Abstract
An easy method is introduced allowing fast polydimethylsiloxane (PDMS) replication of nanofluidic lab-on-chip devices using accurately fabricated molds featuring cross-sections down to 60 nm. A high quality master is obtained through proton beam writing and UV lithography. This master can be used more than 200 times to replicate nanofluidic devices capable of handling single DNA molecules. This method allows to fabricate nanofluidic devices through simple PDMS casting. The extensions of YOYO-1 stained bacteriophage T4 and λ−DNA inside these nanochannels have been investigated using fluorescence microscopy and follow the scaling prediction of a large, locally coiled polymer chain confined in nanochannels.
INTRODUCTION
Fluidics at the micron level is widely used. Further miniaturization down to the 100 nm level and below in at least one of the three dimensions introduces new phenomenon which has to be taken into account.1, 2, 3, 4 The charge selectivity is most pronounced if the Debye screening length is comparable to the smallest dimension of the nanochannel, leading to a predominantly counter-ion containing nanometer-sized aperture. These unique properties contribute to the charge-based partitioning of biomolecules at the micro/nanochannel interface. Additionally, at this free-energy barrier, size-based partitioning can be achieved when biomolecules and nano-constrictions have similar dimensions.5, 6
A considerable amount of research has been reported on nanofluidic phenomenon.7, 8, 9 Furthermore, nanopores and nanochannels are rooted in interesting physical concepts, and since these structures demonstrate sensitive, label-free, and real-time electrical detection of biomolecules, the technologies hold great promise especially in the area of life sciences.2, 10, 11 These developments have been made possible by the advances in micro/nanofabrication technologies.12, 13, 14 In review articles, the current state of the art in nanofluidic device fabrication and their applications are discussed.15, 16
Currently, most nanofluidics devices have typically only one critical dimension below 100 nm, also called nanoslits. Several groups have produced nanoslits in SiO2 for biophysical applications.5, 6, 17, 18, 19 The control of the Si etching requires skilled processing. Even more challenging is the fabrication of nanochannels with two critical dimensions below the 100 nm threshold; several groups have achieved this.2, 20 Because of the advanced processing involved in the fabrication of SiO2 nanofluidic lab on chip devices other groups have looked into alternative ways of producing nanofluidic chips.
Different fabrication techniques have been reported like thermal imprinting in poly(methyl methacrylate) (PMMA),21 electron beam writing in SU-8 followed by poly(dimethylsiloxane) (PDMS) casting,13 focused ion beam etching of SiO2 in combination with PDMS sealing of channels.22 Others have reported methods to fabricate nanofluidic systems in PDMS (Refs. 23, 24, 25) or alternative materials.26 Standard soft lithography uses molds (masters) created from silicon wafers patterned with a photoresist to produce disposable elastomeric replicas,27 most commonly from PDMS, ideally suited for high-throughput applications.
In single DNA molecule studies, a large amount of data is required for reliable analysis. Ideally, a fresh chip is used for every experiment, necessitating the demand for many chips. In standard soft lithography, the master does not support less than 200 nm cross sections,28, 29 the size was thought to be limited by the Young’s modulus of PDMS. Only recently a few groups have succeeded to fabricate PDMS nanoslits (i.e., one dimension at 100 nm) for DNA analysis.4, 30 Up to now only a few groups have managed to make triangular PDMS nanochannels with one dimension below 100 nm.31, 32
Proton beam writing (PBW) is a powerful accurate technique capable of writing details down to the sub 100 nm level in resist material, with sidewall smoothness down to the 2–3 nm root-mean-square level.33, 34, 35, 36 In earlier experiments, proton beam writing has been used to fabricate enclosed nanochannels in PMMA through a double bonding process.37, 38 This process yields high quality nanochannels but every chip has to be written by the proton beam. For DNA analysis at the single molecule level, many samples are required; therefore, we have developed an optimized method for the replication of PDMS nanofluidic chips through proton beam fabricated masters.39, 40
