the GFP tagged bacteria. This proves that PAO1 moves from chambers with low oxygen tension (Fig. 5g) to microaerobic (Fig. 5h) and aerobic chambers (Fig. 5i).
DISCUSSION AND CONCLUSIONS
In this paper, we describe design, fabrication, working principle, and application of a highly complex modular microfluidic system. Integration of different modules, bringing in such important functionalities as multichannel fluid control, bubble trapping, gas control-exchange and bacterial culturing on a microfluidic lab-on-a-chip system, has been shown to be successfully achieved. The modularity allows addition and removal of the different functionalities. The design permits easy reconfiguration and tailoring of the system to match particular needs. In case of malfunctions in a single module, the system benefits from its modular construction and allows uncomplicated exchange of the broken module without the need for fabrication of other essential parts of the system. This is particularly important in the field of life science microfluidic systems, in which not yet all of these components are suitable for mass production. Furthermore, it allows quick prototype testing of different system configurations.
The microfluidic system in its present configuration enables comparison of changes between anatomically driven oxygen tensions in different compartments of the CF airways model, as well as full control and sensing of dissolved oxygen levels. By making the system compatible with common substrates such as microscope slides and multitier plates, it enables research staff to use standard laboratory equipment such as standard microscopes and multitier plate readers.
Furthermore, the microfluidic CF airways model permits to freely reconfigure connections between oxygen rich and oxygen depleted regions without bringing restrictions to the researcher in the design of experiments. It enables to mimic some different conditions and diseases in patients suffering from CF, such as clogging of the ostia in recurrent sinusitis4, 84 or the development of mucus plugs85 in the bronchioles. These experiments were not previously possible to perform in standard invitro flow-cells’ models for biofilm studies. In vivo models will usually not allow precise control of such important conditions.
Moreover, the use of this microfluidic system, instead of a CF airways animal model, is cheaper, safer and easier to handle for researchers. Importantly, it furthermore does not raise any ethical concerns, which is the case for the use of animal models in medical research. We demonstrated the application of our microfluidic airway model for studying P. aeruginosa PAO1 under different oxygen levels in response to treatment with ciprofloxacin. We have in this way explicitly shown that the system is an asset in reliable and controllable biofilm evaluations for treatment with antibiotics at reduced oxygen concentrations. Importantly, such a system allows testing of very small volumes thereby minimizing the use of large amounts of expensive antimicrobials. PAO1 survival was shown to be highly dependent on the amount of oxygen available during the antimicrobial treatment. This corresponds very well with previous studies where it was shown that higher metabolic rate, in nitrate supplemented media under anaerobic conditions, leads to a lower survival rate of the bacteria.86
We have shown that PAO1, under lowered oxygen concentrations, migrates towards higher oxygen concentrations even in nitrate supplemented media. Nitrate, which serves as final electron acceptor for anaerobic nitrate respiration (denitrification), does not seem to be favoured in the presence of oxygen. This scenario can mimic the reinoculation of the lower respiratory tract, previously cleared by the antibiotic treatments, with bacteria from the sinuses. This effect is hypothesised as the main reason for recurrent infections in CF patients.17
Microfluidic systems present an efficient tool in mimicking organ specific functions. The advantage of these systems is the possibility of mimicking key functionalities while omitting non-essential ones. However, researchers should always keep in mind that in general, these systems lack the complexity of real organs. Therefore, careful interpretation of the generated results should always follow. In case of the presented system, the mucociliary clearance was not taken into account due to the general impairment of this mechanism in CF airways. However, if one would like to relate the results obtained in the presented system to a bacterial infection in healthy individuals, the mucociliary clearance mechanism needs to be taken into consideration. Next generation microfluidic systems should focus on invivo-like microenvironmental cues (mucus accumulation, mucus constituents, air-liquid interface, stress factors, etc.) that would lead to more profound understanding of the bacterial role in CF pathology.
ACKNOWLEDGMENTS
We would like to acknowledge for the funding of Ph.D. stipends by The Technical University of Denmark as well as the EU funded FP7 project EXCELL.
