Abstract
The recent explosion of genomic data and technology points to opportunities to redefine lung diseases at the molecular level; to apply integrated genomic approaches to elucidate mechanisms of lung pathophysiology; and to improve early detection, diagnosis, and treatment of lung diseases. Research is needed to translate genomic discoveries into clinical applications, such as detecting preclinical disease, predicting patient outcomes, guiding treatment choices, and most of all identifying potential therapeutic targets for lung diseases. The Division of Lung Diseases in the National Heart, Lung, and Blood Institute convened a workshop, “Genomic Medicine and Lung Diseases,” to discuss the potential for integrated genomics and systems approaches to advance 21st century pulmonary medicine and to evaluate the most promising opportunities for this next phase of genomics research to yield clinical benefit. Workshop sessions included (1) molecular phenotypes, molecular biomarkers, and therapeutics; (2) new technology and opportunity; (3) integrative genomics; (4) molecular anatomy of the lung; (5) novel data and information platforms; and (6) recommendations for exceptional research opportunities in lung genomics research.
Keywords: molecular phenotypes, molecular networks, drug repurposing, epigenetics, data sharing
Background
Most lung diseases are chronic, phenotypically heterogeneous, and often irreversible. These characteristics of lung disease present major challenges for improving early detection and prevention and for developing interventions that will reduce morbidity and mortality. Lung diseases are pervasive worldwide and exert a heavy burden at the individual and societal level. Since 1980, death rates from chronic obstructive pulmonary disease (COPD) and the prevalence of asthma have doubled, but new therapeutics have not altered this progression. Although early diagnostics for lung diseases have not kept pace with other organ systems, and many treatments are predominantly symptom based, new advances in “-omics” technology offer promise for elucidating the pathophysiology of lung diseases and translating this knowledge into improved diagnostic and intervention strategies.
The recent explosion of genomic methods presents an opportunity to redefine lung diseases at the molecular level. Omics research has been efficient because it has taken advantage of existing cohorts and existing analytic technologies. However, few studies are addressing how to use multiple omics approaches in clinical applications, such as detecting preclinical disease, predicting patient outcomes, guiding treatment choices, and most of all identifying potential therapeutic targets. In response to this identified gap in translational research, the Division of Lung Diseases in the National Heart, Lung, and Blood Institute (NHLBI) convened a workshop, “Genomic Medicine and Lung Diseases,” on July 18 and 19, 2011 to discuss how global or systems approaches and omics technologies will contribute to the development of 21st century pulmonary medicine and to evaluate the most promising opportunities for this next phase of genomic research most likely to yield clinical benefit.
Summary of Workshop Discussions
Introduction
The workshop opened with an introductory session, “A holistic view of lung biology at the molecular level,” in which James Kiley charged participants to focus on scientific opportunities that will lead to 21st-century strategies to diagnose and treat lung diseases. David Center introduced the goals of the workshop and challenged the participants to think decades into the future of pulmonary medicine to identify the most exceptional areas of opportunity to focus lung omics research.
David Schwartz presented a vision of genomic medicine that would redefine lung diseases in the context of molecular networks and require innovation to optimally integrate phenotype and omics information. To illustrate the complexity of pulmonary medicine, Dr. Schwartz discussed how seemingly unrelated diseases can be initiated by a common environmental factor (e.g., smoking for COPD, lung cancer, and idiopathic pulmonary fibrosis [IPF]) or that one disease can be caused by a wide variety of factors (e.g., IPF). Gene expression profiles have the potential to identify the molecular pattern of disease pathogenesis, distinguish etiology and prognosis (long versus short survival), and individualize treatments and ways to screen for responses to existing or novel drugs (class and compound specific). He noted that an existing research gap is the lack of integrated systems-level data to delineate specific diagnostic markers related to the underlying and targetable causes of lung diseases. An additional research challenge is to elucidate the complex molecular and cellular processes that couple environmental exposures to disease pathogenesis as well as to adaptive lung functioning; this requires sophisticated omics approaches. The working group concurred on strategies to accomplish these scientific goals, including the need for partnership among academia, industry, and the National Institutes of Health (NIH) to pool resources, integrate infrastructure, invest strategically (faculty, software, hardware, and analysis major computing services), and develop novel training curricula and training programs. The discussion highlighted the need for (1) global electronic medical record (EMR) access, (2) partnering with the Food and Drug Administration (FDA), (3) use of biorepositories, (4) focus on data analysis strategies and methods, and (5) using Clinical and Translational Science Awards supported by the new NIH National Center for Advancing Translational Science.