Here, we report the breakthrough fabrication of functional PDMS lab-on-chip devices with a rectangular cross-section down to 100 × 60 nm2 for single molecule handling. This size reduction has been made possible because of the high quality molds that are available through proton beam writing in hydrogen silsequioxane (HSQ).41 Our approach prevents collapse of the PDMS nanostructures reported by others.42 The size reduction in cross section of the nanochannel coupled with the ease of replication has given us the possibility to quickly fabricate several hundreds of nanofluidic devices, which are actively used for DNA stretching and compaction studies.43
MATERIALS AND METHODS
Mold fabrication
Proton beam writing is performed in the Centre for Ion Beam Applications at the National University of Singapore. In this facility, a 3.5 MV HVEE Singletron accelerator is coupled to a nanoprobe end-station, developed at CIBA.34 This system is capable of focusing proton beams down to 40 × 40 nm2 spot sizes using Oxford Microbeams high demagnification magnetic quadrupole lenses (OM52) in a high excitation triplet configuration. This lens system operates at an object distance of 7 m and a reduced image distance of 70 mm resulting in enhanced system demagnifications (228 × 60 in the X and Y directions, respectively). Resist coated Si wafers are mounted on a Burleigh Inchworm XYZ stage which has a travel of 25 mm for all axes with a 20 nm closed loop resolution. This system is routinely used for mask-less direct write nanolithography.41 During proton exposure, the beam is scanned over the resist in a digitized pattern using a set of electromagnetic scan coils, located directly in front of the quadrupole lens system. In this way, scan fields up to 0.5 × 0.5 mm2 can be achieved. The scan software supports AUTOCAD and Bitmap file format; more details about the scanning system can be found elsewhere.44
In the preparation of proton beam writing, Si wafers are pre-coated with Cr and Au. The metallic layer is applied to guarantee easy release of the PDMS after molding and to ensure good adhesion of the resist to the wafer. Next resist layers of HSQ (Dow Corning Co.); XR 1541 (60 and 105 nm thick layers) and FOX 12 (300 nm thick layer) are spin coated onto the Si wafers followed by a bake at 150 °C for 120 s. In the case of the 60 nm thick mold, 11 parallel channels with a width of 100 nm are written with the proton beam. In the 300 nm tall chip, 13 parallel lines are written; 5 with a width of 300 nm, 4 with a width of 200 nm, and 4 with a width of 150 nm. Through proton exposure the HSQ resist forms a cross-linked network that cannot be easily removed using chemicals like isopropyl alcohol and acetone. More complex shapes can be easily fabricated using PBW (Ref. 45) and will be tested in the future.
After proton beam exposure (Fig. 1a), the samples are developed in a 2.38% tetramethyl ammonium hydroxide solution for 60 s followed by a deionized water rinse (Fig. 1b), in this process the non-exposed parts are removed. The proton exposure was done at 1 MeV using a dose of 300 nC/mm2, the dose is chosen 2–3 times higher than normally required, this to guarantee long term structural integrity of the molds for the PDMS casting process.46
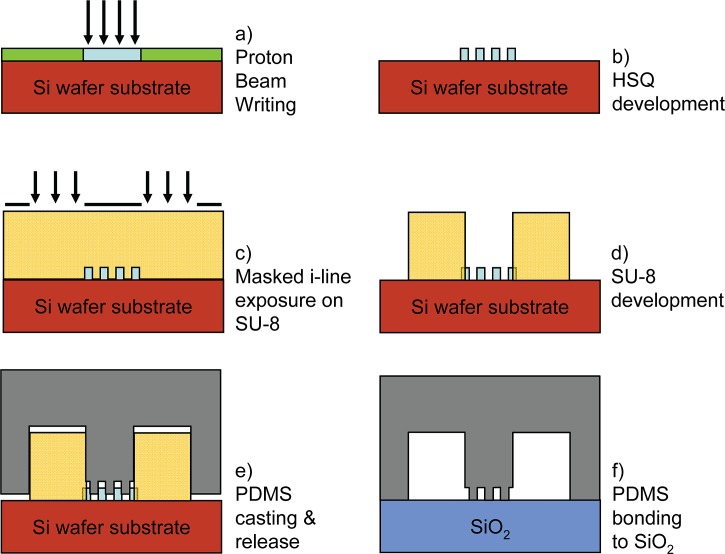
Figure 1.
Steps in the fabrication of PDMS nano fluidic lab on chip devices, (a)–(d) proton beam writing and UV lithography in HSQ and SU-8 respectively, (e)–(f) PDMS casting and bonding.
The final thickness of the HSQ structures is determined by AFM measurements and was 60, 105, and 300 nm for the coated layers. Next the samples are pre-heated at 150 °C for 2 min followed by spin coating at 2000 rpm for 30 s of a 5 μm thick SU-8 layer (SU-8 5, Micro Chem) and exposed using masked UV lithography (Figs. 1c, 1d). Due to the low optical adsorption of SU-8, the micro-channels can be aligned to the underlying nanostructures. Next, the SU-8 is developed in SU-8 developer (Micro Chem) and finally the molds containing nano and micro-structures are baked at 150 °C for 30 min to harden the molds.
