Abstract
Susceptibility to primary biliary cirrhosis (PBC) is strongly associated with HLA region polymorphisms. To determine if associations can be explained by classical HLA determinants we studied Italian 676 cases and 1440 controls with genotyped with dense single nucleotide polymorphisms (SNPs) for which classical HLA alleles and amino acids were imputed. Although previous genome-wide association studies and our results show stronger SNP associations near DQB1, we demonstrate that the HLA signals can be attributed to classical DRB1 and DPB1 genes. Strong support for the predominant role of DRB1 is provided by our conditional analyses. We also demonstrate an independent association of DPB1. Specific HLA-DRB1 genes (*08, *11 and *14) account for most of the DRB1 association signal. Consistent with previous studies, DRB1*08 (p = 1.59 × 10−11) was the strongest predisposing allele where as DRB1*11 (p = 1.42 × 10−10) was protective. Additionally DRB1*14 and the DPB1 association (DPB1*03:01) (p = 9.18 × 10−7) were predisposing risk alleles. No signal was observed in the HLA class 1 or class 3 regions. These findings better define the association of PBC with HLA and specifically support the role of classical HLA-DRB1 and DPB1 genes and alleles in susceptibility to PBC.
Keywords: genetic risk, risk allele, imputation, antigen binding pocket, autoimmune disease
Introduction
The human major histocompatibility complex, HLA, has been implicated in the etiopathogenesis of primary biliary cirrhosis (PBC), similar to many other autoimmune diseases. Genome-wide association studies (GWAS) of PBC including our own find the strongest association with single nucleotide polymorphisms (SNPs) within the HLA region1–3. In these studies the peak association signal is between HLA-DQA1 and HLA-DQB1. Multiple studies of PBC also show association with particular classical HLA alleles in PBC (reviewed in Invernizzi4). These studies have variably implicated different DRB1 alleles in European populations with most studies including all larger cohorts showing association of DRB1*085, 6. Our previous studies in an Italian cohort with PBC showed the association of DRB1*08 as predisposing, and DRB1*11 and DRB1*13 as protective alleles6. A study using a small cohort (32 German PBC cases and 47 controls) suggest that DPB1 associations may also be present in Europeans7. However, a comprehensive study of HLA region associations has not been performed and like other autoimmune diseases it is unclear which determinants are actually causally related to pathogenesis.
To further study HLA associations in PBC, in the current study we used the most recent advances in imputation algorithms and sequence information resources including the 1000 genome database to accurately impute missing SNPs, and importantly HLA classical alleles. Specifically, our investigation rests on recent development and resources for imputing HLA classical alleles including a reference set of European subjects8. For our study we used an inference set of SNP genotypes from both GWAS and a designed chip array, the Immunochip9 that contains a set of SNPs that have been used in multiple studies of HLA10. We perform a series of conditioning analyses that clarify which HLA genes and alleles underlie the major component of the genetic associations of PBC.
Results
Analyses Show Strong Association of Imputed SNPs and HLA Determinants
To further define PBC - HLA region associations we analyzed association using imputed genotypes with high probabilities and information scores (see Materials and Methods). These studies utilized genotypes from both GWAS and Immunochip arrays that contained large numbers of SNPs in the MHC region (Table 1 Supporting Table 1 and see Materials and Methods). Strong association was observed with the peak association (p = 9.83 × 10−17) with rs115721871 at position 32653792 distal to DQB1 (Fig. 1A, Table 2 and Supporting Table 2). Although the strongest associations were with non-coding SNPs, multiple classical genes in HLA show strong association with PBC (Table 2). For the classical HLA genes the strongest association was with DRB*08 (p = 1.59 × 10−11). The DQB1*04:02 and DQA1*04:01 in tight LD with DRB*08 (r2 = 0.84 and 0.89, respectively) showed nearly equivalent signals (1.38 × 10−10, 1.90 × 10−10, respectively). Very strong association was also observed for DRB1*11 (p = 1.42 × 10−10) with a weaker association with the DQB1 allele (DQB1*03:01, p = 6.10×10−9) that is in LD (r2 = 0.75) with DRB1*11. Less strong associations were observed with DRB1*14, DQB1*05:03 (6.89 × 10−7 and 6.21 × 10−7, respectively), and DPB1*03:01 (p =9.18 × 10−7). DQB1*05:03 is in nearly complete LD with DRB1*14 (r2 = 0.97). DPB1*03:01 is not in LD with any of the DRB1, DQB1 or DQA1 classical alleles or AAs (r2 < 0.01). There was no association (p > 10−4) observed for classical alleles in HLA A, B, C or DPA1.