Session I: From Molecular Phenotypes to Molecular Biomarkers and Therapeutics
This session focused on opportunities to leverage molecular profiling data to identify early molecular biomarkers as diagnostic and prognostic tools.
Naftali Kaminski presented gene expression profiles of patients with IPF (1). He described computational approaches to integrate clinical and molecular data that showed clear advantage over methods using clinical data or molecular profiles alone in defining subgroups of patients with IPF with different progression rates (2–4). He demonstrated that when clinical and molecular data from patients with IPF or COPD were analyzed together, patients could be categorized into more distinct subgroups. Dr. Kaminski emphasized that integrative genomics approaches will need to identify the optimal tissue for analysis that is clinically scalable, demonstrate organ and disease specificity, and exhibit efficacy to inform potential drug targets. To this end, surrogates for surgically obtained lung tissue will be important to identify.
Prescott Woodruff discussed genomic approaches to biomarker development using DNA, RNA, and miRNA, which could be applied to stratify patient populations for therapeutic responses and/or to identify subtypes of lung diseases. He presented an example of his work using the mRNA expression profile of airway epithelial cells to predict responsiveness of patients with asthma to inhaled corticosteroids (Th2 low) (5). That profile has now been extended to potential use of a serum protein marker, periostin, to stratify patients in a clinical trial (6). Similar approaches might be used to determine the mRNA profile of peripheral blood cells to differentiate latent from active MTb infection. Another application could be to identify diagnostic subtypes of sarcoidosis from healthy control subjects and other inflammatory lung diseases using whole blood gene expression (7).
Dietrich Stephan described lessons learned from his translational genomics research as part of the NIH Neuroscience Microarray Consortium to support the NIH Blueprint for Neuroscience Research (a cooperative effort among the 16 NIH Institutes, Centers, and Offices). The consortium generated 60,000 RNA expression and 100,000 SNP profiles to identify therapeutic targets for neurological disorders. He emphasized that research challenges in these profiling techniques are study design and the selection of the clinical and molecular phenotype to accurately predict presymptomatic risk assessment, susceptibility to disease, prognosis, and response to therapy (8). Another major challenge is how to store data in the context of access to optimal phenotypes from EMRs. EMR storage will need champions to financially support development and maintenance of deidentified sensitive databases that are constructed on platforms amenable to research inquiries.
Bruce Littman discussed strategies to translate the identification of biomarkers and genetic profiles into drug development (9, 10), using kRAS mutations in cancer treatment as an example. He, and many other participants, stressed the need to integrate biomarker discovery and validation in all new prospective clinical trials, including sample collection designed for biomarker analysis and hypotheses testing; biobanking in the context of EMR could be built into the infrastructure of trials with detailed drug response data fields. Partnering with the FDA would facilitate establishing criteria for drug efficacies as individual variations are defined from outcome studies in biomarker-defined patient subpopulations.
A translational domain wherein molecular profiling data may have immediate impact is drug repurposing. Avrum Spira described his efforts to identify FDA-approved drugs with the potential to treat COPD by mapping gene expression profiling data from patients with COPD to the gene expression signatures from cancer cell lines treated with small molecular drugs (Connectivity Map database at the Broad Institute) Using a similar computational approach, he identified a subgroup of patients with breast cancer who may be responsive to valproic acid, a drug normally used to treat seizures and mood-related conditions (11). This approach to breast cancer will be studied in a recently FDA-approved clinical trial. In two recent articles, Atul Butte’s group demonstrated the potential of using molecular signatures of diseases in drug repurposing studies (12, 13). To facilitate drug repurposing research for lung diseases, drug response gene signatures of lung cell lines that are relevant to lung diseases need to be incorporated into databases, such as Connectivity Map (http://www.broadinstitute.org/genome_bio/connectivitymap.html). In addition, improvements in disease profiling data are needed, including better phenotypes in patient cohorts and dynamic profile changes of disease progression.
The molecular phenotype session concluded with the following recommendations by the working group:
Integrate mRNA, miRNA, protein profiling, and DNA methylation profiling data from patients with lung disease to understand the biological networks behind gene expression patterns unique for the disease.
Standardize and validate transcriptomic, genomic, and epigenomic biomarkers in chronic lung disease in lung and surrogate tissues (e.g., lung epithelium vs. nasal or blood).
Compare lung disease profiling to all other diseases for full identification of targets for drug therapies from existing drug libraries.
Improve the environment for drug and diagnostic discovery and development; establish a biomarker/drug development network that leverages current infrastructure (e.g., existing disease networks).
Incorporate EMR capacity into genomics research for retrospective and longitudinal studies.