PDMS chip replication
PDMS elastomer applied is SylgardTM 184 (Dow Corning Co.). The PDMS base and curing agent are mixed at a ratio of 10:1, and mildly blended to facilitate the mixing of the catalyst into the base polymer. The mixture is then degassed in a desiccator at a pressure of ∼1 Torr. The PDMS-catalyst mixture is poured onto the HSQ/SU-8 master and degassed again to draw out air bubbles trapped inside the PDMS. The pre-polymer is then allowed to cure by placing it in an oven for at least 4 h at 65 °C (see Fig. 1e). After manual delamination, the PDMS sample and a glass substrate are pretreated with an air RF plasma for 30 s at a pressure of ∼0.23 Torr (18 W). To form a reliable bond, the PDMS is immediately brought in contact with the glass substrate. After placing the two pieces together, the lab on chip device is heated on a hot plate at 85 °C for 60 s to improve the bond strength, see Fig. 1f. This plasma oxidation process not only provides good adhesion but also makes the hydrophobic PDMS surface hydrophilic. The bonded chips now have to be filled with buffer solution within 10 min in order not to lose hydrophilicity. Once in contact with buffer solutions the chips remain hydrophilic for tens of hours.47
Nanofluidic chip characterization
In this study, bacteriophage λ-DNA (48 502 bp; BioLabs) and T4 DNA (165 600 bp; Nippon Gene) are diluted from stock solution to a final concentration of 0.003 g/l, and incubated for at least 24 h at 4 °C to obtain a uniform distribution. The so-obtained DNA solution was dialyzed against 5× TBE (445 mM tris, 445 mM boric acid, and 10 mM EDTA, with pH of 8.5), the solution is now ready for application in the chips. DNA is visualized by epi-fluorescence of the bisintercalating dye YOYO-1 (Invitrogen, Carlsbad, CA) using a charged coupled (CCD) camera (Olympus DP 70) and an inverted microscope (Olympus IX71) equipped with a 100 W mercury lamp and a 100x oil immersion objective. At a staining ratio of 1 dye molecule per 23 base pairs, the influence of YOYO-1 bisintercalation is minimal, i.e., the persistence length changes by less than 1% and yet there is enough light intensity for imaging.39 YOYO-1-stained DNA molecules are loaded into the micro-channels via capillary action. Platinum electrode wires are connected to a high-voltage power supply (PowerPAC 300, BioRAD). The DNA molecules are subsequently driven into the nanochannel network by applying a 10–30 V potential across the two loading reservoirs. Once the DNA molecules were localized inside the channels, the electric field was switched off and the molecules were allowed to relax to their equilibrium state for at least 60 s. The extension of every DNA molecule was measured with the help of Image-J software.48
RESULTS
Mold characterization
A scanning electron microscopy (SEM) image obtained for the 300 nm tall nanofluidic channels connected to one of the SU-8 feeding channels is shown in Fig. 2a. The channels have a width of 150, 200, and 300 nm and length of 60 μm. Figure 2b shows an optical micrograph of two SU-8 micro-channel systems connected via a network of nano-channels of the 60 nm tall HSQ lines with a length of 30 μm. We confirmed the cross sections through electrical conductance measurements of a salt solution (1 M KCl, 10 mM TRIS/HCl, pH 8.0) along the nanochannels, following the method used by Chou et al.49
Figure 2.
