Abstract
Background
To determine the association of the A55T and K153R polymorphisms of the Myostatin gene with obesity, abdominal obesity and lean body mass (LBM) in Asian Indians in north India.
Materials and Methods
A total of 335 subjects (238 men and 97 women) were assessed for anthropometry, % body fat (BF), LBM and biochemical parameters. Associations of Myostatin gene polymorphisms were evaluated with anthropometric, body composition and biochemical parameters. In A55T polymorphism, BMI (p = 0.04), suprailiac skinfold (p = 0.05), total skinfold (p = 0.008), %BF (p = 0.002) and total fat mass (p = 0.003) were highest and % LBM (p = 0.03) and total LBM (Kg) were lowest (p = 0.04) in subjects with Thr/Thr genotype as compared to other genotypes. Association analysis of K153R polymorphism showed that subjects with R/R genotype had significantly higher BMI (p = 0.05), waist circumference (p = 0.04), %BF (p = 0.04) and total fat mass (p = 0.03), and lower %LBM (p = 0.02) and total LBM [(Kg), (p = 0.04)] as compared to other genotypes. Using a multivariate logistic regression model after adjusting for age and sex, subjects with Thr/Thr genotype of A55T showed high risk for high %BF (OR, 3.92, 95% Cl: 2.61–12.41), truncal subcutaneous adiposity (OR, 2.9, 95% Cl: 1.57–6.60)] and low LBM (OR, 0.64, 95% CI: 0.33–0.89) whereas R/R genotype of K153R showed high risk of obesity (BMI; OR, 3.2, 95% CI: 1.2–12.9; %BF, OR, 3.6, 95% CI: 1.04–12.4), abdominal obesity (OR, 2.12, 95% CI: 2.71–14.23) and low LBM (OR, 0.61, 95% CI: 0.29–0.79).
Conclusions/Significance
We report that variants of Myostatin gene predispose to obesity, abdominal obesity and low lean body mass in Asian Indians in north India.
Introduction
Asian Indians are predisposed to develop the metabolic syndrome and type 2 diabetes (T2DM). Among other factors, a major factor contributing to metabolic risk is abnormal body composition. Asian Indians tend to have more abdominal adipose tissue, less lean body mass (LBM) and higher magnitude of insulin resistance (IR) despite falling in the normal range of body mass index (BMI) [1]. The high value of waist hip ratio in Asian Indians may be due to less lean mass of the hips and greater fat at the levels of waist [2]. Another study showed that Asian Indian men have low muscle mass and 30% more total body fat (BF) than other ethnic groups [3]. Low lean mass is also evident in Asian Indian neonates as compared to white Caucasian neonates [4]. Whether low muscle mass in Asian Indians is related to high IR and metabolic syndrome is not yet been adequately investigated. Finally, the reason for these characteristics of body composition has been often stated as due to the “genetic predisposition”, “ethnic factors”, and “chronic protein deprivation” but remains largely uninvestigated [5].
Muscle is the major target for insulin-stimulated glucose uptake, the key determinant of total body insulin sensitivity. Skeletal muscle is the major constituent of LBM and the major determinant of energy expenditure both at rest and during physical activity [6]. Skeletal muscle IR, due to decreased muscle glycogen synthesis, promotes atherogenic dyslipidemia by diverting energy derived from ingested carbohydrates away from muscle glycogen synthesis into increased hepatic de novo lipogenesis. These findings are important for understanding the mechanism by which skeletal muscle IR promotes the development of the IR, the metabolic syndrome and T2DM [7].
Myostatin [growth/differentiation factor-8 (GDF-8)], belongs to the transforming growth factor β super family of secreted proteins that control growth and differentiation [8]. Myostatin is expressed uniquely in human skeletal muscle as a 26 kDa mature glycoprotein and secreted into the plasma [8], [9]. Since the first report of human Myostatin gene in 1998 [9], six polymorphisms and one intronic mutation have been identified in the Myostatin gene [9], [10], [11].
A few studies are available regarding Myostatin gene and its functions in animals and humans. Zhiliang et al [12] reported that Myostatin gene in chickens not only plays an important role in controlling skeletal muscle growth and differentiation, but also appears to have some regulatory function on adipose tissue. In transgenic animal models, a lack of myostatin appears to reduce age-related sarcopenia and loss of muscle regenerative capacity [13]. In human studies, Ferrel et al. [14] studied A55T and K153R allelic variants in 49 Caucasians or African Americans, but did not find any significant impact of these polymorphisms in either races, however, a small effect could not be excluded. A loss-of-function mutation (G378A) in the Myostatin gene has been associated with muscle hypertrophy in the humans [15]. Gonzalez et al [16] studied 41 nonagenarians (33 women, age range, 90, 97 yrs) for A55T, E164K, I225T, K153R and P198A variants of Myostatin gene. According to these authors heterozygosity for the K153R polymorphism did not seem to exert any negative influence on the muscle phenotypes of women but homozygosity may do so. Further, Myostatin K153R polymorphism was investigated in 281 non-athletic young adults, where it was shown to be associated with the ability to produce ‘peak’ power during muscle contractions [17]. Overall, the genetic basis of muscle mass, and specifically the Myostatin gene, has been sparsely investigated in human beings.
We hypothesized that the A55T and K153R polymorphisms in the Myostatin gene may have an association with obesity, abdominal and truncal adiposity and LBM in non-diabetic Asian Indians. In this study, we assessed the polymorphic status of the Myostatin gene and its correlation with the measures of generalized and abdominal obesity and LBM in Asian Indians residing in north India.