References
- Smith D. J., Gaffney E. A., and Blake J. R., Respir. Physiol. Neurobiol. 163(1–3), 178–188 (2008). 10.1016/j.resp.2008.03.006 [DOI] [PubMed] [Google Scholar]
- Campodónico V. L., Gadjeva M., Paradis-Bleau C., Uluer A., and Pier G. B., Trends Mol. Med. 14(3), 120–133 (2008). 10.1016/j.molmed.2008.01.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moskowitz S. M., Chmiel J. F., Sternen D. L., Cheng E., Gibson R. L., Marshall S. G., and Cutting G. R., Genet. Med. 10(12), 851–868 (2008). 10.1097/GIM.0b013e31818e55a2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aust R. and Drettner B., Acta Otolaryngol. 78(3–4), 264–269 (1974). 10.3109/00016487409126354 [DOI] [PubMed] [Google Scholar]
- Matsui H., Randell S. H., Peretti S. W., Davis C. W., and Boucher R. C., J. Clin. Invest. 102(6), 1125–1131 (1998). 10.1172/JCI2687 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hekiert A. M., Kofonow J. M., Doghramji L., Kennedy D. W., Chiu A. G., Palmer J. N., Leid J. G., and Cohen N. A., Otolaryngol.-Head Neck Surg. 141(4), 448–453 (2009). 10.1016/j.otohns.2009.06.090 [DOI] [PubMed] [Google Scholar]
- Jih K.-Y., Li M., Hwang T.-C., and Bompadre S. G., J. Physiol. 589(11), 2719–2731 (2011). 10.1113/jphysiol.2010.202861 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antunes M. and Cohen N., Curr. Opin. Allergy Clin. Immunol. 7(1), 5 (2007). 10.1097/ACI.0b013e3280114eef [DOI] [PubMed] [Google Scholar]
- Leal T., Fajac I., Wallace H. L., Lebecque P., Lebacq J., Hubert D., Dall’Ava J., Dusser D., Ganesan A. P., Knoop C., Cumps J., Wallemacq P., and Southern K. W., Clin. Biochem. 41(10–11), 764–772 (2008). 10.1016/j.clinbiochem.2008.03.013 [DOI] [PubMed] [Google Scholar]
- Bjarnsholt T., Jensen P. O., Fiandaca M. J., Pedersen J., Hansen C. R., Andersen C. B., Pressler T., Givskov M., and Hoiby N., Pediatr. Pulmonol. 44(6), 547–558 (2009). 10.1002/ppul.21011 [DOI] [PubMed] [Google Scholar]
- Bjarnsholt T., Biofilm Infections (Springer, New York, 2010), p. 251. [Google Scholar]
- Costerton J. W., Stewart P. S., and Greenberg E. P., Science (N.Y.) 284(5418), 1318–1322 (1999). 10.1126/science.284.5418.1318 [DOI] [PubMed] [Google Scholar]
- Bjarnsholt T., Biofilm Infections (Springer, New York, 2010), p. 216. [Google Scholar]
- Jedlicka S. S., Rickus J. L., and Zemlyanov D., J. Phys. Chem. C 114(1), 342–344 (2010). 10.1021/jp907551t [DOI] [Google Scholar]
- Rasmussen K. and Lewandowski Z., Biotechnol. Bioeng. 59(3), 302–309 (1998). [DOI] [PubMed] [Google Scholar]
- Stewart P. S., Antimicrob. Agents Chemother. 40(11), 2517–2522 (1996). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aanaes K., Rickelt L. F., Johansen H. K., von Buchwald C., Pressler T., Høiby N., and Jensen P. Ø., J. Cyst. Fibros. 10(2), 114–120 (2011). 10.1016/j.jcf.2010.12.002 [DOI] [PubMed] [Google Scholar]