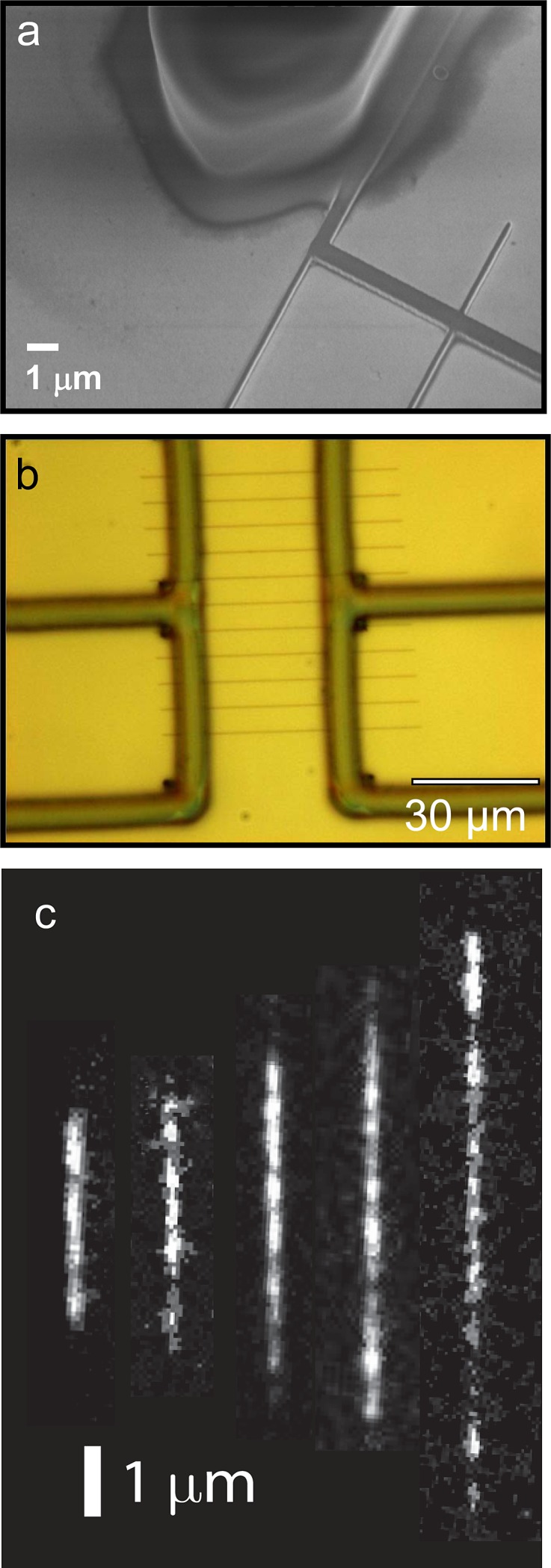
(a) SEM photograph of the HSQ and SU-8 mold featuring 300 nm tall HSQ lines. (b) Optical micrograph of two SU-8 micro-channel systems connected via a network of nano-channels of the 60 nm tall HSQ lines, with a length 30 μm. (c) Montage of fluorescence images of λ-DNA in 5x TBE buffer (pH 8.5). The channel cross-sections are 300 × 300, 200 × 300, 150 × 300, 100 × 105, 100 × 60 nm2 channels from left to right.
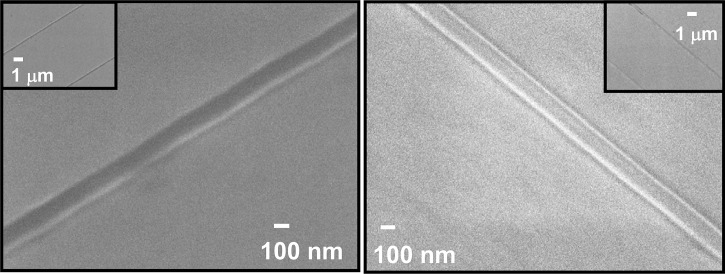
The fact that the proton beam produces vertical and smooth sidewalls guarantees easy delamination and structural integrity of the PDMS chips down to the 60 nm level for long nanochannels of at least 30 μm in length. The channels produced are of high quality providing a uniform cross-section along the channel, which guarantees an opening of the same size and allows DNA to enter under relatively high electric field (up to 1 V/μm for the 100 × 60 nm2, compared to 0.1 V/μm for the 300 × 300 nm2 channels). The integrated molds typically can be used to replicate high quality copies until the Si substrate breaks or the HSQ delaminates. Careful handling of the molds has allowed more than 200 replicas. No degradation of the 100 × 60 nm2 mold has been observed even after more than 200 times casting; see Figs. 3a, 3b, also no change in relative extension has been observed irrespective of the number of replicas made.
Figure 3.
High resolution SEM photograph of the 100 × 60 nm2 HSQ lines, inset showing a lower magnification image of two of the HSQ lines, (a) before PDMS casting, (b) after more than 200 times PDMS casting.
DNA stretching results
Figure 2c shows fluorescence images of single λ-DNA molecules confined in nanochannels with cross sections of 300 × 300, 200 × 300, 150 × 300, 100 × 105, and 100 × 60 nm2 channels from left to right. The expected extension of T4 DNA is on the order of the channel length for the 30 μm long 100 × 60 nm2 channels which renders measurement of the T4 extension in the current nanofluidic chip prone to errors. As cross-sectional diameters drop below the radius of gyration Rg of around 500 and 300 nm for T4 and λ-DNA, respectively, the relative extension of biopolymers is independent on contour length and depends solely on the local properties of the nanochannel. In our earlier research, the extensions of the DNA molecules were measured as a function of the ionic strength and composition of the buffer as well as the intercalation level by the YOYO-1 dye.39 From an applications point of view, the results obtained in channels with a cross-section of around 100 nm or less offer an opportunity to investigate the properties of biopolymers in relatively strong confinement.