Leverage existing clinical research networks and data sets to gather data/samples to support biomarker discovery.
Standardize phenotyping across NHLBI trials, and reprobe with translationally useful questions for therapies and proof of concept. This will require adding healthy control subjects in data sets, promoting precompetitive sharing and ongoing accumulation of data concerning these samples, and using EMR to monitor variation over time.
Advance research in drug repurposing for lung diseases by generating genomic data sets of human lung cell lines treated with FDA-approved drug libraries.
Session II: New Technology and Opportunity
The technology session focused on the development and application of new genomics technologies to improve our ability to diagnose and treat lung diseases. Rasika Mathias began the session by discussing the latest DNA sequencing technologies that will lead to new ways of carrying out biomedical research and create new scientific opportunities for advancing lung disease research. She described the NHLBI’s lung cohort exome sequencing project, which aims to identify common and rare exonic variants; most are synonymous, fewer are nonsynonymous or missense mutations. She stressed the advantages of using large cohorts with many related (e.g., first cousin) subjects to detect rare mutations/variants. She discussed a cost-effective strategy to conduct genetic studies for lung diseases by adding exome sequencing data to NHLBI’s cohort studies with existing genome-wide association study (GWAS) data.
Pierre Chaurand described imaging mass spectrometry (MS) technology (14) that permits imaging in thin tissue sections or whole small animals of ion concentrations in formalin-fixed, paraffin-embedded tissue stored at room temperature. These techniques can be applied to lipid spectral imaging to predict presence/absence of cancers and to identify targets for therapy. In proof-of-concept studies in whole animals, drug tissue distribution can be determined by ex vivo analysis from frozen sections using imaging matrix-assisted laser desorption/ionization MS based on the ringed structure of molecules of interest. The technology allows simultaneous mapping of the location of hundreds of different biocompounds present in thin tissue sections. The system is suitable for analyzing 25 to 100 samples in imaging mode per day on one machine; thus, it can be applied to large collections of tissue samples and is suitable to be used for the development of novel genomic pathology tools.
Irfan Rahman described the potential of using epigenomics to elucidate the contributions of histone methylation and acetylation in chromatin remodeling in lung cells in response to environmental factors (pollution, cigarette smoke, oxidants, foods, drugs, radiation, infections other stresses) (15). He noted that the genetic circadian clock is present in lung cells and that a high proportion of the lung transcriptome exhibits significant diurnal variation. The genetic feedback loops composing the circadian clock are directly coupled to mechanisms regulating histone modification and chromatin remodeling in the lung. These findings introduce a new layer of molecular control and complexity to lung genomic function and may have significant implications for lung diseases in which symptom exacerbations, molecular markers of disease, and responses to therapeutic agents vary with the time of day. There is relatively little information on how the genetic circadian clock intersects with transcription, miRNA regulation, post-translational modifications, cell cycle, cell regeneration, repair, or immune responses in normal or diseased lung cells.
Nirinjini Naidoo discussed the potential role of protein misfolding in lung diseases associated with aging and sleep disturbance (16). Changes in cellular redox, glucose, calcium, pH, and viral infections contribute to endoplasmic reticular stress and changes in protein folding. Protein misfolding has been described in the pathophysiology of α1 antitrypsin deficiency and cystic fibrosis, and pharmaceutical compounds targeting protein misfolding in these diseases have been developed. Sleep disorders (e.g., sleep apnea) and impaired sleep quality are common in patients with lung diseases, and recent studies have shown that unfolded protein responses occur in sleep deprivation and sleep apnea associated with intermittent hypoxia. These findings raise the possibility that sleep disorders and age-related sleep alterations are involved in post-translational mechanisms of lung disease and that sleep may be an important phenotype to delineate in lung disease cohorts.
Kathleen Stringer described her metabolomic research on lung injury (17) and stressed how infrequently metabolomics technology has been applied to the lung. Specifically, the basic definition of the animal or human metabolome (e.g., small molecules, peptides, sugars, lipids, oligonucleotides, ketones, and amino acids) in normal lung cells is unknown, let alone in the context of lung diseases (18). Following an understanding of the metabolomes of the various lung cells, we would next need to integrate this information in complex lung diseases that involve multiple cell types. Dr. Stringer emphasized that metabolomic technologies might hold greatest promise for biomarker discovery in lung disease, especially for systemic conditions, for example defining susceptibility, diagnosis, disease severity, prognosis, and selection of therapy in acute lung injury and sepsis. Any systems approach to lung metabolomics must involve establishing the baseline or normal metabolomic profiles in the lung and determining how they are related to profiles in blood, plasma, and urine.