Table 1.
Summary of Subject Genotyping Information
| Genotypinga | ||||||
| GWASb | ICc | GWAS- only |
IC-only | Bothd | Total | |
| Italian PBC Cases | 453 | 622 | 54 | 223 | 399 | 676 |
| Italian Controls | 1042 | 597 | 843 | 398 | 199 | 1440 |
| MHC region SNP genotypes | ||||||
| GWAS | IC | GWAS- only |
IC-only | Both | Total | |
| Number of SNPse | 1548 | 4885 | 604 | 3941 | 944 | 5489 |
| SNPs overlaping 1000 Genome referencef | 1435 | 4386 | 595 | 3546 | 840 | 4981 |
| SNPs overlaping HLA referencef | 648 | 1444 | 166 | 962 | 482 | 1610 |
Additional details on subject set genotyping is in Supplemental Table 1.
GWAS: Genotyped using Illumina platforms with >550K genome-wide association study (GWAS) SNPs. The samples included in previous GWAS study.
IC: Subjects genotyped using Immunochip (IC) array.
Subjects genotyped using both GWAS platform and Immunochip array
Number of SNPs within the region [chromosome 6 bps 28911802 – 33813043 (HG19 map)] used for analyses.
Number of SNPs genotyped in region that overlap the reference sets used for imputation.
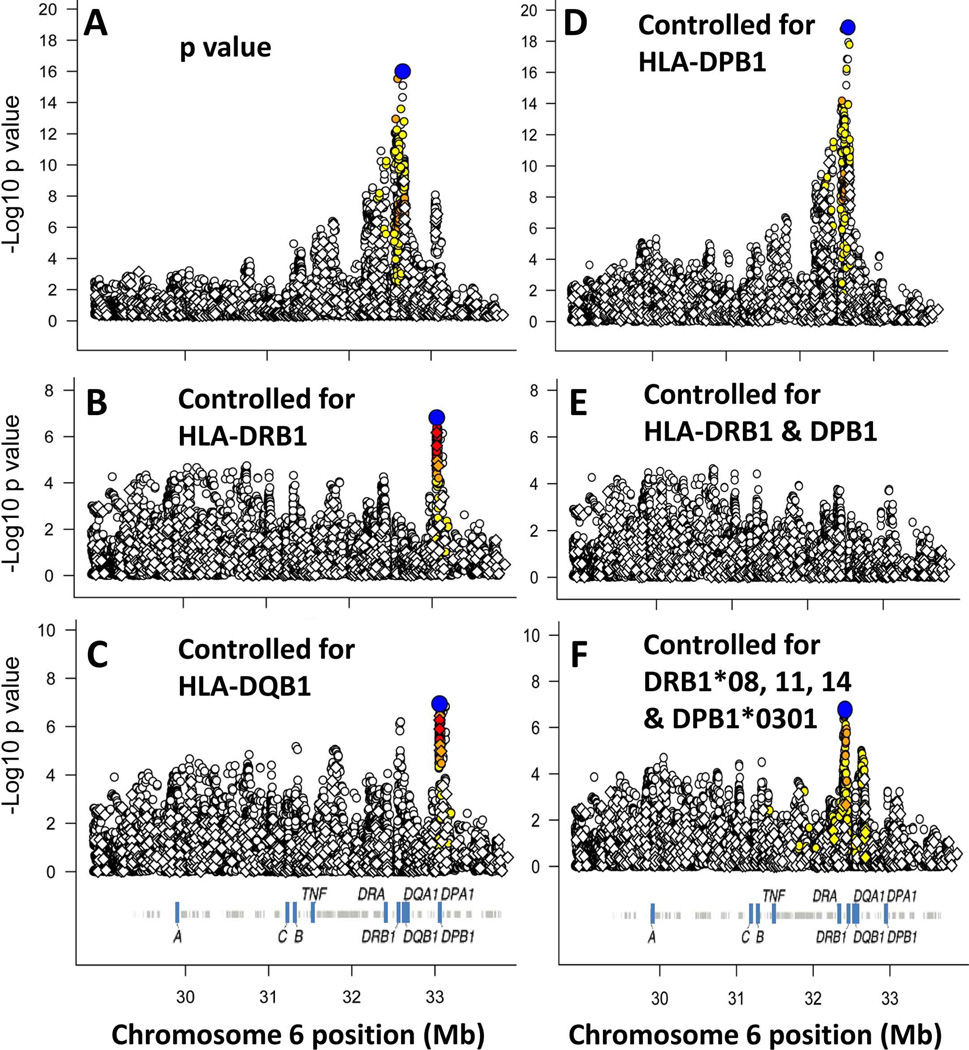
Figure 1. Analysis of the HLA region association signals in PBC.
In each panel, the symbols show the strength of the association signal (ordinate) for the corresponding position (Mb, HG19) on chromosome 6 (abscissa). For panel A, the p value before conditioning is shown. For panels B-F the p values are shown after conditioning on the HLA determinant(s) indicated in the panel. The blue color coded symbols denotes the strongest associated marker with p value <10−6, and the other markers are color coded to indicate marker LD with the strongest associated marker: Markers with strong LD (r2 > 0.8) (red); moderate LD (r2 > 0.5) (orange), weak LD (r2 > 0.2) (yellow) and little or no LD (open symbols) are shown. The SNPs with the strongest associations were rs115721871 at bp 32653792 (panels A and D), rs9277558 at bp 33056711 (panels B and C), and rs9268668 at bp 32413889 (panel F).