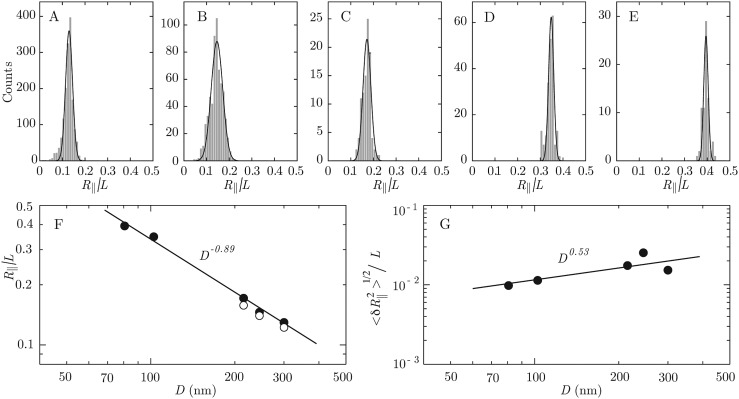
For the different channel cross-sections, the distributions in relative extensions (i.e., the measured extension along the channel divided by the contour length of the molecule) of many λ-DNA molecules are shown in Figs. 4a, 4b, 4c, 4d, 4e (for T4 DNA similar distributions are obtained). The distributions were obtained by using several casted PDMS chips. Typically, around 150 DNA molecules can be measured with a single chip in one experiment, limited by photo bleaching of the DNA and hydrophobicity of the chip. Next a new chip is casted and typically up to 8 chips are used to obtain the data for a channel with a certain cross-sectional diameter (i.e., for each panel Figs. 4a, 4b, 4c, 4d, 4e). The solid lines in the distributions denote Gaussian fits. We have verified that measurements of individual DNA molecules within the time frame of the camera (50 ms) yield the same distribution as obtained from many different molecules. Furthermore, we observed the same relative extension for different channels with the same width, replicated using the 300 nm tall mold. Multiple molds at a cross section of 300 × 300 nm2 have been fabricated which also yield the same relative extension. The fitted average values of the extensions and standard deviations resulting from the Gaussian fits to the distributions in Figs. 4a, 4b, 4c, 4d, 4e versus the geometric average of the cross-sections are shown in Figs. 4f, 4g, respectively. In the scaling laws for relative extension and width of the distribution based on Daoud and de Gennes blob model,50 the chain statistics is described as a linearly packed array of sub-coils (blobs) of uniform size. The extension in the longitudinal direction is then simplythe number of blobs times the blob size, which results in: , is the DNA extension; L is the contour length; D is the average channel diameter; Lp is the persistence length. Each blob fluctuates with a second moment of its distribution of mass on the order of the square of the channel diameter. For a string of independently fluctuating blobs, the second moment of the total extension is given by the square root of the number of blobs times the second moment of the blob size: .51, 52 We have verified that the prediction of the blob model agrees with the stretch of DNA obtained with Monte Carlo simulation within 6% for D > 2.5 Lp (there is perfect agreement for D > 5 Lp, unpublished results). Accordingly, the blob model is applicable for DNA confined in channels of larger tube diameters D exceeding, say, 125 nm (2.5 Lp). A fit of a power law including all channel sizes results in exponents −0.89 ± 0.04 and 0.5 ± 0.2, for the stretch and distribution width, respectively. The scaling exponents deviate from the predictions based on the blob model, which is related to its range of applicability to the wider channels only (a fit of the stretch to the wider channels with D > 200 nm yields an exponent of −0.8 ± 0.1). For smaller cross-sectional diameters, the values of extensions deviate from the scaling predictions based on the blob model. However, we do not cover the transition to Odijk’s deflection regime, where the chain is undulating inside the channel by deflection from the walls. This is because the geometric cross section of the narrowest channel (around 80 nm) exceeds the DNA bending persistence length of around 50 nm in the present buffer condition, which allows looping due to the formation of hairpins.53 However, we point out that the channels are in principle narrow enough to investigate this phenomenon, once the bending rigidity is increased by a decrease in ionic strength of the supporting medium.
Figure 4.
(a)–(e) Distributions in relative extension of λ-DNA in 5x TBE buffer (pH 8.5). The channel cross-sections are 300 × 300, 200 × 300, 150 × 300, 100 × 105, and 100 × 60 nm2 from A to E. The solid lines denote Gaussian fits. (f) Mean values of the relative extension. The closed symbols refer to λ-DNA, whereas the open symbols denote values pertaining to T4 DNA. The solid line represents a fit to the data. (g) As in panel (f), but for the width of the distribution.