The new technology session concluded with the following recommendations by the working group:
Advance the application of metabolomics, epigenomics, protein unfolding/misfolding, and imaging MS technologies in human lung diseases by defining the profiles in normal and diseased lung cells and tissues.
Collect biospecimens from participants of ongoing clinical studies using methods that are compatible with metabolomic, epigenomic, and protein unfolding/misfolding analyses.
Promote sequencing based genetic studies of lung diseases using case/control or cohort designs that include familial risk profiles.
Session III: Integrative Genomics
Scott Weiss chaired a session on integrative genomics aimed at identifying opportunities to better integrate different types of molecular measurements (e.g., DNAs, RNAs, proteins, metabolites) to untangle the complexity of lung biology. Benjamin Raby described mapping of (epi)genetic regulatory networks in lung diseases by integrating genetic polymorphisms, gene expression, CpG methylation marks, and altered chromatin structure data with clinical phenotypes (19, 20). Various approaches are currently available for this effort, including population-based approaches, such as expression and methylation quantitative trait locus mapping (eQTL and mQTL, respectively), and cellular approaches, including genome-wide surveys of allelic imbalance, DNA–protein interactions (ChIP-Chip and ChIP-Seq), and DNA–DNA interactions (Chromosome Conformational Capture approaches). He stressed that we are currently at a transitional phase, moving beyond simple GWAS and gene expression analysis of one gene or variant at a time toward developing holistic models of disease pathogenesis (i.e., models that simultaneously factor genetic sequence variants, epigenetic markers, and environmental risk factors).
Paolo Sassone-Corsi presented an integrative analysis of circadian clock genomics with gene expression, chromatin remodeling, and metabolomic profiling data (21) that complemented the emphasis from an earlier session by Dr. Rahman. He noted that at least 10 to 15% of the transcriptome in tissues throughout the body is under the control of the genetic circadian clock. Clock genes are coupled to mechanisms of cellular energetics, redox, histone modification, cell cycle, and nuclear receptor function. His recent work demonstrated that circadian clock genes are coupled to the histone deacetylase, sirtuin, and confer diurnal rhythmicity in chromatin remodeling. The link between the circadian clock and epigenetic modification has broad implications for regulating genomic responses to cellular and environmental signals, including regulation of cellular metabolism and thus the metabolome. An opportunity exists now to apply circadian genomic discoveries that have been made in tissues such as the liver, pancreas, vasculature, and brain to the lung, to improve our understanding of cellular dynamics involved in lung disease pathophysiology and symptomatology. The cancer field has made significant advances in chronopharmacology, which entails the specific timing of chemotherapy based on knowledge of the circadian cycle of tumor cells. Therefore, circadian genomics may inform drug development by revealing new targets for drug development as well as informing treatment regimens. There are now open access sources for searching for genes and metabolites under circadian control. Attention to the time of day of collection of phenotypes and biospecimens may be critical for optimizing integrated analysis of genomic, epigenomic, and metabolomic data.
Nathan Price discussed application of complex biomolecular networks and systems biology approaches to studying lung diseases. He used single-cell organisms to model metabolic pathways to delineate mechanistic interactions, nodal points, and potential drug targets (22). This systems approach permits one to determine the consequences or importance of gene/protein expression in the context of gene networks in specific types of cells, which could be used, for example, in tissue-specific cells derived from iPS cells.
Mariano Alvarez and Mukesh Bansal presented a computational model for the analysis of gene networks in cancer cells to elucidate master regulators that predict malignancy. They described the concept of “interactomes” defined by nodes versus edges of a gene regulatory network. The interactome is built with integrated data ranging from environmental stimuli, cell-specific signal transduction, cellular responses, and the combined elements of genomics/epigenomics/transcriptomics (23). A new algorithm, MAster Regulator INference algorithm (MARINa), is designed to infer transcription factors controlling the transition between two phenotypes that permits use of nodal points or regulatory modules as biomarkers for disease stratification or therapeutic targets (e.g., with a vector-containing transcription factors). Interactomes or nodal analyses can be applied for small-molecule targeting, like the examples from Dr. Spira’s presentation on repurposing FDA-approved drugs. Eventually, this would expand from single cell to synergistic pairs of master regulators for multiple cell lines under basal and stimulated conditions as a proof of concept for responsiveness of small molecules, existing drugs, or new drug target development ex vivo. The goal would be to use human cells to help predict therapeutic responses and potential toxicities before human trials are initiated. The summary discussion, led by Dr. Weiss, emphasized the need in all of these methodologies to determine the differences between susceptibility genes and severity genes, as both are essential in targeting prevention and therapy.