Materials and Methods
Study subjects
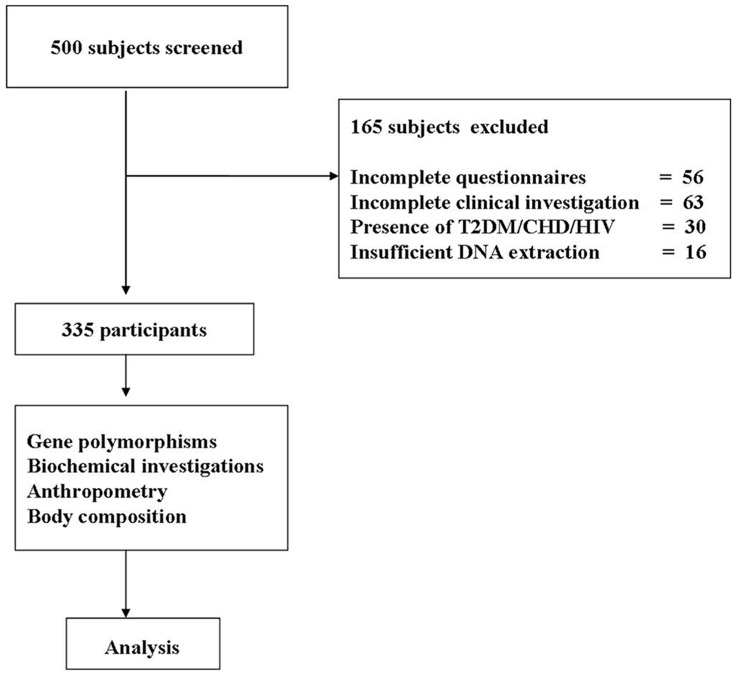
This cross-sectional population-based study was conducted in New Delhi (North India) from May 2006 to October 2011 and was approved by the institute ethics committees of the All India Institute of Medical Sciences, and Fortis hospital, Vasant Kunj, New Delhi, India. A total of 335 subjects (238, males; 97 females) were enrolled in this study after obtaining informed written consent. Subjects were randomly selected from residential colonies to have approximate representation from each income group (high income group∼10%, middle income group∼65–70%, and low income group∼15–20%) according to the proportion living in a metropolitan city. First, a list of total number of houses in each locality with the number of adult subjects in each household was obtained. Subsequently a random number list was generated to select the household that was approached for the participation in the study. Only one individual from one household was selected. Subjects with known T2DM, cardiovascular disease (CVD), severe end organ damage, HIV infection, pregnancy and lactation, were excluded from the study ( figure 1 ).
Figure 1. Flow diagram explaining the subject recruitment, selection and reasons for exclusion.
Clinical and anthropometric measurements
Body weight (to the nearest 0.1 kg) and height (to the nearest 0.1 cm) were recorded without shoes and allowing only light indoor clothes. BMI was calculated using the formula; weight (Kg)/height (m2). Circumferences of the waist, hip, mid-thigh, mid arm and neck were recorded to the nearest 0.1 cm. A mean of three readings of each circumference was taken for the calculation of waist-hip ratio. Biceps, triceps, sub-scapular, anterior axillary, supra-iliac, lateral thoracic and thigh skinfolds were measured by using Lange skinfold calipers as described previously [18]. Ratios of sub-scapular and triceps skinfold, and central (sum of sub-scapular and supra-iliac), peripheral skinfolds (sum of biceps and triceps) and sum of all four skinfolds (TSF) were calculated.
Biochemical analysis
Fasting insulin levels were measured using commercially available radioimmunoassay insulin kits (Immunotech, France) as described previously [19]. The intra- and inter-assay percentage variation was 2.1% and 3.3%.
Dual Energy X-ray Absorptiometry (DEXA)
Body composition was estimated by DEXA [QDR-2000; Hologic, Waltham, MA, USA] and analyzed using the software version 7.20 as previously described (1, 5). The subject was asked to lie on a padded table, provided with an X-ray generator below the table and a detector above, and was asked to remain still and breathe quietly for a short time while the scanner of the machine passed over the body. The data were collected at 0.5-cm intervals and reported in the standard format. Fat mass (FM), LBM, fat-free mass (FFM) and bone mineral content (BMC) of the whole body and at specific anatomical regions (trunk, arm and legs) were obtained. Total body fat (%BF) was calculated by dividing the FM by the sum of FM, LBM and BMC. Total LBM (%) was calculated by dividing the LBM by the sum of FM, LBM and BMC.
Genetic analyses
Genomic DNA was extracted from peripheral blood leukocytes by rapid non-enzymatic method [20]. Polymerase chain reaction and restriction fragment length polymorphism of the Myostatin gene (K153R) was performed by standard protocols [14]. DNA amplification of the Myostatin A55T polymorphism was performed using the forward and reverse primers (New England Biolab, MA, USA) 5′-CCTGTTTATGCTGATTGTTG-3′, 5′-TACTGA GGAT TTGT ACT TAATAG-3′ respectively. The PCR reaction contained 500 ng genomic DNA, 1× PCR buffer, 2.5 µM MgCl2, 200 µM dNTP ((dATP, dCTP, dGTP, and dGTP), 0.2 µM each primer, and 1.25 U Taq DNA polymerase (Sigma, St Louis, MO, USA) in a total volume of 25 µl. Initial denaturation was at 94°C for 5 min followed by 34 subsequent cycles of denaturation at 94°C for 1 minute, annealing at 55°C for 1 minute; extension at 72°C for 1 minute and final extension at 72°C for 10 minutes. The 154 bp PCR (10 µl) product was digested with AluI (New England BioLabs, Beverly, MA, USA) restriction enzyme (0.1 µl) at 37°C overnight. The restriction fragments (134 and 20) were electrophoresed in 20% polyacrylamide gels and were visualized using silver staining.