CONCLUSIONS
We have developed a unique fabrication scheme for the production of PDMS lab-on-chip devices with details down to 60 nm using high quality molds HSQ molds. The replicated structures have high pattern fidelity, and no signs of mold degradation have been observed after more than 200 imprints, featuring details down to 60 nm.
The entropically induced stretching of YOYO-1 stained bacteriophage, T4 and λ-DNA inside these PDMS nanochannels was investigated. A Gaussian DNA extension distribution is obtained from the measurement of over 1000 independent DNA molecules. No difference in relative extension has been observed for chips of the same size. The standard deviation in the measured extension lengths of multiple confined DNA molecules is relatively large compared to the mean value, but agrees with fluctuation in extension on the order of the width of the channel. For larger channel diameters, the extensions follow the scaling prediction for the statistics of a large, locally coiled polymer chain confined in a channel. We do not cover the transition to the deflection regime, because the diameter of the narrowest channel is still about twice the DNA’s persistence length.
Although proton beam writing can achieve details down to the 20 nm level in HSQ, PDMS has a too low Young’s modulus to support details at this level. In order to achieve smaller nanochannels with high throughput nanoimprinting techniques or novel materials will have to be tested. From the present results, it is clear that inexpensive and disposable polymer biochips represent a competitive alternative to conventional fused silica based applications when it comes to investigating DNA molecules confined inside nanofluidic structures. Due to the high quality molds produced using proton beam writing, we are now able to replicate polymer biochips at the 60 nm level, previously deemed impossible due to the low Young’s modulus of PDMS. Using a simple imprint-and-bond scheme, it is now possible to produce PDMS lab-on-chip devices with details down to 60 nm at a large-scale.
ACKNOWLEDGMENTS
We acknowledge the financial support from ASTAR (Singapore) R-144-000-261-305 and the MOE Academic Research Fund R-144-000 270-112 as well as the AOARD (Asian Office of Aerospace Research and Development).
NOMENCLATURE
- AFM
Atomic force microscopy
- CCD
Charge coupled device
- EDTA
Ethylenediaminetetraacetic acid
- HSQ
Hydrogen silsesquioxane; Dow Corning Co.
- PBW
Proton beam writing
- PDMS
Polydimethylsiloxane
- PMMA
Poly(methyl methacrylate)
- SEM
Scanning electron microscopy
- SU-8
Photoresist; Micro Chem
- T4-DNA
165 600 bp; Nippon Gene
- TBE
Solution of Tris base, boric acid, EDTA, and water
- TRIS
(2,3-dibromopropyl) phosphate
- YOYO-1
Intercalating fluorescence dye
- λ-DNA
48 502 bp; BioLabs
References
- Schoch R. B., Han J., and Renaud P., Rev. Mod. Phys. 80, 839 (2008). 10.1103/RevModPhys.80.839 [DOI] [Google Scholar]
- Reisner W., Morton K. J., Riehn R., Wang Y. M., Yu Z. N., Rosen M., Sturm J. C., Chou S. Y., Frey E., and Austin R. H., Phys. Rev. Lett. 94, 196101 (2005). 10.1103/PhysRevLett.94.196101 [DOI] [PubMed] [Google Scholar]
- Riehn R., Lu M. C., Wang Y. M., Lim S. F., Cox E. C., and Austin R. H., Proc. Natl. Acad. Sci. U.S.A. 102, 10012 (2005). 10.1073/pnas.0503809102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jo K., Dhingra D. M., Odijk T., de Pablo J. J., Graham M. D., Runnheim R., Forrest D., and Schwartz D. C., Proc. Natl. Acad. Sci. U.S.A. 104, 2673 (2007). 10.1073/pnas.0611151104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han J. and Craighead H. G., Science 288, 1026 (2000). 10.1126/science.288.5468.1026 [DOI] [PubMed] [Google Scholar]