The integrative genomics session concluded with the following recommendations:
Improve translatable experimental models and phenotyping/characterization of those models (e.g., iPS cells, body on a chip, cocultures), taking into account time scale and other feature mapping; improve translational technologies.
Advance computational modeling to discern emergent behaviors across multiple biological mechanisms/components relevant to human respiratory disease.
Facilitate integrative genomics research in lung diseases by integrating omics measurements (e.g., exome/whole-genome sequencing, DNA methylation profile, metabolomics, etc.) with well-phenotyped cohorts that have existing GWAS or gene expression data.
Session IV: Molecular Anatomy of the Lung
A recurring theme throughout the workshop discussion concerned the paucity of knowledge about the molecular anatomy of the lung. Based on light and electron microscopic studies (24) it is estimated that the lung is composed of more than 40 different types of cells. Molecular data have been collected from only a small number of these lung cells using omic technologies, but we need to move toward establishing a comprehensive molecular anatomy of the lung, including profiles/signatures of metabolites, epigenomic modifications, and proteins. RNA expression has received the most focus in lung research; however, these data are predominantly from cross-sectional studies and are less informative about how gene regulatory networks in the lung change in response to internal (i.e., genetic) and external perturbations. Whereas existing lung disease categories are defined based on lung anatomy at the tissue/cellular level, the definition of lung diseases in the 21st century will need to be built on the molecular anatomy of the lung and individual cell types. Ronald Crystal conveyed the complexity of respiratory systems by demonstrating that environmental exposure (e.g., smoke) could lead the researcher to analyze lung biology using a wide variety of approaches (25). A reductionist approach would entail the entire systems biology of a single cell of interest, a hierarchical approach would include regions or types of lung structures (e.g., airways versus vascular), or a holistic approach would include the whole lung in all diseases. Given limitations in access to lung samples, the first two approaches are likely to be the most informative, as a whole lung would never be available in premorbid states when diagnostic and prognostic information is needed to guide therapy. Dr. Crystal emphasized the need to identify entire omic networks and to develop a complete atlas of responses in all primary lung cell types as fundamentally necessary to build a holistic understanding of lung biology. He proposed that it is time to conduct systematic investigation of the molecular changes in lung cells and tissues in response to environmental perturbations and during lung development and growth.
Session V: From Data to Information, Knowledge, and Application
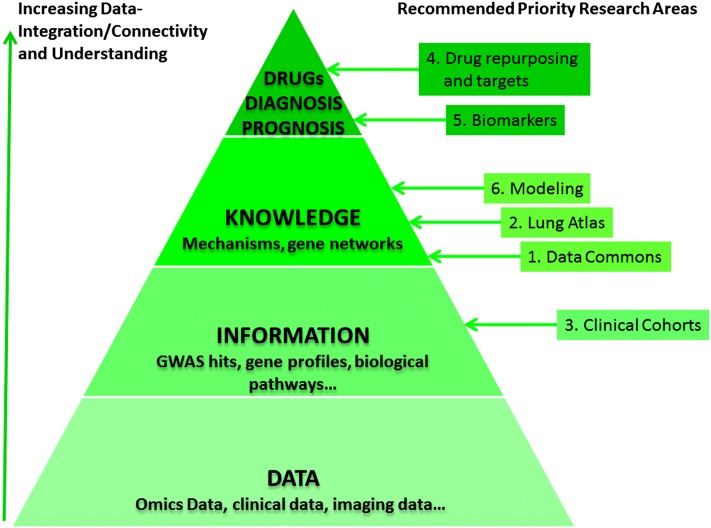
Another central theme of the workshop was how to facilitate the transition from data collection to information, to knowledge, and then to application (wisdom). We are now focusing more on data collection through omic technologies and just starting to integrate associations among data points (e.g., GWAS hits and gene signatures for diseases); we should be planning our current methods of data collection in ways that will permit us to integrate this information into knowledge of how the lung functions in health and disease and how to manipulate reparative responses to recapitulate normal growth and development. As shown in Figure 1, the data-to-knowledge process is driven by integrating seemingly unrelated data points into organized and interconnected molecular networks that change dynamically in response to internal and external perturbations and provide us the insight of biology at the molecular level. The last session (chaired by Julian Solway and David Center) used the discussion points from the first day to explore how the lung community should work together to leverage each other’s efforts and existing infrastructures.
Figure 1.