- Walters M. C., Roe F., Bugnicourt A., Franklin M. J., and Stewart P. S., Antimicrob. Agents Chemother. 47(1), 317–323 (2003). 10.1128/AAC.47.1.317-323.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Worlitzsch D., Tarran R., Ulrich M., Schwab U., Cekici A., Meyer K. C., Birrer P., Bellon G., Berger J., Weiss T., Botzenhart K., Yankaskas J. R., Randell S., Boucher R. C., and Doring G., J. Clin. Invest. 109(3), 317–325 (2002). 10.1172/JCI13870 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreau-Marquis S., Stanton B. A., and O’Toole G. A., Pulm. Pharmacol. Ther. 21(4), 595–599 (2008). 10.1016/j.pupt.2007.12.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guss A. M., Roeselers G., Newton I. L. G., Young C. R., Klepac-Ceraj V., Lory S., and Cavanaugh C. M., ISME J. 5(1), 20–29 (2011). 10.1038/ismej.2010.88 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skolimowski M., Nielsen M. W., Emnéus J., Molin S., Taboryski R., Sternberg C., Dufva M., and Geschke O., Lab Chip 10(16), 2162–2169 (2010). 10.1039/c003558k [DOI] [PubMed] [Google Scholar]
- Kim J., Hahn J.-S., Franklin M. J., Stewart P. S., and Yoon J., J. Antimicrob. Chemother. 63(1), 129–135 (2009). 10.1093/jac/dkn462 [DOI] [PubMed] [Google Scholar]
- Hansen S. K., Rau M. H., Johansen H. K., Ciofu O., Jelsbak L., Yang L., Folkesson A., Jarmer H. O., Aanaes K., von Buchwald C., Hoiby N., and Molin S., ISME J. 6(1), 31–45 (2012). 10.1038/ismej.2011.83 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kukavica-Ibrulj I. and Levesque R. C., Lab. Anim. 42(4), 389–412 (2008). 10.1258/la.2007.06014e [DOI] [PubMed] [Google Scholar]
- Pedersen S. S., Shand G. H., Hansen B. L., and Hansen G. N., Acta Pathol. Microbiol. Immunol. Scand. 98(3), 203–211 (1990). [PubMed] [Google Scholar]
- Cash H. A., Woods D. E., McCullough B., W. G.Johanson, Jr., and Bass J. A., Am. Rev. Respir. Dis. 119(3), 453–459 (1979). [DOI] [PubMed] [Google Scholar]
- Sato A., Kitazawa H., Chida K., Hayakawa H., and Iwata M., Drugs 49(Suppl. 2), 253–255 (1995). 10.2165/00003495-199500492-00059 [DOI] [PubMed] [Google Scholar]
- Pennington J. E., Hickey W. F., Blackwood L. L., and Arnaut M. A., J. Clin. Invest. 68(5), 1140–1148 (1981). 10.1172/JCI110358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomassen M. J., Klinger J. D., Winnie G. B., Wood R. E., Burtner C., Tomashefski J. F., Horowitz J. G., and Tandler B., Infect. Immun. 45(3), 741–747 (1984). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheung A. T., Moss R. B., Leong A. B., and W. J.Novick, Jr., J. Med. Primatol. 21(7–8), 357–362 (1992). [PubMed] [Google Scholar]
- Cheung A. T., Moss R. B., Kurland G., Leong A. B., and W. J.Novick, Jr., J. Med. Primatol. 22(4), 257–262 (1993). [PubMed] [Google Scholar]
- Starke J. R., Edwards M. S., Langston C., and Baker C. J., Pediatr. Res. 22(6), 698–702 (1987). 10.1203/00006450-198712000-00017 [DOI] [PubMed] [Google Scholar]
- Morissette C., Skamene E., and Gervais F., Infect. Immun. 63(5), 1718–1724 (1995). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stevenson M. M., Kondratieva T. K., Apt A. S., Tam M. F., and Skamene E., Clin. Exp. Immunol. 99(1), 98–105 (1995). 10.1111/j.1365-2249.1995.tb03478.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stotland P. K., Radzioch D., and Stevenson M. M., Pediatr. Pulmonol. 30(5), 413–424 (2000). [DOI] [PubMed] [Google Scholar]