Statistical analysis
Data were recorded on a pre-designed excel sheet (Microsoft Corp, USA). The allelic and genotypic frequencies were determined by manual counting. Statistical analysis was performed using STATA Version 9 (Stata Corp, Texas, USA). After confirming the normality aspect of quantitative variables, descriptive statistics were computed using Mean ±SD and student t test. Difference between proportions was tested using Chi-square test. The influence of the genotype on the clinical parameters was estimated by the Analysis of covariance (ANCOVA) test with multiple comparisons. RFLP of Myostatin gene with its respective restriction sites was performed and studied for the genotypes. Allelic and genotypic frequencies were estimated by gene counting, and the observed and expected allele and genotype counts were compared by the Chi-square test to check for Hardy Weinberg Equilibrium. Significant levels for multiple comparisons were corrected by the Bonferroni method. Myostatin alleles and genotypes were checked for their association with the clinical, biochemical, anthropometric and body composition profiles. Forward stepwise logistical regression was applied, first adjusting for confounding factors (age, sex and BMI), and to see the independent effect of gene on phenotype variables. Multivariate logistic regression analysis was used for relative risk for obesity (BMI, %BF), abdominal obesity, truncal subcutaneous adiposity and low LBM. The odds ratio (OR) and 95% confidence interval were used as a measure of strength for the association between different A55T and K153R genotypic combinations. Statistical significance was established at a P value of 0.05.
Definitions
Overweight was defined as BMI≥23 kg/m2. WC cut-offs of >90 cm for males and >80 cm for females were considered an indicator of abdominal obesity. Other cutoffs were; FBG≧100 mg/dl, serum TG≧150 mg/dl, blood pressure≧130/85 mmHg and HDL-C; males≤40 mg/dl, and females≤50 mg/dl [21]. IR was measured by two surrogate measures: fasting hyperinsulinemia and HOMA-IR. Hyperinsulinemia was defined as values in the highest quartile as described previously [19]. The value of HOMA denoting IR was termed as HOMA-IR and was calculated as = {fasting insulin ( µU/ml) × fasting glucose (mmol/l)/22·5, [22]}.
Results
Clinical, biochemical, anthropometric and body composition profiles (table 1)
Table 1. Clinical, biochemical, anthropometric and body composition profiles.
| Variables | Total | Males | Females | p-value |
| Numbers (n) | 335 | 238 | 97 | |
| Age (yrs) | 38.0±6.9 | 38.2±7.0 | 38.0±6.9 | 0.2 |
| Body Mass Index (kg/m2) | 28.5±7.9 | 27.1±3.2 | 29.8±3.2 | 0.02 |
| Insulin (µU/ml) | 9.7±3.2 | 9.6±3.0 | 7.6±2.3 | 0.01 |
| HOMA | 2.2±1.02 | 2.2±0.98 | 1.9±0.8 | 0.04 |
| Circumferences (cm) | ||||
| Waist | 92.2±9.6 | 93.9±9.4 | 88.8±9.1 | 0.001 |
| Hip | 95.7±7.8 | 95.1±7.2 | 97.0±8.2 | 0.005 |
| Waist hip ratio | 0.96±0.1 | 0.97±0.1 | 0.86±0.1 | 0.001 |
| MTC | 54.6±8.1 | 57.2±7.7 | 48.4±8.4 | 0.001 |
| MAC | 27.8±4.7 | 27.1± 3.3 | 28.1±5.7 | 0.1 |
| Neck | 34.3±3.7 | 35.7 ± 4.2 | 31.2±3.1 | 0.01 |
| Skinfolds (mm) | ||||
| Biceps | 13.1±4.3 | 13.2±6.1 | 12.9±5.9 | 0.3 |
| Triceps | 20.6±7.1 | 18.2±7.0 | 25.2±7.1 | 0.001 |
| Subscapular | 39.9±10.8 | 42.8±11.1 | 33.2±9.7 | 0.001 |
| Anterior axillary | 10.6±3.7 | 11.0±4.0 | 8.2±3.4 | 0.05 |
| Suprailiac | 40.0±11.2 | 43.7±11.3 | 30.2±9.9 | 0.001 |
| Thigh | 38.9±9.2 | 39.8±9.3 | 37.3±8.8 | 0.05 |
| Lateral thoracic | 40.4±11.5 | 44.1±11.3 | 30.0±10.6 | 0.001 |
| Dual Energy X-ray Absorptiometry | ||||
| Total body fat (%) | 34.7±7.8 | 35.1±6.7 | 46.3±8.8 | 0.002 |
| Total body fat (kg) | 26.5±9.6 | 25.8±7.9 | 28.2±6.9 | 0.01 |
| Total lean mass (%) | 68.35±12.6 | 75.20±15.8 | 54.9±12.8 | 0.03 |
| Total lean mass (kg) | 46.2±8.5 | 49.4±8.2 | 39.0±7.7 | 0.002 |
| Total BMD (g cm−2) | 1.19±0.14 | 1.21±0.1 | 1.10±0 .1 | 0.001 |
Values are given as the mean ± standard deviation. HOMA-homoeostasis model assessment for insulin resistance. MTC, mid thigh circumference; MAC, mid arm circumference; BMD, bone mineral density. Statistical significance was established at a P value of 0.05.
The total number of subjects was 335 (238, males; 97 females: mean age: 38.2±7.0 years and 38.0±6.9 years, respectively). There were gender differences in BMI, other body composition parameters, biochemical profile and, insulin and HOMA-IR values.