From data to information, knowledge, and application. Recommended priority research areas are mapped to knowledge management schemes to show that a collection of (omic) data is not information until, for instance, disease-associated patterns are identified; a collection of information (profiling patterns) is not knowledge until, for example, the patterns are systematically integrated and analyzed to understand how they work together in disease development. Further analysis of knowledge to understand how the biology works in certain ways will lead to clinical applications in the prediction of disease progression and treatment. GWAS = genome-wide association study.
Stephen H. Friend discussed the importance of developing bioinformatics networks and making data publically available to the research community. Dr. Friend developed Sage Bionetworks, a nonprofit bioinformatics organization, based on his experiences in building cancer drug networks at Rosetta Pharmaceuticals and Merck. The overarching goal of the bioinformatics approach is to build better maps of disease (e.g., by linking clinical presentations with specific pathways/pathogenesis), host data repositories, and generate computational platforms by which various scientists working on a given project can share their data, models, and tools, which can interface with existing databases (e.g., the Framingham Heart Study) (26). A critical component of this bioinformatics model is to make data from industry and academia available in the public domain. He provided an example of the role that the immune response plays in breast cancer models that Sage scientists generated from published studies. Primary data were converted into models and targets. Dr. Friend defined omics of the present/future as evolving to provide complete collaborative networks with a continuum from patients and their data through e-networks for both discovery and therapy validation. Proximity is defined by open source data (www.sagebase.org). His presentation prompted an extended discussion within a scientific framework, as presented in Figure 1.
Recommended Priority Research Areas
Progress in lung genomics research and medicine will require strategic thinking about short- and long-term scientific priorities and opportunities as well as innovative ways to create and use ever-growing resources and technologies. It will be essential to identify exceptional scientific questions in domains of pulmonary medicine, where promising advances are at hand, and to identify the technologies most needed to accomplish the continuum of basic, translational, and implementation science.
Discussions during the workshop clarified some key emerging areas for future research in genomic discovery for lung diseases. During the next 3 to 5 years, genomic data derived from patient biospecimens will become increasingly available. A growing understanding of the important role of gene and environment interactions in the etiology and pathogenesis of lung diseases will require carefully generated disease-specific functional genomic profiles (transcriptomics, epigenomics, proteomics, metabolomics). The lung community should be encouraged to pursue collaborative efforts to generate disease-specific functional genomic datasets and share them with the broad pulmonary community. Computational biology technologies should be used to integrate genetics, genomics, and clinical data into new platforms for integrative lung omics research. Integrating omics approaches with data derived from virtual patient cohorts using electronic health information is a future effort that may yield powerful new information. It is increasingly important for the lung community to share and analyze omics datasets across many lung diseases to build a knowledge base or atlas of lung molecular function. Within 5 years, omics research should be leading discovery in translational lung disease research, especially in the areas of redefining lung diseases, identifying molecular biomarkers, and generating new ideas for drug development. The workshop recommended the following research priorities:
Data commons: Build a virtual “Pulmonary (Gen)omics Workstation” by encouraging/incentivizing collaboration throughout the lung genetics/genomics community through data sharing in computing spaces like Synapse of Sage Bionetworks to generate a common data repository suitable for multilevel analysis. This workstation would serve as the model for a full “Omics” collaborative lung disease database.
Lung atlas: Construct cell-type specific molecular/functional network atlas of human lung cells relevant to respiratory diseases (epithelial, fibroblast, endothelial, and smooth muscle) using integrative genomic approaches. This is to enable the discovery of dynamic changes of gene networks in response to relevant perturbations and deviations from normal states in disease, and develop network analyses of these to reveal functionally important modules and potential therapeutic targets.
Clinical cohorts: Apply pan-omics analysis to clinical cohorts to delineate functional molecular network components that have direct impact on the development of lung diseases and response to therapeutic treatments. Identify cost-effective strategies, such as adding complementary omics measurements to the cohorts already collecting some omics data (e.g., GWAS or gene expression), and make these data widely available.
Drug repurposing and targets: Promote research to investigate the intersection between the molecular phenotype of lung diseases and the molecular signatures of drug compounds (e.g., Connectivity Map) for the purpose of drug repurposing for lung diseases and delineating the interactions between genes and drugs.
Biomarkers: Standardize and validate transcriptomic, genomic, and epigenomic biomarkers in chronic lung disease in surrogate tissues across disease boundaries, and use integrated genomic and phenomic profiles to reclassify chronic lung disease with complex phenotypes. Encourage investigation of mechanisms underlying heterogeneity of response to drugs.
Modeling: Advance computational biology (e.g., multiscale modeling) to build molecular networks from omics data to discern emergent behaviors across multiple biological mechanisms/components relevant to human respiratory disease and to systematically study how dynamic interactions among genes and environmental perturbations are involved in disease development.