- Creton S., Billington R., Davies W., Dent M. R., Hawksworth G. M., Parry S., and Travis K. Z., Toxicology 262(1), 10–11 (2009). 10.1016/j.tox.2009.04.007 [DOI] [PubMed] [Google Scholar]
- Hendriksen C. F. M., Expert Rev. Vaccines 8(3), 313–322 (2009). 10.1586/14760584.8.3.313 [DOI] [PubMed] [Google Scholar]
- Guilbault C., Saeed Z., Downey G. P., and Radzioch D., Am. J. Respir. Cell Mol. Biol. 36(1), 1–7 (2007). 10.1165/rcmb.2006-0184TR [DOI] [PubMed] [Google Scholar]
- Bangel N., Dahlhoff C., Sobczak K., Weber W. M., and Kusche-Vihrog K., J. Cyst. Fibros. 7(3), 197–205 (2008). 10.1016/j.jcf.2007.07.012 [DOI] [PubMed] [Google Scholar]
- Zhou Z., Duerr J., Johannesson B., Schubert S. C., Treis D., Harm M., Graeber S. Y., Dalpke A., Schultz C., and Mall M. A., J. Cyst. Fibros. 10(Suppl. 2), S172–S182 (2011). 10.1016/S1569-1993(11)60021-0 [DOI] [PubMed] [Google Scholar]
- Weiss Nielsen M., Sternberg C., Molin S., and Regenberg B., J. Vis. Exp. (47), e2383 (2011). 10.3791/2383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitesides G. M., Ostuni E., Takayama S., Jiang X., and Ingber D. E., Annu. Rev. Biomed. Eng. 3, 335–373 (2001). 10.1146/annurev.bioeng.3.1.335 [DOI] [PubMed] [Google Scholar]
- El-Ali J., Sorger P. K., and Jensen K. F., Nature 442(7101), 403–411 (2006). 10.1038/nature05063 [DOI] [PubMed] [Google Scholar]
- Shin M., Matsuda K., Ishii O., Terai H., Kaazempur-Mofrad M., Borenstein J., Detmar M., and Vacanti J. P., Biomed. Microdevices 6(4), 269–278 (2004). 10.1023/B:BMMD.0000048559.29932.27 [DOI] [PubMed] [Google Scholar]
- Jang K., Sato K., Igawa K., Chung U. I., and Kitamori T., Anal. Bioanal. Chem. 390(3), 825–832 (2008). 10.1007/s00216-007-1752-7 [DOI] [PubMed] [Google Scholar]
- Lam M. T., Huang Y. C., Birla R. K., and Takayama S., Biomaterials 30(6), 1150–1155 (2009). 10.1016/j.biomaterials.2008.11.014 [DOI] [PubMed] [Google Scholar]
- Carraro A., Hsu W. M., Kulig K. M., Cheung W. S., Miller M. L., Weinberg E. J., Swart E. F., Kaazempur-Mofrad M., Borenstein J. T., Vacanti J. P., and Neville C., Biomed. Microdevices 10(6), 795–805 (2008). 10.1007/s10544-008-9194-3 [DOI] [PubMed] [Google Scholar]
- Lee P. J., Hung P. J., and Lee L. P., Biotechnol. Bioeng. 97(5), 1340–1346 (2007). 10.1002/bit.21360 [DOI] [PubMed] [Google Scholar]
- Powers M. J., Domansky K., Kaazempur-Mofrad M. R., Kalezi A., Capitano A., Upadhyaya A., Kurzawski P., Wack K. E., Stolz D. B., Kamm R., and Griffith L. G., Biotechnol. Bioeng. 78(3), 257–269 (2002). 10.1002/bit.10143 [DOI] [PubMed] [Google Scholar]
- Khetani S. R. and Bhatia S. N., Nat. Biotechnol. 26(1), 120–126 (2008). 10.1038/nbt1361 [DOI] [PubMed] [Google Scholar]
- Park J. W., Vahidi B., Taylor A. M., Rhee S. W., and Jeon N. L., Nat. Protoc. 1(4), 2128–2136 (2006). 10.1038/nprot.2006.316 [DOI] [PubMed] [Google Scholar]
