Abstract
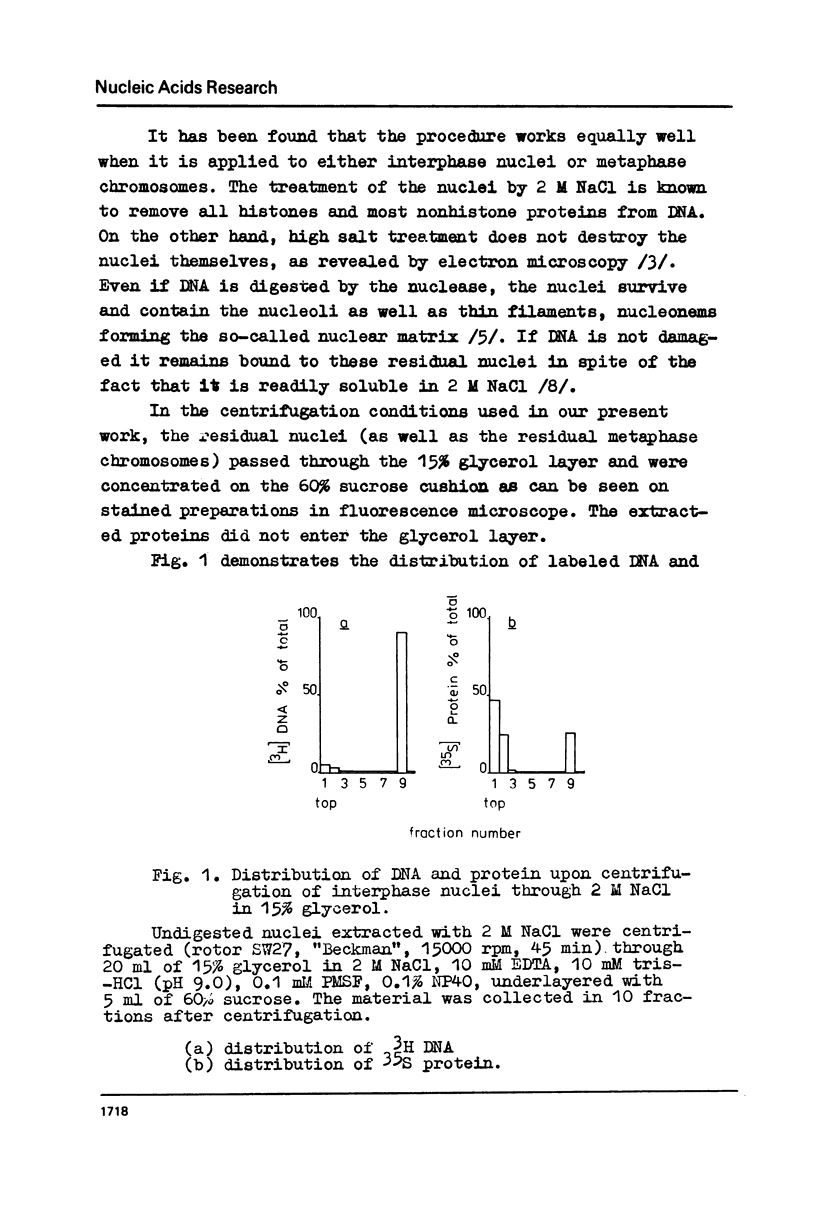
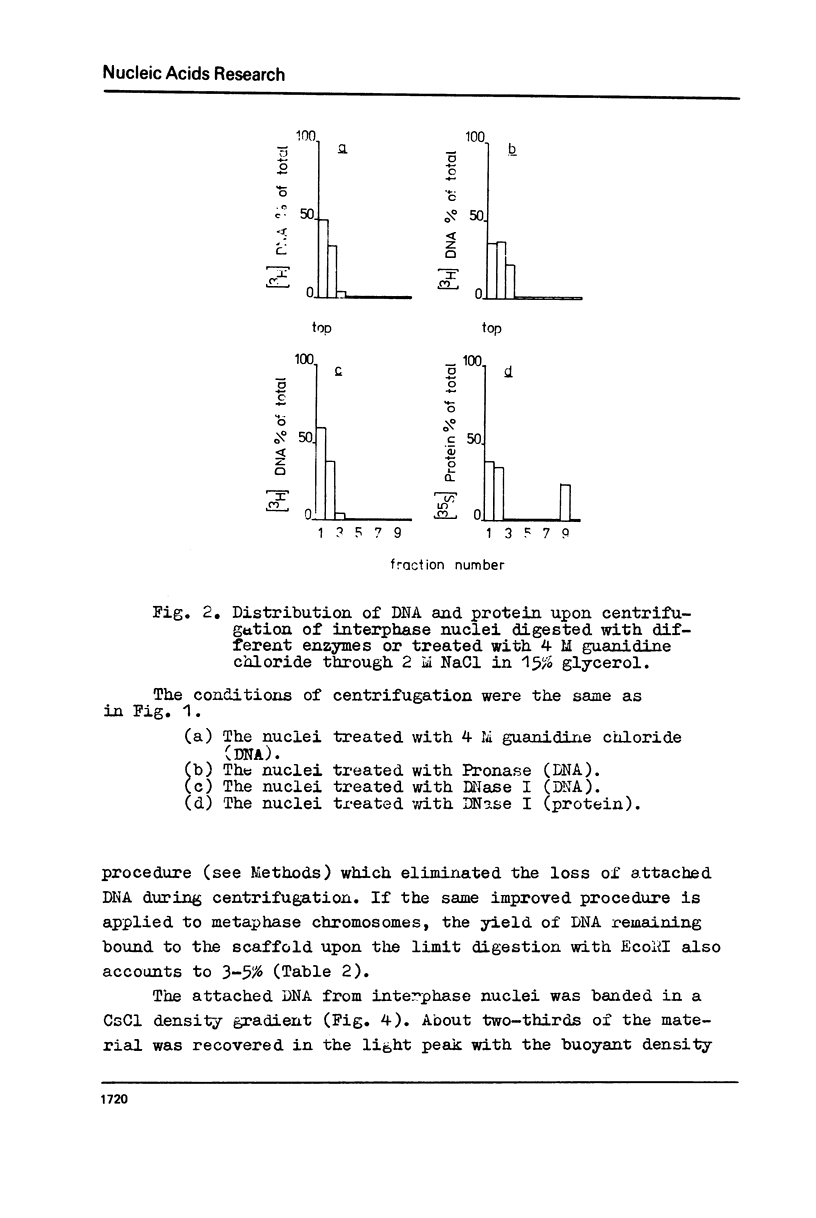
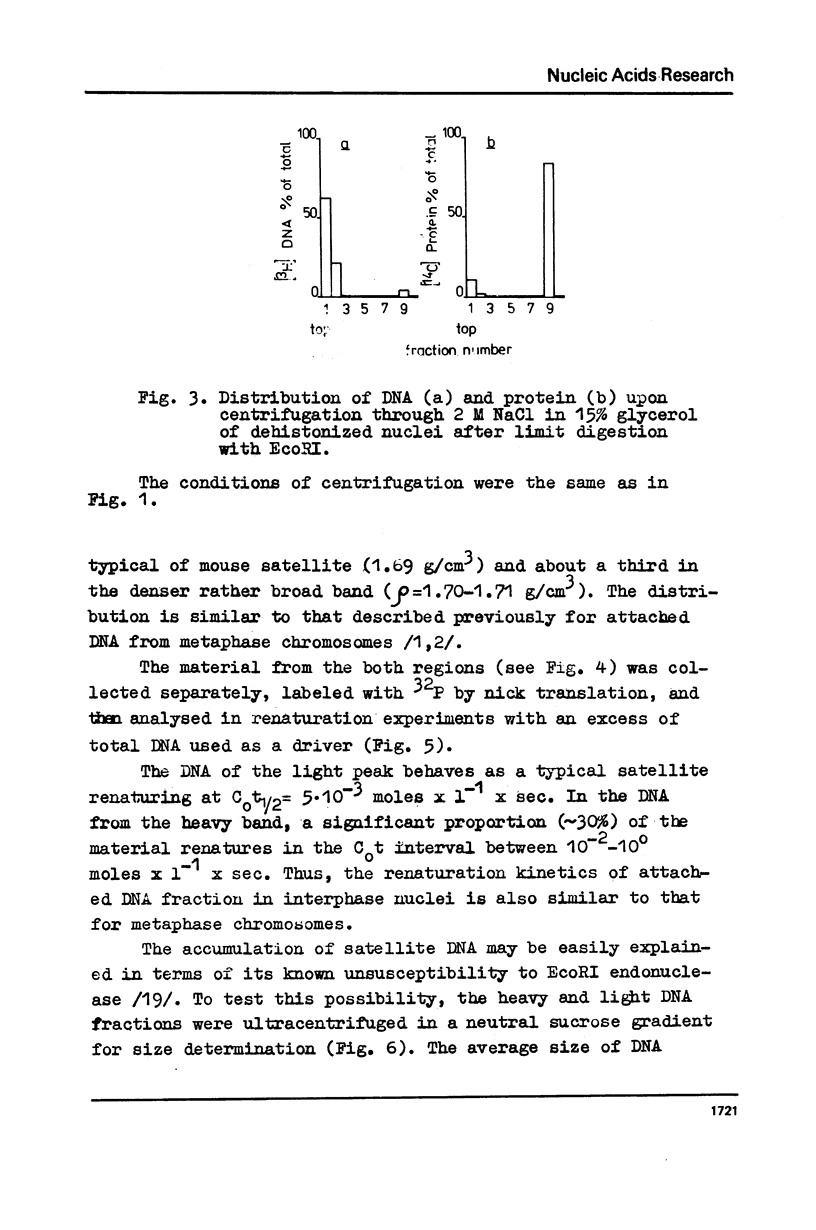
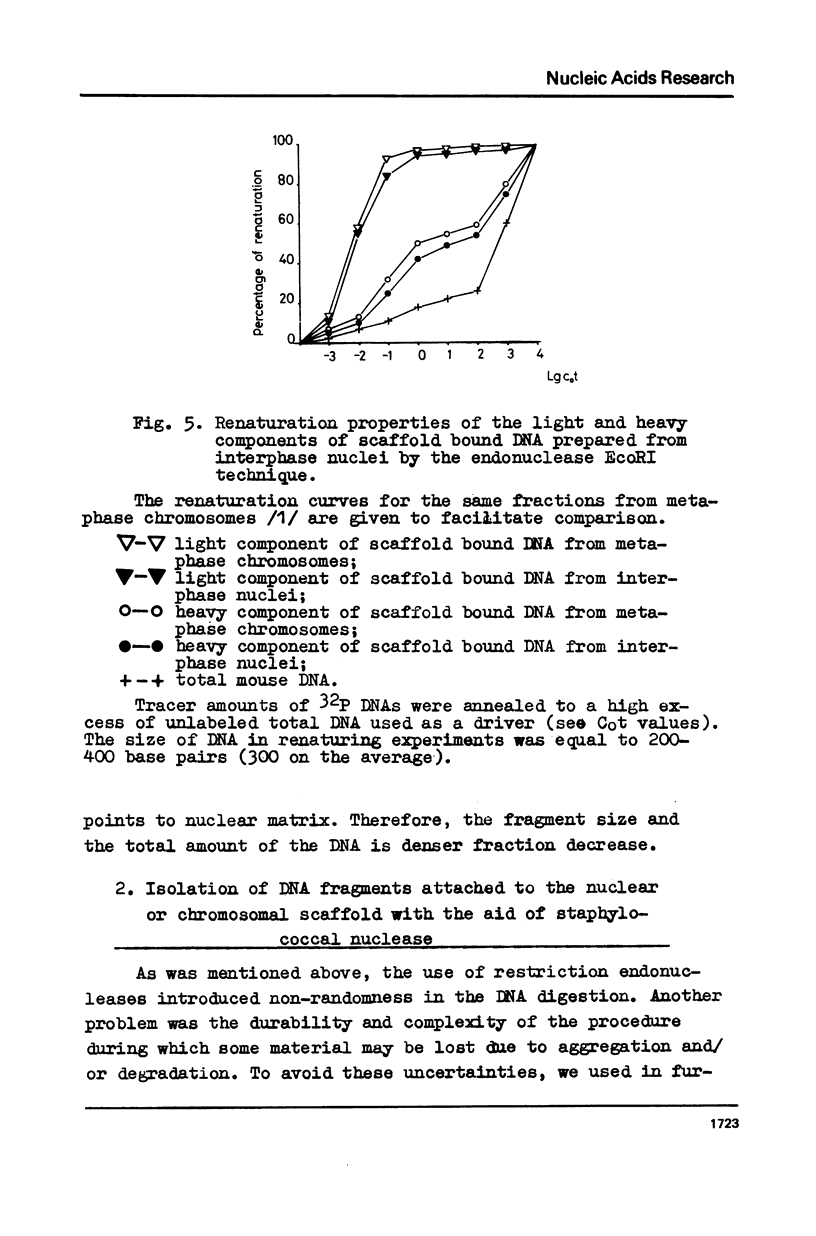
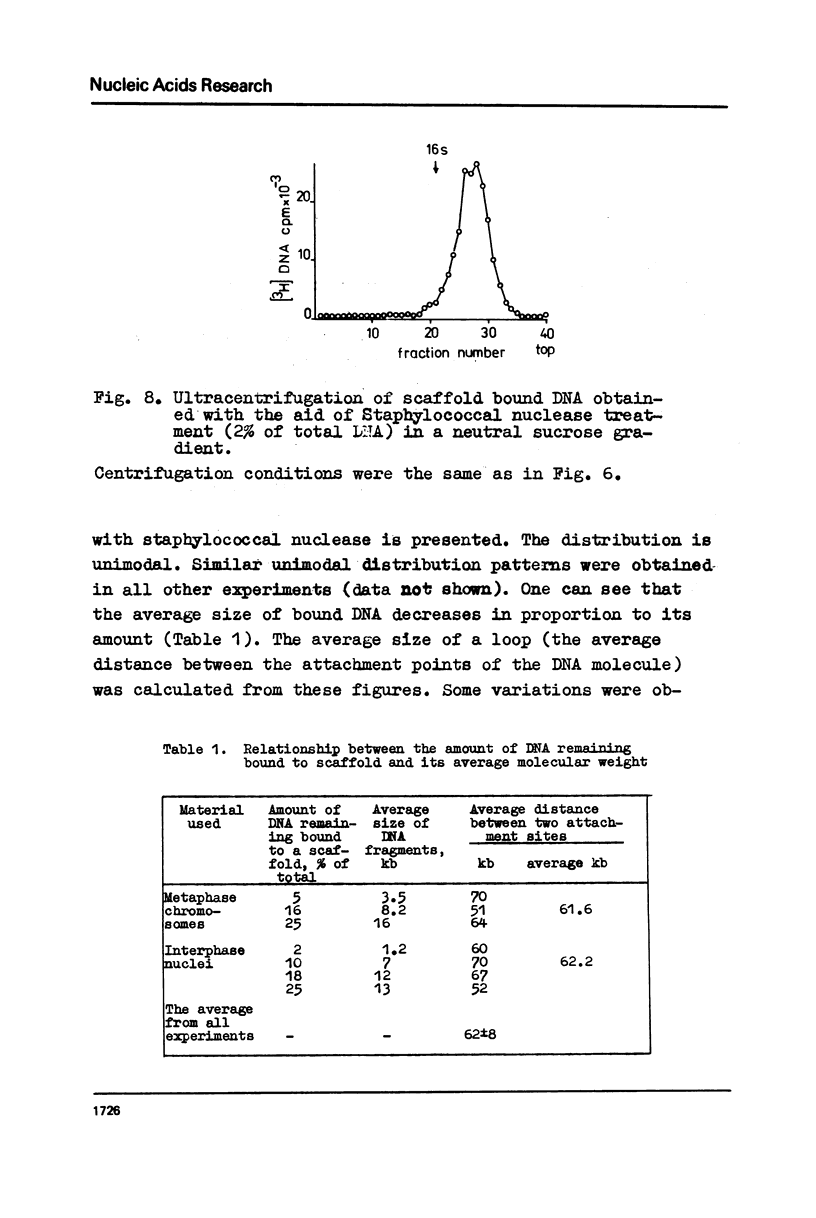
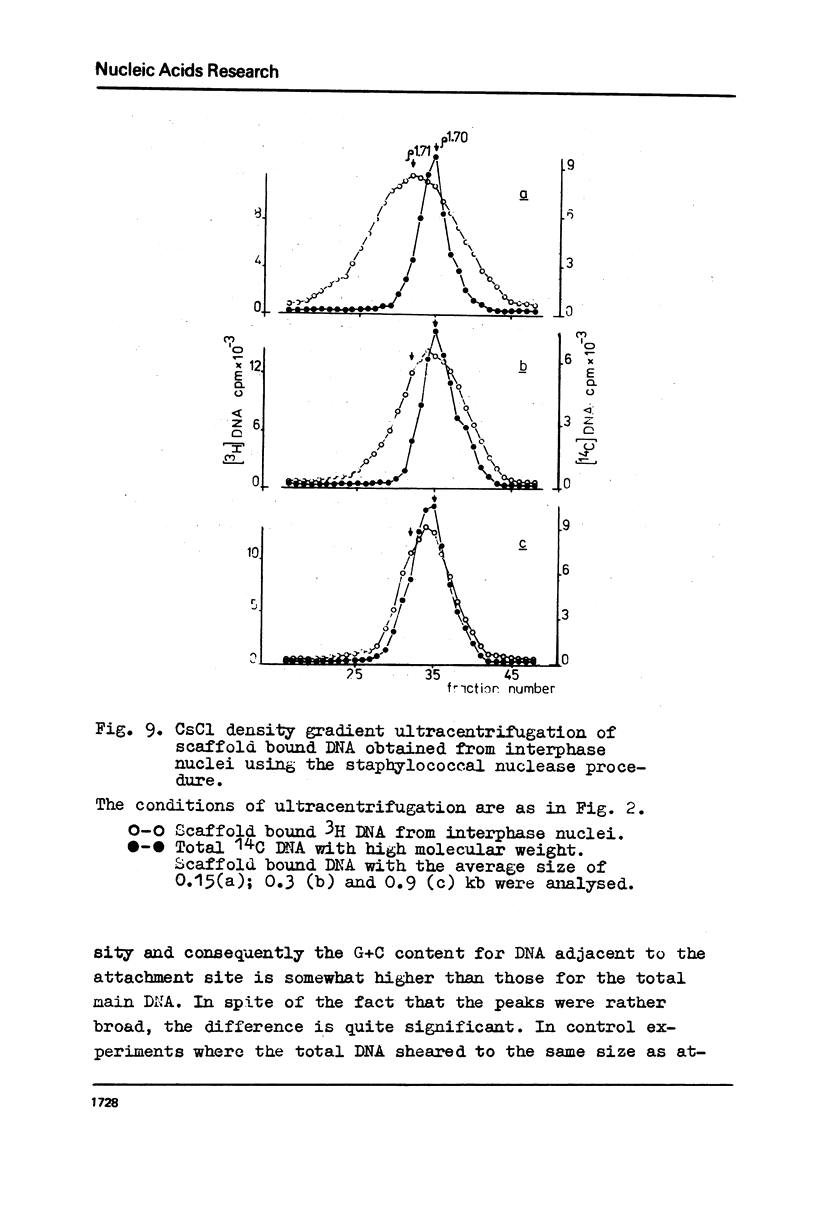
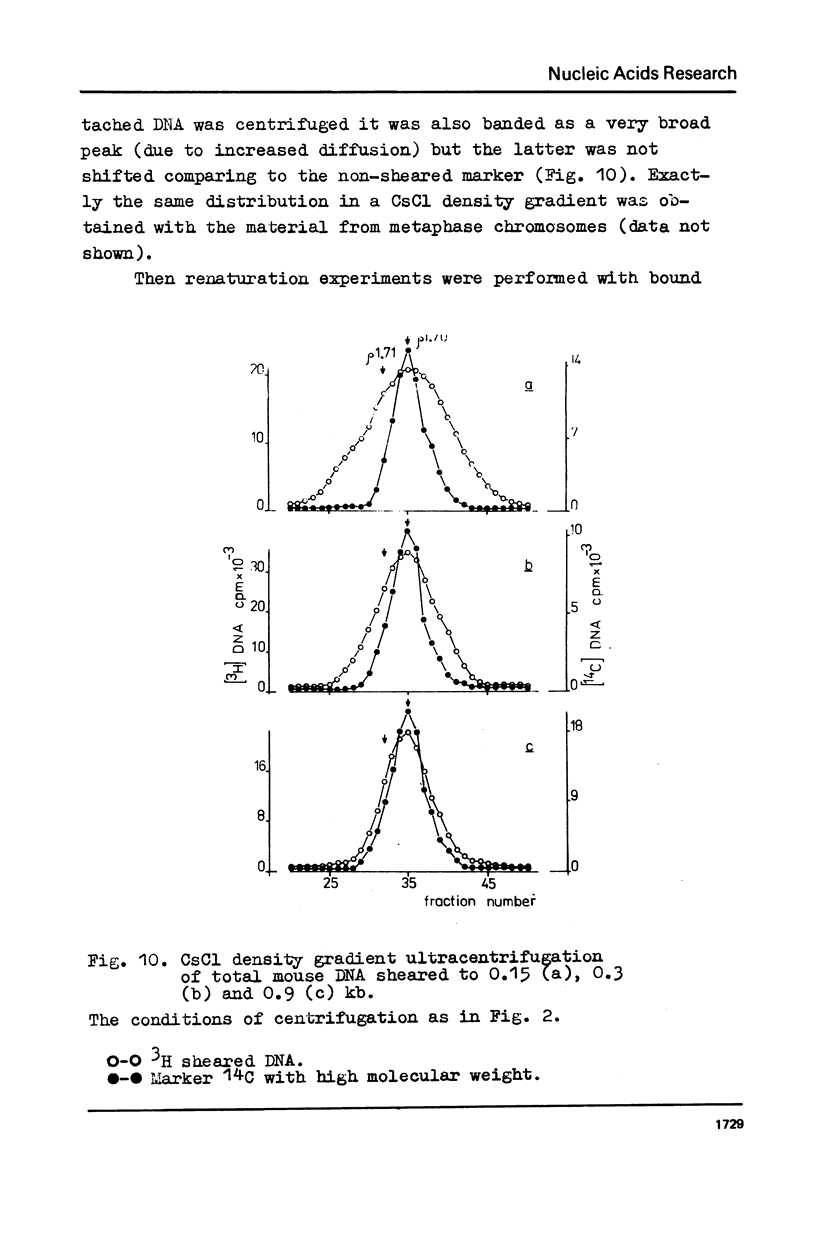
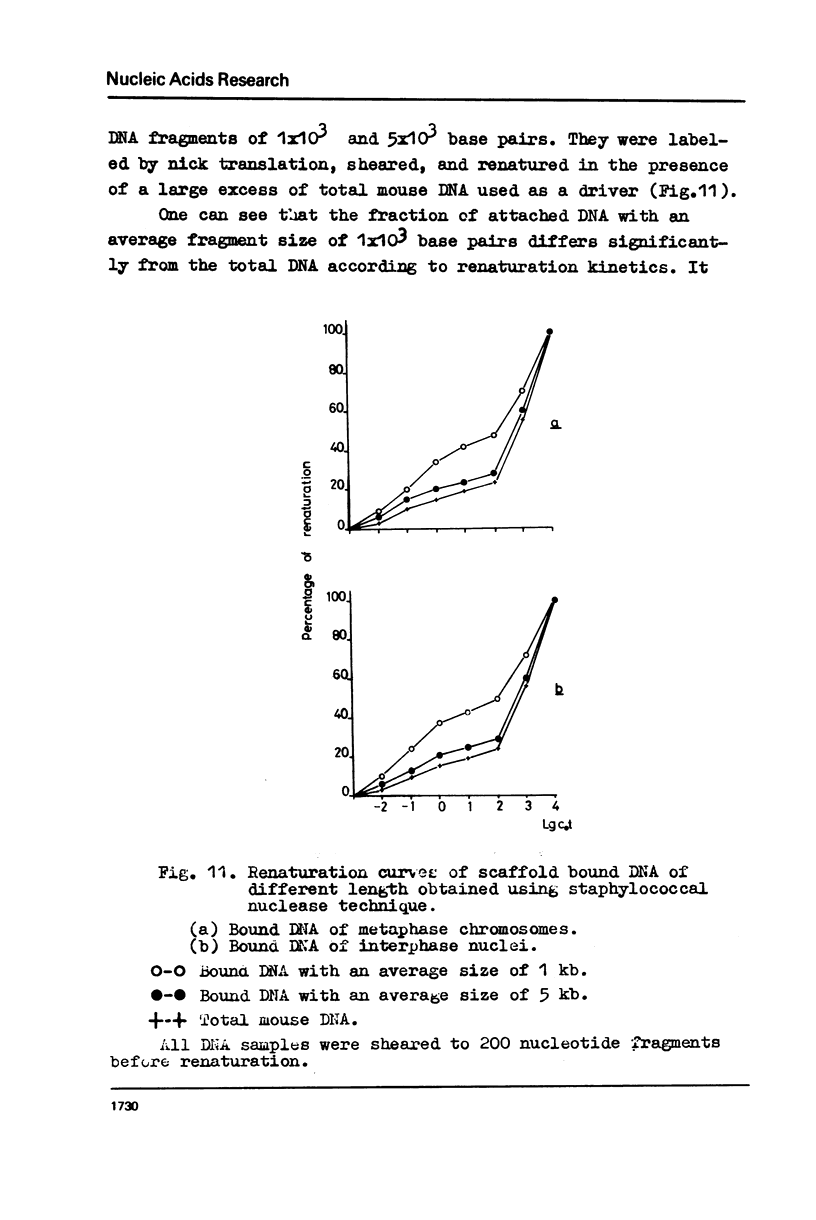
The fragments of DNA attached to protein skeleton of interphase nuclei or metaphase chromosomes were obtained. Both the method involving restriction