- Fu F., Schoch R. B., Stevens A. L., Tannenbaum S. R., and Han J., Nat. Nanotechnol. 2, 121 (2007). 10.1038/nnano.2006.206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S. J., Wang Y. C., Lee J. H., Jang H., and Han J., Phys. Rev. Lett. 99, 045501 (2007). 10.1103/PhysRevLett.99.045501 [DOI] [PubMed] [Google Scholar]
- Stone H. A., Stroock A. D., and Ajdari A., Annu. Rev. Fluid Mech. 36, 381 (2004). 10.1146/annurev.fluid.36.050802.122124 [DOI] [Google Scholar]
- Squires T. M. and Quake S. R., Rev. Mod. Phys. 77, 977–1026 (2005). 10.1103/RevModPhys.77.977 [DOI] [Google Scholar]
- Craighead H., Nature 442, 387 (2006). 10.1038/nature05061 [DOI] [PubMed] [Google Scholar]
- Min S. K., Kim W. Y., Cho Y., and Kim K. S., Nat. Nanotechnol. 6, 162 (2011). 10.1038/nnano.2010.283 [DOI] [PubMed] [Google Scholar]
- Lee C., Yang E. H., Myung N. V., and George T., Nano Lett. 3, 1339 (2003). 10.1021/nl034399b [DOI] [Google Scholar]
- Saleh O. A. and Sohn L. L., Nano Lett. 3, 37 (2003). 10.1021/nl0255202 [DOI] [Google Scholar]
- Stern M. B., Geis M. W., and Curtin J. E., J. Vac. Sci. Technol. B 15, 2887 (1997). 10.1116/1.589750 [DOI] [Google Scholar]
- Abgrall P. and Nguyen N. T., Anal. Chem. 80, 2326 (2008). 10.1021/ac702296u [DOI] [PubMed] [Google Scholar]
- Persson F. and Tegenfeldt J. O., Chem. Soc. Rev. 39, 985 (2010). 10.1039/b912918a [DOI] [PubMed] [Google Scholar]
- Hsieh C. C., Balducci A., and Doyle P. S., Nano Lett. 8, 1683 (2008). 10.1021/nl080605+ [DOI] [PubMed] [Google Scholar]
- Gu J., Gupta R., Chou C. F., Wei Q., and Zenhausern F., Lab Chip 7, 1198 (2007). 10.1039/b704851c [DOI] [PubMed] [Google Scholar]
- Bonthuis D. J., Meyer C., Stein D., and Dekker C., Phys. Rev. Lett. 101, 108303 (2008). 10.1103/PhysRevLett.101.108303 [DOI] [PubMed] [Google Scholar]
- Mannion J. T., Reccius C. H., Cross J. D., and Craighead H. G., Biophys. J. 90, 4538 (2006). 10.1529/biophysj.105.074732 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thamdrup L. H., Klukowska A., and Kristensen A., Nanotechnology 19, 125301 (2008). 10.1088/0957-4484/19/12/125301 [DOI] [PubMed] [Google Scholar]
- Campbell L. C., Wilkinson M. J., Manz A., Camilleri P., and Humphreys C. J., Lab Chip 4, 225 (2004). 10.1039/b312592k [DOI] [PubMed] [Google Scholar]
- Bellan L. M., Strychalski E. A., and Craighead H. G., J. Vac. Sci. Technol. B26, 1728 (2008). 10.1116/1.2975199 [DOI] [Google Scholar]
- Park S. M., Huh Y. S., Szeto K., Joe D. J., Kameoka J., Coates G. W., Edel J. B., Erickson D., and Craighead H. G., Small 6, 2420 (2010). 10.1002/smll.201000884 [DOI] [PubMed] [Google Scholar]
- Kim T. N., Campbell K., Groisman A., Kleinfeld D., and Schaffer C. B., Appl. Phys. Lett. 86, 201106 (2005). 10.1063/1.1926423 [DOI] [Google Scholar]
- De Marco C., Mele F., Camposeo A., Stabile R., Cingolani R., and Pisignano D., Adv. Mater. 20, 4158 (2008). 10.1002/adma.200703033 [DOI] [Google Scholar]
- Whitesides G. M., Ostuni E., Takayama S., Jiang X. Y., and Ingber D. E., Annu. Rev. Biomed. Eng. 3, 335 (2001). 10.1146/annurev.bioeng.3.1.335 [DOI] [PubMed] [Google Scholar]
- Viero Y., He Q., Mazenq L., Ranchon H., Fourniols J. Y., and Bancaud A., Microfluid. Nanofluid. 12, 465 (2011). 10.1007/s10404-011-0888-0 [DOI] [Google Scholar]
- Xu S. and Zhao Y., Microfluid. Nanofluid. 11, 359 (2011). 10.1007/s10404-011-0801-x [DOI] [Google Scholar]