- Shuler M. L., Mahler G. J., Esch M. B., and Glahn R. P., Biotechnol. Bioeng. 104(1), 193–205 (2009). 10.1002/bit.22366 [DOI] [PubMed] [Google Scholar]
- Baudoin R., Griscom L., Monge M., Legallais C., and Leclerc E., Biotechnol. Prog. 23(5), 1245–1253 (2007). 10.1021/bp0603513 [DOI] [PubMed] [Google Scholar]
- Jang K. J. and Suh K. Y., Lab Chip 10(1), 36–42 (2010). 10.1039/b907515a [DOI] [PubMed] [Google Scholar]
- Song J. W., Gu W., Futai N., Warner K. A., Nor J. E., and Takayama S., Anal. Chem. 77(13), 3993–3999 (2005). 10.1021/ac050131o [DOI] [PubMed] [Google Scholar]
- Harris S. G. and Shuler M. L., Biotechnol. Bioprocess Eng. 8(4), 246–251 (2003). 10.1007/BF02942273 [DOI] [Google Scholar]
- Booth R. and Kim H., Lab Chip 12(10), 1784–1792 (2012). 10.1039/c2lc40094d [DOI] [PubMed] [Google Scholar]
- Huh D., Fujioka H., Tung Y. C., Futai N., R.Paine, 3rd, Grotberg J. B., and Takayama S., Proc. Natl. Acad. Sci. U.S.A. 104(48), 18886–18891 (2007). 10.1073/pnas.0610868104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tavana H., Kuo C. H., Lee Q. Y., Mosadegh B., Huh D., Christensen P. J., Grotberg J. B., and Takayama S., Langmuir 26(5), 3744–3752 (2010). 10.1021/la903038a [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huh D., Matthews B. D., Mammoto A., Montoya-Zavala M., Hsin H. Y., and Ingber D. E., Science 328(5986), 1662–1668 (2010). 10.1126/science.1188302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pacella H. E., Eash H. J., Frankowski B. J., and Federspiel W. J., J. Membr. Sci. 382(1–2), 238–242 (2011). 10.1016/j.memsci.2011.08.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polk A. A., Maul T. M., McKeel D. T., Snyder T. A., Lehocky C. A., Pitt B., Stolz D. B., Federspiel W. J., and Wagner W. R., Biotechnol. Bioeng. 106(3), 490–500 (2010). 10.1002/Bit.22675 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kniazeva T., Hsiao J. C., Charest J. L., and Borenstein J. T., Biomed. Microdevices 13(2), 315–323 (2011). 10.1007/s10544-010-9495-1 [DOI] [PubMed] [Google Scholar]
- Hoganson D. M., Pryor H. I., Bassett E. K., Spool I. D., and Vacanti J. P., Lab Chip 11(4), 700–707 (2011). 10.1039/c0lc00158a [DOI] [PubMed] [Google Scholar]
- Kniazeva T., Epshteyn A. A., Hsiao J. C., Kim E. S., Kolachalama V. B., Charest J. L., and Borenstein J. T., Lab Chip 12(9), 1686–1695 (2012). 10.1039/c2lc21156d [DOI] [PMC free article] [PubMed] [Google Scholar]
- Figure modified from: http://www.medical-exam-essentials.com/respiratory-system-diagram.html.
- Skafte-Pedersen P., Sabourin D., Dufva M., and Snakenborg D., Lab Chip 9(20), 3003–3006 (2009). 10.1039/b906156h [DOI] [PubMed] [Google Scholar]
- Sabourin D., Snakenborg D., Skafte-Pedersen P., Kutter J. P., and Dufva M., in Proceedings of the Fourteenth International Conference on Miniaturized Systems for Chemistry and Life Sciences, 2010, pp. 1433–1435.
- See supplementary material at http://dx.doi.org/10.1063/1.4742911 for details about design and fabrication of the microfluidic system.