Table 2.
Summary of Association Results and Conditioning Studies of HLA alleles in PBCa
| Marker/Amino Acid/HLA determinantb |
base pair position (HG19)c |
Allele 1 (Minor Allele)d |
Allele 2 | MAF (Case)e |
MAF (Control) |
p-valuef | Odds Ratio minor allele |
Cond. p- value DRB1g |
Cond. p- value DPB1g |
Cond. p- value DRB1 and DPB1g |
|---|---|---|---|---|---|---|---|---|---|---|
| rs115721871 | 32653792 | A | G | 0.17 | 0.08 | 9.8E-17 | 2.61 | 9.4E-02 | 1.3E-19 | 1.2E-01 |
| rs4246055 | 32592360 | C | T | 0.19 | 0.10 | 2.9E-16 | 2.47 | 2.1E-02 | 1.6E-19 | 2.8E-02 |
| rs114183935 | 32655730 | A | G | 0.59 | 0.43 | 8.1E-16 | 1.80 | 1.9E-02 | 1.2E-18 | 3.1E-02 |
| rs114432443 | 32652278 | T | C | 0.39 | 0.53 | 4.6E-15 | 0.55 | 2.3E-02 | 1.4E-17 | 2.8E-02 |
| rs116493712 | 32669767 | T | C | 0.57 | 0.46 | 1.1E-09 | 1.56 | 2.6E-01 | 5.2E-10 | 1.8E-01 |
| rs114796881 | 32664960 | A | G | 0.30 | 0.41 | 1.2E-09 | 0.63 | 3.9E-01 | 1.8E-10 | 3.7E-01 |
| rs114327274 | 32289594 | A | G | 0.24 | 0.17 | 4.1E-08 | 1.66 | 1.2E-01 | 1.9E-08 | 1.2E-01 |
| rs116348417 | 32389648 | A | G | 0.26 | 0.37 | 4.9E-08 | 0.66 | 5.2E-01 | 6.2E-11 | 5.7E-01 |
| HLA-DRB1*08 | 32552064 | SPEC | OTHER | 0.08 | 0.03 | 1.6E-11 | 3.22 | 1.0E+00 | 7.3E-13 | 1.0E+00 |
| HLA-DQB1*04:02 | 32631061 | SPEC | OTHER | 0.07 | 0.03 | 1.4E-10 | 3.16 | 6.8E-01 | 9.0E-12 | 4.5E-01 |
| HLA-DRB1*11 | 32552064 | SPEC | OTHER | 0.15 | 0.26 | 1.4E-10 | 0.55 | 1.0E+00 | 9.4E-13 | 8.3E-01 |
| HLA-DQA1*04:01 | 32608306 | SPEC | OTHER | 0.07 | 0.02 | 1.9E-10 | 0.32 | 6.5E-01 | 9.9E-12 | 5.0E-01 |
| HLA-DQB1*03:01 | 32631061 | SPEC | OTHER | 0.21 | 0.31 | 6.1E-09 | 0.61 | 8.9E-01 | 3.8E-10 | 4.5E-01 |
| HLA-DQB1*05:03 | 32631061 | SPEC | OTHER | 0.09 | 0.05 | 6.2E-07 | 2.14 | 7.6E-01 | 8.2E-09 | 1.0E+00 |
| HLA-DRB1*14 | 32552064 | SPEC | OTHER | 0.09 | 0.05 | 6.9E-07 | 2.12 | 1.0E+00 | 8.8E-09 | 1.0E+00 |