endonuclease treatment/1,2/and a novel procedure based on mild staphylococcal nuclease digestion were used. In the latter case, DNA fragments remaining bound to nuclei or chromosomes are not enriched in satellite but only in abundant middle repetitive DNA. The shorter the fragments of attached DNA, the higher the content of middle repetitive DNA in the fraction. It has a slightly higher density in a CsCl gradient comparing to the main DNA. The yield of attached DNA, its distribution in a CsCl density gradient, and its renaturation properties are essentially the same for interphase and metaphase chromosomes. The average size of DNA loops was found to be equal to approximately 60 kb for both metaphase chromosomes and interphase nuclei. The conclusion has been drawn that the bulk of attachment sites of DNP fibrils to axial chromosomal structures remains unchanged during the cell cycle.
Full text
PDF














Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Adolphs K. W., Cheng S. M., Paulson J. R., Laemmli U. K. Isolation of a protein scaffold from mitotic HeLa cell chromosomes. Proc Natl Acad Sci U S A. 1977 Nov;74(11):4937–4941. doi: 10.1073/pnas.74.11.4937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benyajati C., Worcel A. Isolation, characterization, and structure of the folded interphase genome of Drosophila melanogaster. Cell. 1976 Nov;9(3):393–407. doi: 10.1016/0092-8674(76)90084-2. [DOI] [PubMed] [Google Scholar]