Supplementary Material
Acknowledgments
Workshop RosterCochairs and Session Chairs: David M. Center, M.D., Boston University; David A. Schwartz, M.D., University of Colorado; Julian Solway, M.D., University of Chicago; Scott T. Weiss, M.D., Channing Laboratory of the Brigham and Women’s Hospital, Harvard Medical School. Participants: Mariano Alvarez, Ph.D., Columbia University; Mukesh Bansal, Ph.D., Columbia University; Pierre Chaurand, Ph.D., Université de Montréal; Ronald G. Crystal, M.D., Cornell University; Stephen H. Friend, M.D., Ph.D., Sage Bionetworks; Naftali Kaminski, M.D., University of Pittsburgh; Bruce Littman, M.D., Translational Medicine Associates, LLC; Rasika Mathias, Ph.D., Johns Hopkins University; Nirnjini Naidoo, Ph.D., University of Pennsylvania; Nathan D. Price, Ph.D., University of Illinois; Benjamin A. Raby, M.D., M.P.H., Channing Laboratory of the Brigham and Women’s Hospital, Harvard Medical School; Irfan Rahman, Ph.D., University of Rochester; Paolo Sassone-Corsi. Ph.D., University of California, Irvine; Avrum Spira, M.D., M.Sc., Boston University; Dietrich Stephan, Ph.D., Navigenics, Inc; Kathleen A. Stringer, Pharm.D., University of Michigan; Jennifer Wambach, M.D., Washington University; Prescott Woodruff, M.D., M.P.H., University of California San Francisco. NHLBI Staff: Weiniu Gan, Ph.D., Division of Lung Diseases, Organizer; Sandra Colombini-Hatch, M.D., Division of Lung Diseases; Thomas Croxton, M.D., Ph.D., Division of Lung Diseases; Dorothy Gail, Ph.D., Division of Lung Diseases; James P. Kiley, Ph.D., Division of Lung Diseases; Aaron D. Laposky, Ph.D., Division of Lung Diseases; Qing S. Lin, Ph.D., Division of Lung Diseases.
Footnotes
Supported by the Division of Lung Diseases, National Heart, Lung, and Blood Institute, National Institutes of Health.
A complete workshop roster may be found before the beginning of the References.
Author Contributions: D.M.C., D.A.S., J.S. (workshop co-chairs, presenters, and manuscript preparation); D.G., A.D.L., S.L., W.G. (workshop organizers and manuscript preparation).
Originally Published in Press as DOI: 10.1164/rccm.201203-0569WS on May 31, 2012
Author disclosures are available with the text of this article at www.atsjournals.org.
References
- 1.Konishi K, Gibson KF, Lindell KO, Richards TJ, Zhang Y, Dhir R, Bisceglia M, Gilbert S, Yousem SA, Song JW, et al. Gene expression profiles of acute exacerbations of idiopathic pulmonary fibrosis. Am J Respir Crit Care Med 2009;180:167–175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Pandit KV, Milosevic J, Kaminski N. MicroRNAs in idiopathic pulmonary fibrosis. Transl Res 2011;157:191–199 [DOI] [PubMed] [Google Scholar]
- 3.Kass DJ, Kaminski N. Evolving genomic approaches to idiopathic pulmonary fibrosis: moving beyond genes. Clin Transl Sci 2011;4:372–379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Richards TJ, Kaminski N, Baribaud F, Flavin S, Brodmerkel C, Horowitz D, Li K, Choi J, Vuga LJ, Lindell KO, et al. Peripheral blood proteins predict mortality in idiopathic pulmonary fibrosis. Am J Respir Crit Care Med 2012;185:67–76 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Woodruff PG, Modrek B, Choy DF, Jia G, Abbas AR, Ellwanger A, Koth LL, Arron JR, Fahy JV. T-helper type 2-driven inflammation defines major subphenotypes of asthma. Am J Respir Crit Care Med 2009;180:388–395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Corren J, Lemanske RF, Hanania NA, Korenblat PE, Parsey MV, Arron JR, Harris JM, Scheerens H, Wu LC, Su Z, et al. Lebrikizumab treatment in adults with asthma. N Engl J Med 2011;365:1088–1098 [DOI] [PubMed] [Google Scholar]
- 7.Koth LL, Solberg OD, Peng JC, Bhakta NR, Nguyen CP, Woodruff PG. Sarcoidosis blood transcriptome reflects lung inflammation and overlaps with tuberculosis. Am J Respir Crit Care Med 2011;184:1153–1163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Coon KD, Dunckley TL, Stephan DA. A generic research paradigm for identification and validation of early molecular diagnostics and new therapeutics in common disorders. Mol Diagn Ther 2007;11:1–14 [DOI] [PubMed] [Google Scholar]
- 9.Littman BH, Di Mario L, Plebani M, Marincola FM. What’s next in translational medicine? Clin Sci (Lond) 2007;112:217–227 [DOI] [PubMed] [Google Scholar]