Genotype distribution
In A55T polymorphism, the observed frequency of the Ala allele was 0.84 and of the Thr allele was 0.16. Overall, 74.93% subjects were Ala/Ala homozygous, 18.81% were Ala/Thr heterozygous and 6.27% were Thr/Thr homozygous. In K153R polymorphism, the observed frequency of the K allele was 0.84 and of the R allele was 0.16. Overall, 72.84% subjects were K/K homozygous, 22.9% were K/R heterozygous and 5.07% were R/R homozygous. The reproducibility of the genotyping data was checked by 75 duplicate samples.
Genotype association
Combined data for both genders were analyzed and presented in table 2 for each of the genotypes (Ala/Ala, Ala/Thr and Thr/Thr) of Ala55Thr. BMI (p = 0.04), suprailiac skinfold (p = 0.05), TSF (p = 0.008), %BF (p = 0.002) and total fat mass (p = 0.003) were highest and %LBM (p = 0.03) and total LBM (Kg) were lowest (p = 0.04) in subjects with Thr/Thr genotype as compared to other genotypes. Association analysis of K153R polymorphism (table 3) showed that R/R genotype was associated with significantly higher levels of BMI (p = 0.05), WC (p = 0.04), %BF (p = 0.04), total fat mass (p = 0.03), %LBM (p = 0.02) and total LBM (Kg) were lowest (p = 0.04) in subjects with R/R genotype as compared to other genotypes.
Table 2. Association of Myostatin (A55T) genotypes with clinical, biochemical, anthropometric and body composition profiles.
| Parameters | A/A (1) (n = 251) | A/T (2) (n = 63) | T/T (3) (n = 21) | p value (1&2) | p value (2&3) | p value (1&3) | p value (1, 2&3) |
| Age (yrs) | 37.5±8.7 | 38.5±9.9 | 37.0±6.8 | 0.4 | 0.5 | 0.6 | 0.6 |
| Body mass index (kg/m2) | 26.4±4.4 | 27.2±3.9 | 28.3±4.2 | 0.3 | 0.7 | 0.3 | 0.04 |
| Insulin (µU/ml) | 8.7±3.2 | 8.4±3.0 | 9.1±2.3 | 0.7 | 0.5 | 0.8 | 0.2 |
| HOMA | 2.1±1.02 | 2.2±0.98 | 2.3±0.8 | 0.7 | 0.9 | 0.5 | 0.5 |
| Circumferences (cm) | |||||||
| Waist | 91.4±12.0 | 93.4±11.5 | 94.4±10.8 | 0.2 | 0.4 | 0.4 | 0.3 |
| Hip | 95.2±9.7 | 96.7±8.7 | 97.2±7.9 | 0.8 | 0.5 | 0.7 | 0.3 |
| WH-R | 0.96±0.09 | 0.97±0.08 | 0.97±0.05 | 0.6 | 0.6 | 0.8 | 0.3 |
| Mid thigh | 54.2±10.4 | 55.3±9.7 | 56.4±12.2 | 0.4 | 0.7 | 0.5 | 0.4 |
| Mid arm | 27.1±3.3 | 27.9±3.3 | 28.1±5.7 | 0.9 | 0.6 | 0.4 | 0.3 |
| Neck | 35.7±4.2 | 37.1±3.1 | 33.2±3.1 | 0.09 | 0.08 | 0.2 | 0.5 |
| Skinfolds thickness (mm) | |||||||
| Biceps | 17.0±8.1 | 17.6±9.4 | 19.4±7.5 | 0.7 | 0.4 | 0.6 | 0.2 |
| Triceps | 20±9.3 | 21.1±9.1 | 21.5±6.4 | 0.6 | 0.4 | 0.4 | 0.2 |
| Subscapular | 39.1±8.6 | 41.2±8.9 | 40.1±10.2 | 0.1 | 0.3 | 0.4 | 0.09 |
| Suprailiac | 38.2±13.9 | 41.3±10.3 | 45.6±11.5 | 0.1 | 0.06 | 0.04 | 0.05 |
| Lateral thoracic | 39.3±11.2 | 40.4±12.3 | 41.1±9.5 | 0.5 | 0.7 | 0.9 | 0.2 |
| TSF | 101.1±30.6 | 104.1±30.9 | 126.7±24.9 | 0.06 | 0.009 | 0.001 | 0.008 |
| Dual Energy X-ray Absorptiometry | |||||||
| Body fat (%) | 35.1±6.7 | 36.3±8.8 | 45.2±9.8 | 0.4 | 0.02 | 0.001 | 0.002 |
| Body fat (%) | 35.1±6.7 | 36.3±8.8 | 45.2±9.8 | 0.4 | 0.02 | 0.001 | 0.002 |
| Total lean mass (%) | 72.2±12.3 | 65.2±11.8 | 51.9±10.8 | 0.02 | 0.01 | 0.002 | 0.03 |
| Total lean mass (kg) | 49.6±8.2 | 46.8±7.7 | 45.6±9.6 | 0.6 | 0.7 | 0.06 | 0.04 |
| Total BMD (g cm−2) | 1.21±0.15 | 1.21±0.08 | 1.23±0.09 | 0.9 | 0.6 | 0.6 | 0.8 |
All values are given as mean ± standard deviation; n, number of subjects. Due to multiple comparisons within each genotype, a Bonferroni correction was applied. Statistical significance was established at a P value of 0.05. HOMA-homoeostasis model assessment; WH-R, Waist-to-hip ratio; TSF, total skinfolds (sum of four skinfold thickness); BMD, bone mineral density.