- Yoon T. H., Hong L. Y., Lee C. S., and Kim D. P., J. Phys. Chem. Solids 69, 1325 (2008). 10.1016/j.jpcs.2007.10.135 [DOI] [Google Scholar]
- Huh D., Mills K. L., Zhu X., Burns M. A., Thouless M. D., and Takayama S., Nature Mater. 6, 424 (2007). 10.1038/nmat1907 [DOI] [PubMed] [Google Scholar]
- Park S. M., Huh Y. S., Craighead H. G., and Erickson D., Proc. Natl. Acad. Sci. 106, 15549 (2009). 10.1073/pnas.0904004106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watt F., Breese M. B. H., Bettiol A. A., and van Kan J. A., Mater. Today 10, 20 (2007). 10.1016/S1369-7021(07)70129-3 [DOI] [Google Scholar]
- van Kan J. A., Bettiol A. A., and Watt F., Appl. Phys. Lett. 83, 1629 (2003). 10.1063/1.1604468 [DOI] [Google Scholar]
- Whitlow H. J., Ng M. L., Auželyté V., Maximov I., Montelius L., van Kan J. A., Bettiol A. A., and Watt F., Nanotechnology 15, 223 (2004). 10.1088/0957-4484/15/1/040 [DOI] [Google Scholar]
- van Kan J. A., Sum T. C., Osipowicz T., and Watt F., Nucl. Instrum. Methods B161–163, 366 (2000). 10.1016/S0168-583X(99)00862-9 [DOI] [Google Scholar]
- Wang L. P., Shao P. G., van Kan J. A., Ansari K., Bettiol A. A., Pan X. T., Wohland T., and Watt F., Nucl. Instrum. Methods B260, 450 (2007). 10.1016/j.nimb.2007.02.062 [DOI] [Google Scholar]
- Shao P. G., van Kan J. A., Wang L. P., Ansari K., Bettiol A. A., and Watt F., Appl. Phys. Lett. 88, 093515 (2006). 10.1063/1.2181631 [DOI] [Google Scholar]
- Zhang C., Zhang F., van Kan J. A., and van der Maarel J. R. C., J. Chem. Phys. 128, 225109 (2008). 10.1063/1.2937441 [DOI] [PubMed] [Google Scholar]
- Zhang C., Shao P. G., van Kan J. A., and van der Maarel J. R. C., Proc. Natl. Acad. Sci. 106, 16651 (2009). 10.1073/pnas.0904741106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Kan J. A., Bettiol A. A., and Watt F., Nano Lett. 6, 579 (2006). 10.1021/nl052478c [DOI] [PubMed] [Google Scholar]
- Odom T. W., Thalladi V. R., Love J. C., and Whitesides G. M., J. Am. Chem. Soc. 124, 12112 (2002). 10.1021/ja0209464 [DOI] [PubMed] [Google Scholar]
- Zhang C., Gong Z., Guttula D., Malar P. P., van Kan J. A., Doyle P. S., and van der Maarel J. R. C., J. Phys. Chem. B 116, 3031 (2012). 10.1021/jp2124907 [DOI] [PubMed] [Google Scholar]
- Bettiol A. A., van Kan J. A., Sum T. C., and Watt F., Nucl. Instrum. Methods B181, 49 (2001). 10.1016/S0168-583X(01)00554-7 [DOI] [Google Scholar]
- van Kan J. A., Shao P. G., Wang Y. H., and Malar P. P., Microsyst. Technol. 17, 1519 (2011). 10.1007/s00542-011-1321-z [DOI] [Google Scholar]
- van Kan J. A., Zhang F., Zhang C., Bettiol A. A., and Watt F., Nucl. Instrum. Methods B266, 1676 (2008). 10.1016/j.nimb.2007.12.081 [DOI] [Google Scholar]
- Hong S. M., Kim S. H., Kim J. H., and Hwang H. I., J. Phys.: Conf. Ser. 34, 656 (2006). 10.1088/1742-6596/34/1/108 [DOI] [Google Scholar]
- Abramoff M. D., Magalhaes P. J., and Ram S. J., Biophotonics Int. 11, 36 (2004). [Google Scholar]
- Liang X., Morton K. J., Austin R. H., and Chou S. Y., Nano Lett. 7, 3774 (2007). 10.1021/nl072253x [DOI] [PubMed] [Google Scholar]
- Daoud M. and de Gennes P. G., J. Phys. (France) 38, 85 (1977). 10.1051/jphys:0197700380108500 [DOI] [Google Scholar]
- de Gennes P. G., Scaling Concepts in Polymer Physics (Cornell University Press, Ithaca, New York, 1979). [Google Scholar]
- van der Maarel J. R. C., Introduction to Biopolymer Physics (World Scientific, Singapore, 2008). [Google Scholar]
- Odijk T., Macromolecules 16, 1340 (1983). 10.1021/ma00242a015 [DOI] [Google Scholar]