- 10.Littman BH, Marincola FM. Create a translational medicine knowledge repository-research downsizing, mergers and increased outsourcing have reduced the depth of in-house translational medicine expertise and institutional memory at many pharmaceutical and biotech companies: how will they avoid relearning old lessons? J Transl Med 2011;9:56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cohen AL, Soldi R, Zhang H, Gustafson AM, Wilcox R, Welm BE, Chang JT, Johnson E, Spira A, Jeffrey SS, et al. A pharmacogenomic method for individualized prediction of drug sensitivity. Mol Syst Biol 2011;7:513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sirota M, Dudley JT, Kim J, Chiang AP, Morgan AA, Sweet-Cordero A, Sage J, Butte AJ. Discovery and preclinical validation of drug indications using compendia of public gene expression data. Sci Transl Med 2011;3:96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dudley JT, Sirota M, Shenoy M, Pai RK, Roedder S, Chiang AP, Morgan AA, Sarwal MM, Pasricha PJ, Butte AJ. Computational repositioning of the anticonvulsant topiramate for inflammatory bowel disease. Sci Transl Med 2011;3:96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chaurand P, Cornett DS, Angel PM, Caprioli RM. From whole-body sections down to cellular level, multiscale imaging of phospholipids by MALDI mass spectrometry. Mol Cell Proteomics 2011;10:O110.004259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sundar IK, Mullapudi N, Yao H, Spivack SD, Rahman I. Lung cancer and its association with chronic obstructive pulmonary disease: update on nexus of epigenetics. Curr Opin Pulm Med 2011;17:279–285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Naidoo N. Cellular stress/the unfolded protein response: relevance to sleep and sleep disorders. Sleep Med Rev 2009;13:195–204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Stringer KA, Serkova NJ, Karnovsky A, Guire K, Paine R, III, Standiford TJ. Metabolic consequences of sepsis-induced acute lung injury revealed by plasma 1H-nuclear magnetic resonance quantitative metabolomics and computational analysis. Am J Physiol Lung Cell Mol Physiol 2011;300:L4–L11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Serkova NJ, Standiford TJ, Stringer KA. The emerging field of quantitative blood metabolomics for biomarker discovery in critical illnesses. Am J Respir Crit Care Med. 2011;184:647–655 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Murphy A, Chu JH, Xu M, Carey VJ, Lazarus R, Liu A, Szefler SJ, Strunk R, Demuth K, Castro M, et al. Mapping of numerous disease-associated expression polymorphisms in primary peripheral blood CD4+ lymphocytes. Hum Mol Genet 2010;19:4745–4757 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chu JH, Lazarus R, Carey VJ, Raby BA. Quantifying differential gene connectivity between disease states for objective identification of disease-relevant genes. BMC Syst Biol 2011;5:89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bellet MM, Sassone-Corsi P. Mammalian circadian clock and metabolism – the epigenetic link. J Cell Sci 2010;123:3837–3848 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Milne CB, Eddy JA, Raju R, Ardekani S, Kim PJ, Senger RS, Jin YS, Blaschek HP, Price ND. Metabolic network reconstruction and genome-scale model of butanol-producing strain Clostridium beijerinckii NCIMB 8052. BMC Syst Biol 2011;5:130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lefebvre C, Rajbhandari P, Alvarez MJ, Bandaru P, Lim WK, Sato M, Wang K, Sumazin P, Kustagi M, Bisikirska BC, et al. A human B-cell interactome identifies MYB and FOXM1 as master regulators of proliferation in germinal centers. Mol Syst Biol 2010;6:377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Franks TJ, Colby TV, Travis WD, Tuder RM, Reynolds HY, Brody AR, Cardoso WV, Crystal RG, Drake CJ, Engelhardt J, et al. Resident cellular components of the human lung: current knowledge and goals for research on cell phenotyping and function. Proc Am Thorac Soc 2008;5:763–766 [DOI] [PubMed] [Google Scholar]
- 25.Hackett NR, Shaykhiev R, Walters MS, Wang R, Zwick RK, Ferris B, Witover B, Salit J, Crystal RG. The human airway epithelial basal cell transcriptome. PLoS ONE 2011;6:e18378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Friend SH. Something in common. Sci Transl Med 2010;2:40ed6. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.