Table 3. Association of Myostatin (K153R) genotypes with clinical, biochemical, anthropometric and body composition profiles.
| Parameters | K/K (1) (n = 244) | K/R (2) (n = 74) | R/R (3) (n = 17) | p value (1&2) | p value (2&3) | p value (1&3) | p value (1, 2&3) |
| Age (yrs) | 37.0±6.9 | 37.5±8.05 | 38.5±5.1 | 0.9 | 0.7 | 0.6 | 0.5 |
| Body mass index (kg/m2) | 24.4±2.8 | 25.5±3.9 | 28.2±4.6 | 0.7 | 0.06 | 0.05 | 0.05 |
| Insulin (µU/ml) | 7.6±4.2 | 7.7±5.5 | 8.0±2.7 | 0.6 | 0.5 | 0.8 | 0.1 |
| HOMA | 1.9±1.02 | 2.4±0.88 | 2.3±0.8 | 0.07 | 0.4 | 0.08 | 0.07 |
| Circumferences (cm) | |||||||
| Waist | 90.4±13.0 | 91.3±9.8 | 96.4±10.1 | 0.4 | 0.05 | 0.03 | 0.04 |
| Hip | 96.2±9.2 | 97.2±8.7 | 98.4±8.1 | 0.6 | 0.8 | 0.4 | 0.3 |
| WH-R | 0.97±0.09 | 0.98±0.08 | 0.98±0.05 | 0.7 | 0.6 | 0.4 | 0.8 |
| Mid thigh | 54.2±10.4 | 55.3±9.7 | 56.4±12.2 | 0.3 | 0.6 | 0.2 | 0.4 |
| Mid arm | 26.3±3.2 | 27.3±3.2 | 28.4±5.6 | 0.1 | 0.7 | 0.7 | 0.5 |
| Neck | 34.7±4.1 | 36.2±3.1 | 36.8±3.1 | 0.9 | 0.4 | 0.6 | 0.7 |
| Skinfolds thickness (mm) | |||||||
| Biceps | 16.9±7.1 | 17.8±9.4 | 18.9±7.5 | 0.6 | 0.5 | 0.6 | 0.9 |
| Triceps | 21±8.3 | 21.4±8.9 | 21.7±7.4 | 0.6 | 0.3 | 0.7 | 0.3 |
| Subscapular | 39.5±8.7 | 40.5±8.9 | 41.7±9.7 | 0.3 | 0.6 | 0.4 | 0.09 |
| Suprailiac | 39.3±11.9 | 41.3±10.3 | 41.8±9.5 | 0.09 | 0.5 | 0.07 | 0.08 |
| Lateral thoracic | 39.3±11.2 | 41.2±12.3 | 41.8±9.5 | 0.4 | 0.7 | 0.3 | 0.4 |
| TSF | 103.1±23.6 | 105.1±28.9 | 107±24.9 | 0.5 | 0.1 | 0.09 | 0.9 |
| Dual Energy X-ray Absorptiometry | |||||||
| Body fat (%) | 36.4±7.1 | 38.5±7.9 | 43.6±8.8 | 0.4 | 0.05 | 0.03 | 0.04 |
| Body fat (%) | 24.9±8.0 | 26.8±7.3 | 30.6±7.8 | 0.3 | 0.05 | 0.02 | 0.03 |
| Total lean mass (%) | 75.7±13.4 | 63.9±11.3 | 49.5±10.3 | 0.01 | 0.04 | 0.01 | 0.02 |
| Total lean mass (kg) | 51.6±9.2 | 45.6±7.3 | 44.4±9.1 | 0.2 | 0.2 | 0.05 | 0.04 |
| Total BMD (g cm−2) | 1.25±0.15 | 1.26±0.08 | 1.28±0.1 | 0.5 | 0.4 | 0.6 | 0.8 |
All values are given as mean ± standard deviation; n, number of subjects. Due to multiple comparisons within each genotype, a Bonferroni correction was applied. Statistical significance was established at a P value of 0.05. HOMA-homoeostasis model assessment; WH-R, Waist-to-hip ratio; TSF, total skinfolds (sum of four skinfold thickness); BMD, bone mineral density.
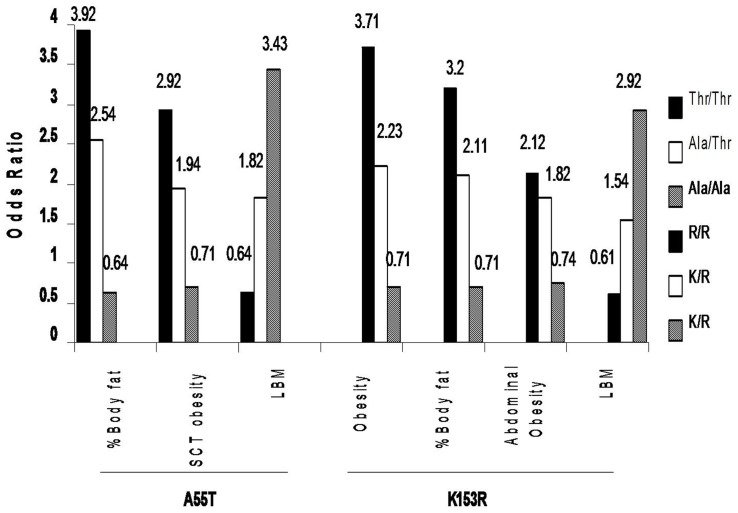
Logistic regression analysis (figure 2)
Figure 2. Odds Ratios for obesity, abdominal obesity, subcutaneous adiposity and LBM according to Myostatin (A55T & K153R) genotypes.