| HLA-DPB1*03:01 | 33049368 | DET | OTHER | 0.16 | 0.10 | 9.2E-07 | 1.78 | 4.2E-06 | 1.0E+00 | 1.0E+00 |
| DRB1-AA74 | 32551948 | OTHER | A | 0.43 | 0.31 | 1.2E-11 | 1.70 | 5.9E-01 | 3.2E-12 | 9.9E-01 |
| DRB1-AA74 | 32551948 | L | OTHER | 0.08 | 0.03 | 1.3E-11 | 3.23 | 2.9E-01 | 6.1E-13 | 2.0E-01 |
| DQA1-AA69 | 32609279 | T | OTHER | 0.08 | 0.03 | 2.7E-11 | 3.21 | 6.0E-01 | 8.5E-13 | 6.4E-01 |
| DRB1-AA58 | 32551996 | E | A | 0.15 | 0.26 | 1.5E-10 | 0.55 | 1.0E+00 | 9.4E-13 | 1.0E+00 |
This Table provides a summary of results before and after conditioning on particular HLA determinants and includes: 1) the top four imputed SNPs (rows 1–4); 2) top four genotyped SNPs (rows 5–8); 3) all classical HLA determinants with p-values < 10–6 (rows 9–16); and 4) the top four HLA amino acid (AA) associations (rows 17–20). P values after conditioning on particular HLA genes (see methods) are also shown. The p values were determined using SNPTEST V2 (see Patients and Methods) and used the genotype probabilities. A complete summary of strongly association markers is provided in Supplementary Table 1.
The SNP name, classical HLA determinant or AA is shown.
The position corresponding to the marker is shown (chromosomal base pair positions from HG19).
Allele 1 is the minor allele used for the analysis. For sequences designated with amino acid (AA) numbers the symbol refers to the allele symbol corresponds to standard AA nomenclature. Where the AA is compared with multiple other AAs, "Other" is used to represent all alternative AAs. The AA position includes the leader sequence (e.g. DRB1* position 104 is the same as AA position 74 without the leader sequence). Where HLA determinants are provided, the specificity is compared with not having the specificity (OTHER).
MAF: Minor allele frequencies were estimated from probabilities (SNPTEST V2).
p values before conditioning using population substructure and gender as covariates.
p value after conditioning on the allele(s) listed in the heading line.