- Chen S.-H., Ruaan R.-C., and Lai J.-Y., J. Membr. Sci. 134(2), 143–150 (1997). 10.1016/S0376-7388(97)00132-4 [DOI] [Google Scholar]
- Ogonczyk D., Wegrzyn J., Jankowski P., Dabrowski B., and Garstecki P., Lab Chip 10(10), 1324–1327 (2010). 10.1039/b924439e [DOI] [PubMed] [Google Scholar]
- Holloway B. W., J. Gen. Microbiol. 13(3), 572–581 (1955). 10.1099/00221287-13-3-572 [DOI] [PubMed] [Google Scholar]
- Klausen M., Heydorn A., Ragas P., Lambertsen L., Aaes-Jorgensen A., Molin S., and Tolker-Nielsen T., Mol. Microbiol. 48(6), 1511–1524 (2003). 10.1046/j.1365-2958.2003.03525.x [DOI] [PubMed] [Google Scholar]
- Pamp S. J. and Tolker-Nielsen T., J. Bacteriol. 189(6), 2531–2539 (2007). 10.1128/JB.01515-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fercher A., Borisov S. M., Zhdanov A. V., Klimant I., and Papkovsky D. B., ACS Nano 5, 5499–5508 (2011). 10.1021/nn200807g [DOI] [PubMed] [Google Scholar]
- Fercher A., O’Riordan T. C., Zhdanov A. V., Dmitriev R. I., and Papkovsky D. B., Methods Mol. Biol. 591, 257–273 (2010). 10.1007/978-1-60761-404-3_16 [DOI] [PubMed] [Google Scholar]
- O’Donovan C., Hynes J., Yashunski D., and Papkovsky D. B., J. Mater. Chem. 15(27–28), 2946–2951 (2005). 10.1039/b501748c [DOI] [Google Scholar]
- O’Riordan T. C., Zhdanov A. V., Ponomarev G. V., and Papkovsky D. B., Anal. Chem. 79, 9414–9419 (2007). 10.1021/ac701770b [DOI] [PubMed] [Google Scholar]
- See http://mindstorms.lego.com/en-us/default.aspx for LEGO® Mindstorms® NXT 2.0.
- Mehta G., Mehta K., Sud D., Song J. W., Bersano-Begey T., Futai N., Heo Y. S., Mycek M.-A., Linderman J. J., and Takayama S., Biomed. Microdevices 9(2), 123–134 (2007). 10.1007/s10544-006-9005-7 [DOI] [PubMed] [Google Scholar]
- Papkovsky D. B., in Methods in Enzymology, edited by Chandan K. S. and Gregg L. S. (Academic, 2004), Vol. 381, pp. 715–735. [DOI] [PubMed] [Google Scholar]
- Yoon S. S., Hennigan R. F., Hilliard G. M., Ochsner U. A., Parvatiyar K., Kamani M. C., Allen H. L., DeKievit T. R., Gardner P. R., Schwab U., Rowe J. J., Iglewski B. H., McDermott T. R., Mason R. P., Wozniak D. J., Hancock R. E. W., Parsek M. R., Noah T. L., Boucher R. C., and Hassett D. J., Dev. Cell 3(4), 593–603 (2002). 10.1016/S1534-5807(02)00295-2 [DOI] [PubMed] [Google Scholar]
- Carenfelt C. and Lundberg C., Acta Otolaryngol. 84(1–2), 138–144 (1977). 10.3109/00016487709123952 [DOI] [PubMed] [Google Scholar]
- Odry B. L., Kiraly A. P., Novak C. L., Naidich D. P., Ko J., and Godoy M. C. B., Proc. SPIE 7260(1), 72603B–72608 (2009). 10.1117/12.811923 [DOI] [Google Scholar]
- Major T. A., Panmanee W., Mortensen J. E., Gray L. D., Hoglen N., and Hassett D. J., Antimicrob. Agents Chemother. 54(11), 4671–4677 (2010). 10.1128/AAC.00379-10 [DOI] [PMC free article] [PubMed] [Google Scholar]