SCT Obesity, subcutaneous obesity; LBM, lean body mass.
Using a multivariate logistic regression model after adjusting for age, sex and BMI, subjects with Thr/Thr genotype of A55T showed high risk for high %BF (OR, 3.92, 95% Cl: 2.61–12.41), truncal subcutaneous adiposity [(based on subscapular skinfold thickness) (OR, 2.9, 95% Cl: 1.57–6.60)] and low LBM (OR, 0.64, 95% Cl: 0.33–0.89) whereas R/R genotype of K153R showed high risk of obesity(BMI; OR, 3.2, 95% CI: 1.2–12.9; %BF, OR, 3.6, 95% CI: 1.04–12.4), abdominal obesity [(based on WC) (OR, 2.12, 95% CI: 2.71–14.23)], and low LBM (OR, 0.61, 95% Cl: 0.29–0.79).
Discussion
In the present study, K153R and A55T polymorphisms of the Myostatin gene was associated with generalized obesity, abdominal obesity and low LBM in non-diabetic Asian Indians. Genetic predisposition to develop excess adiposity and low muscle mass in Asian Indians are important findings, since these may explain the high prevalence of hyperinsulinemia and greater risk for T2DM in this ethnic group [23].
Considering that low muscle mass in Asian Indians may determine insulin resistance, we previously showed that three months of supervised progressive resistance exercises for skeletal muscles significantly improved insulin sensitivity and glycemia and decreased truncal and peripheral subcutaneous adipose tissue in Asian Indians with T2DM [1], [23]. Further, Unni et al. [24] have reported that %BF was negatively associated with insulin sensitivity, whereas muscle mass was positively associated with insulin sensitivity in Asian Indian men. Possible reasons for low LBM in Asian Indians remains unclear, and genetic susceptibility remains uninvestigated. Since Asian Indians have been exposed frequently to chronic protein deficiency due to exposure to famines, food shortage, and vegetarian food, it is likely that these factors may have affected the skeletal muscle mass.
The functional role of myostatin in controlling muscle mass has been well documented, however the mechanism by which myostatin controls the muscle fiber number is poorly understood. Myostatin enters the bloodstream as a latent precursor protein and then undergoes a proteolytic process to become a mature peptide that binds to extracellular activin type II receptor (ActRIIB) [25]. Binding of myostatin to ActRIIB induces intracellular activation of Smad proteins; through this pathway, myostatin modulates myoblast proliferation and differentiation [26], and thus ultimately muscle mass. Jiang et al [27] showed that the inhibitory domain was located in the region between 42–115 amino acid sequence of myostatin precursor and exerts an important role in the stability of the GDF-8 propeptide inhibitory activity on myostatin activity. Therefore the A55T polymorphism could affect GDF8 activity.
Knowledge about negative regulation of skeletal muscle mass by myostatin was based on loss-of-function models, where myostatin expression and activity was blocked from gestation all throughout life by spontaneous or experimental mutations [26]. The Lys(K)153Arg(R) amino acid replacement was found within the active mature peptide of the myostatin protein and theoretically influenced (a) proteolytic processing with its propeptide (b) affinity to bind with ActRIIB [28]. Reisz et al, [29] reported that increased expression of myostatin in skeletal muscle is associated with lower muscle mass and decreased fiber size and myonuclear number, decreased cardiac muscle mass, and increased fat mass in male mice, consistent with its role as an important inhibitor of skeletal muscle mass, in addition to its influences on adipose tissue. Naturally occurring myostatin mutations in cattle have been shown to lead to pronounced hyper muscularity [26].
Data suggest an important role of the myostatin gene in muscle anatomy. McFarlane et al [30], suggested that myostatin regulation of postnatal myogenesis involves interactions with numerous downstream signaling mediators (MyoD, via canonical Smad signaling), and notch signaling pathway during inhibition of human myoblast differentiation. Xiao et al [31] reported that the negative role of myostatin during muscle regeneration and the increased expression of myostatin observed in Smad3-null muscle may contribute to the regeneration defects. Sartori et al [32] showed that the muscle fiber atrophy induced by Smad2/3 activation was prevented by the presence of constitutively active Akt. He also suggested that muscle fibers transfected with a gene encoding dominant negative activin receptor 2B (which inhibits myostatin signaling) are ∼30% larger than normal fibers; this hypertrophy occurs without recruitment of new myonuclei, is diminished by blocking mTOR activity with either rapamycin or mTOR siRNAs, and is enhanced in transgenic mice with constitutively active Akt. Finally, during embryogenesis, myostatin is exclusively expressed in skeletal muscle to control the differentiation and proliferation of the myoblast [8].
Some studies in human beings have suggested that Myostatin gene has effects on muscle mass and function. A loss-of-function mutation in the myostatin gene has been associated with muscle hypertrophy in a child [15], Seibert et al. [33] reported lower muscle strength (hip and knee flexion and handgrip strength combined) in those who carried the 153R allele in subjects in USA. Santiago et al. [17] reported that K153R polymorphism was associated with the ability to produce peak power during muscle contractions, in young non-athletic men in Spain. Han et al. [34] reported that higher serum myostatin levels were associated with lower muscle function in subjects from Taiwan. Another study also showed a lower muscle mass/function in a 96 year old woman from Madrid, Spain, with the very rare Myostatin R/R genotype compared to her age-matched referents with the 153KK genotype [16]. Further, myostatin expression levels have been shown to be inversely correlated with muscle mass in healthy and HIV-infected subjects [7].