- Berezney R., Coffey D. S. Nuclear matrix. Isolation and characterization of a framework structure from rat liver nuclei. J Cell Biol. 1977 Jun;73(3):616–637. doi: 10.1083/jcb.73.3.616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Igó-Kemenes T., Zachau H. G. Domains in chromatin structure. Cold Spring Harb Symp Quant Biol. 1978;42(Pt 1):109–118. doi: 10.1101/sqb.1978.042.01.012. [DOI] [PubMed] [Google Scholar]
- Laemmli U. K., Cheng S. M., Adolph K. W., Paulson J. R., Brown J. A., Baumbach W. R. Metaphase chromosome structure: the role of nonhistone proteins. Cold Spring Harb Symp Quant Biol. 1978;42(Pt 1):351–360. doi: 10.1101/sqb.1978.042.01.036. [DOI] [PubMed] [Google Scholar]
- Razin S. V., Mantieva V. L., Georgiev G. P. DNA adjacent to attachment points of deoxyribonucleoprotein fibril to chromosomal axial structure is enriched in reiterated base sequences. Nucleic Acids Res. 1978 Dec;5(12):4737–4751. doi: 10.1093/nar/5.12.4737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- STUDIER F. W. SEDIMENTATION STUDIES OF THE SIZE AND SHAPE OF DNA. J Mol Biol. 1965 Feb;11:373–390. doi: 10.1016/s0022-2836(65)80064-x. [DOI] [PubMed] [Google Scholar]
- Wanka F., Mullenders L. H., Bekers A. G., Pennings L. J., Aelen J. M., Eygensteyn J. Association of nuclear DNA with a rapidly sedimenting structure. Biochem Biophys Res Commun. 1977 Jan 24;74(2):739–747. doi: 10.1016/0006-291x(77)90364-3. [DOI] [PubMed] [Google Scholar]