Myostatin may have a strong role in the regulation of adipose tissue mass [35]. Specifically, myostatin-deficient mice have a significant associated to a 70% reduction in fat accumulation [36]. Further, a recent study in mice model suggested that contribution to low fat mass in mice lacking myostatin may be due to increased energy expenditure together with increased leptin sensitivity [37]. It is also possible that the absence of myostatin results in enhanced peripheral tissue fatty acid oxidation and increased thermogenesis, which result in increased fat utilization and reduced adipose tissue mass [35]. Based on our findings, we have a reason to believe that these polymorphisms may have lead to gain of function of myostatin, leading to observed changes in LBM and BF.
Conclusion
To conclude, we showed association of polymorphisms of Myostatin gene with increased adiposity and low LBM in Asian Indians. These findings may have implications for the development of IR and T2DM in Asian Indians, and should be further confirmed in larger studies.
Acknowledgments
The authors acknowledge the contribution of Mr. Kirti Pratap who performed many of the biochemical investigations. Finally, the cooperation of the subjects who took part in the study is greatly appreciated.
Funding Statement
This study was fully supported by a grant from the Indian Council of Medical Research (No. 5/9/70/2008-RHM) Government of India. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Misra A, Alappan NK, Vikram NK, Goal K, Gupta N, et al. (2008) Effect of supervised progressive resistance-exercise training protocol on insulin sensitivity, glycemia, lipids, and body composition in Asian Indians with type 2 diabetes. Diabetes Care 31 7:1282–1287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Chowdhury B, Lantz H, Sjostrom L (1996) Computed tomography-determined body composition in relation to cardiovascular risk factors in Indian and matched Swedish males. Metabolism 45 5:634–44. [DOI] [PubMed] [Google Scholar]
- 3. Banerji MA, Faridi N, Atluri R, Chaiken RL, Lebovitz HE (1999) Body composition, visceral fat, leptin and insulin resistance in Asian Indian men. J Clin Endocrinol Metab 84: 137–144. [DOI] [PubMed] [Google Scholar]
- 4. Yajnik CS, Fall CH, Coyaji KJ, Hirve SS, Rao S, et al. (2003) Neonatal anthropometry: the thin fat Indian baby. The Pune maternal nutrition study. Int J Obes Relat Metab Disord. 27: 173–80. [DOI] [PubMed] [Google Scholar]
- 5. Goel K, Misra A, Vikram NK, Poddar P, Gupta N (2010) Subcutaneous abdominal adipose tissue is associated with the metabolic syndrome in Asian Indians independent of intra-abdominal and total body fat. Heart 96 8:579–583. [DOI] [PubMed] [Google Scholar]
- 6. Jørgensen JO, Rubeck KZ, Nielsen TS, Clasen BF, Vendelboe M, et al. (2010) Effects of GH in human muscle and fat. Pediatr Nephrol 25 4:705–9. [DOI] [PubMed] [Google Scholar]
- 7. Petersen KF, Dufour S, Savage DB, Bilz S, Solomon G, et al. (2007) The role of skeletal muscle IR in the pathogenesis of the metabolic syndrome. Proc Natl Acad Sci USA 104 31:12587–12594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. McPherron AC, Lawler AM, Lee SJ (1997) Regulation of skeletal muscle mass in mice by a new TGF-beta superfamily member. Nature 387 6628:83–90. [DOI] [PubMed] [Google Scholar]
- 9. Gonzalez-Cadavid NF, Taylor WE, Yarasheski K, Sinha-Hikim I, Ma K, et al. (1998) Organization of the human myostatin gene and expression in healthy men and HIV-infected men with muscle wasting. Proc Natl Acad Sci USA 95: 14938–14943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Nishiyama A, Takeshima Y, Saiki K, Narukage A, Oyazato Y, et al. (2007) Two novel missense mutations in the myostatin gene identified in Japanese patients with duchenne muscular dystrophy. BMC Med Genet 8: 19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Thomis MA, Huygens W, Heuninckx S, Chagnon M, Maes HH, et al. (2004) Exploration of myostatin polymorphisms and the angiotensin-converting enzyme insertion/deletion genotype in responses of human muscle to strength training. Eur J Appl Physiol 92 3:267–274. [DOI] [PubMed] [Google Scholar]
- 12. Zhiliang G, Dahai Z, Ning L, Hui L, Xuemei D, et al. (2004) The single nucleotide polymorphisms of the chicken myostatin gene are associated with skeletal muscle and adipose growth. Sci China C Life Sci 47 1:25–30. [DOI] [PubMed] [Google Scholar]
- 13. Siriett V, Platt L, Salerno MS, Ling N, Kambadur R, et al. (2006) Prolonged absence of myostatin reduces sarcopenia. J Cell Physiol 209 3:866–73. [DOI] [PubMed] [Google Scholar]
- 14.Ferrell RE, Conte V, Lawrence EC, Roth SM, Hagberg JM, et al.. (1999) Frequent sequence variation in the human Myostatin gene as a marker for analysis of muscle-related phenotypes. 62; 2: : 203–207. [DOI] [PubMed]
- 15. Schuelke M, Wagner KR, Stolz LE, Hübner C, Riebel T, et al. (2004) Myostatin mutation associated with gross muscle hypertrophy in a child. N Engl J Med 350: 2682–2688. [DOI] [PubMed] [Google Scholar]
- 16. González-Freire M, Rodríguez-Romo G, Santiago C, Bustamante-Ara N, Yvert T, et al. (2010) The K153R variant in the myostatin gene and sarcopenia at the end of the human lifespan. Age (Dordr) 32: 405–409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Santiago C, Ruiz JR, Rodríguez-Romo G, Fiuza-Luces C, Yvert T, et al. (2011) The K153R Polymorphism in the Myostatin Gene and Muscle Power Phenotypes in Young, Non-Athletic Men. PLoS ONE 6 1:e16323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Misra A, Pandey RM, Devi JR, Sharma R, Vikram NK, et al. (2001) High prevalence of diabetes, obesity and dyslipidaemia in urban slum population in northern India. Int J Obes Relat Metab Disord 25 11:1722–9. [DOI] [PubMed] [Google Scholar]
- 19. Vikram NK, Misra A, Pandey RM, Dwivedi M, Luthra K, et al. (2006) Association between subclinical inflammation & fasting insulin in urban young adult north Indian males. Indian J Med Res 124 6:677–682. [PubMed] [Google Scholar]
- 20. Debomoy K, Lahiri and John I, et al. (1991) A rapid non-enzymatic method for the preparation of HMW DNA from blood for RFLP studies. Nucleic Acids Research 19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Misra A, Chowbey P, Makkar BM, Vikram NK, Wasir JS, et al. (2009) Consensus statement for diagnosis of obesity, abdominal obesity and the metabolic syndrome for Asian Indians and recommendations for physical activity, medical and surgical management. J Assoc Physicians India 57: 163–70. [PubMed] [Google Scholar]
- 22. Matthews DR, Hosker JP, Rudenski AS, Naylor BA, Treacher DF, et al. (1985) Homeostasis model assessment: insulin resistance and beta-cell function from fasting plasma glucose and insulin concentrations in man. Diabetologia 28: 412–9. [DOI] [PubMed] [Google Scholar]
- 23. Misra A, Khurana L (2008) Obesity and the Metabolic Syndrome in Developing Countries. J Clin Endocrinol Metab 93: 11 Supplement 1 s9–s30. [DOI] [PubMed] [Google Scholar]
- 24. Unni US, Ramakrishnan G, Raj T, Kishore RP, Thomas T, et al. (2009) Muscle mass and functional correlates of insulin sensitivity in lean young Indian men. Eur J Clin Nutr 63 10:1206–12. [DOI] [PubMed] [Google Scholar]
- 25. Rios R, Carneiro I, Arce VM, Devesa J (2002) Myostatin is an inhibitor of myogenic differentiation. Am J Physiol Cell Physiol 282: C993–999. [DOI] [PubMed] [Google Scholar]
- 26. Kambadur R, Sharma M, Smith T, Bass JJ (1997) Mutations in myostatin (GDF8) in double-muscled Belgian Blue and Piedmontese cattle. Genome Res 7 9:910–6. [DOI] [PubMed] [Google Scholar]
- 27. Jiang MS, Liang LF, Wang S, Ratovitski T, Holmstrom J, et al. (2004) Biochemical and Biophysical Research Communications. 315: 525–531. [DOI] [PubMed] [Google Scholar]
- 28. Lee SJ, McPherron AC (2001) Regulation of myostatin activity and muscle growth. Proc Natl Acad Sci U S A 98: 9306–9311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Reisz-Porszasz S, Bhasin S, Artaza JN, Shen R, Sinha-Hikim I, et al. (2003) Lower skeletal muscle mass in male transgenic mice with muscle-specific overexpression of myostatin. Am J Physiol Endocrinol Metab 285 4:E876–88. [DOI] [PubMed] [Google Scholar]
- 30. McFarlane C, Hui GZ, Amanda WZ, Lau HY, Lokireddy S, et al. (2011) Human myostatin negatively regulates human myoblast growth and differentiation. Am J Physiol Cell Physiol 301 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Ge X, Vajjala A, McFarlane C, Wahli W, Sharma M, et al. (2012) Lack of Smad3 signaling leads to impaired skeletal muscle regeneration. J Physiol Endocrinol Metab 303 1:E90–E102. [DOI] [PubMed] [Google Scholar]
- 32. Sartori R, Milan G, Patron M, Mammucari C, Blaauw B, et al. (2009) Smad2 and 3 transcription factors control muscle mass in adulthood. Am J Physiol Cell Physiol 296: C1248–C1257. [DOI] [PubMed] [Google Scholar]
- 33. Seibert MJ, Xue QL, Fried LP, Walston JD (2001) Polymorphic variation in the human myostatin (GDF-8) gene and association with strength measures in the Women's Health and Aging Study II cohort. J Am Geriatr Soc 49: 1093–1096. [DOI] [PubMed] [Google Scholar]
- 34. Han DS, Chen YM, Lin SY, Chang HH, Huang TM, et al. (2011) Serum myostatin levels and grip strength in normal subjects and patients on maintenance haemodialysis. Clin Endocrinol (Oxf) 1; 75 6:857–63. [DOI] [PubMed] [Google Scholar]
- 35. Zhang C, McFarlane C, Lokireddy S, Masuda S, Ge X, et al. (2012) Inhibition of myostatin protects against diet-induced obesity by enhancing fatty acid oxidation and promoting a brown adipose phenotype in mice. Diabetologia 55 1:183–93. [DOI] [PubMed] [Google Scholar]
- 36. McPherron and Se-Jin Lee (2012) Suppression of body fat accumulation in myostatin-deficient mice. J Clin Invest 109 5:595–601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Choi SJ, Yablonka-Reuveni Z, Kaiyala KJ, Ogimoto K, Schwartz MW, et al. (2011) Increased energy expenditure and leptin sensitivity account for low fat mass inmyostatin-deficient mice. Am J Physiol Endocrinol Metab 300 6:E1031–7. [DOI] [PMC free article] [PubMed] [Google Scholar]