As expected from the analysis of classical HLA alleles, PBC also showed strong association with specific AAs in these genes. Most of the HLA AA showing association signals corresponded to the key residues that distinguish the specific classical alleles which for DRB1, included lysine (L) at AA74 in DRB1 (DRB*08), glutamate (E) at AA58 (DRB*11), alanine (A) at AA57 (DRB1*14), and histidine (H) at AA60 (DRB1*14). Similar results were observed for specific DQB1, and DQA1 AAs that are in strong LD with specific DRB1 alleles and AAs (Supporting Table 2).
Conditioning Studies Using Classical HLA Genes
To examine whether these associations could be explained for by known coding differences in genes we next performed a series of conditional analyses. These studies were done by conditioning on a combination of the alleles from an HLA gene (e.g. DRB1) to control for the association that might be attributable to each gene, albeit some of the effect may not be directly attributed to that gene due to extensive LD across this region. The residual signals after controlling for the effect of various combinations of classical alleles and AA residues in these HLA genes show that both DRB1 and DQB1 could account for most of the association signal (Fig. 1 B-E, Table 2 and Supporting Table 3). In addition, the signal in the DPB1 region was only marginally decreased conditioning on DRB1, DQB1 or DQA1. Conditioning on DPB1 eliminated the signal in the DPB1 region and showed a modest increase in the signal in the across DRB1, DQA1 and DQB1.
To further assess these conditional analyses, we also examined the relative difference of the conditioning by different HLA genes by examining beta estimates and their differences. The beta estimate is the measure of the increase in log-odds that can be attributed to each copy of a given minor allele. The largest effect is from the composite of DRB1 alleles as shown by the residual beta estimates (and odds ratios) after conditioning and the mean change in the beta estimates (Table 3). This is most evident examining the SNPs with the strongest signals from association (original signal < 5 × 10−8). For example, the DRB1 conditioning had a much larger mean change in beta estimate (−0.424) compared with DQB1 (−0.236) (p value < 10−10, paired T test). Additional conditional analyses using combinations with DQA1 demonstrated that DQA1 could not substitute for DRB1 or DPB1 in any of the combinations tested (data not shown).
Table 3.
Comparison of Residual Association Beta Estimates and Odds Ratios after Conditioning Studies
| All Markersa | Associationb | Controlled DRB1c |
Controlled DQB1 |
Controlled DQA1 |
Controlled DPB1 |
Controlled DRB1 & DPB1 |
Controlled DQB1 & DPB1 |
Controlled DRB1*8, 11, 14, & DPB1*03:01 |
|---|---|---|---|---|---|---|---|---|
| mean B estimate | 0.242 | 0.188 | 0.205 | 0.211 | 0.237 | 0.180 | 0.197 | 0.190 |
| mean change in beta estimate | Reference | −0.055 | −0.037 | −0.026 | −0.005 | −0.062 | −0.045 | −0.052 |
| mean OR | 1.274 | 1.207 | 1.228 | 1.234 | 1.268 | 1.197 | 1.218 | 1.209 |
| SNPs < 5E-8d | Association |
Controlled DRB1 |
Controlled DQB1 |
Controlled DQA1 |
Controlled DPB1 |
Controlled DRB1 & DPB1 |
Controlled DQB1 & DPB1 |
Controlled DRB1*8, 11, 14, & DPB1*03:01 |
| mean B estimate | 0.710 | 0.165 | 0.297 | 0.340 | 0.585 | 0.159 | 0.295 | 0.179 |
| mean change in beta estimate | Reference | −0.424 | −0.236 | −0.229 | −0.005 | −0.430 | −0.240 | −0.410 |
| mean OR | 2.03 | 1.18 | 1.35 | 1.41 | 1.79 | 1.17 | 1.34 | 1.20 |
All markers including SNPs (MHC region), classical HLA determinants and HLA Amino Acids.
The beta estimate, change in beta estimate and odds ratios from SNPTESTV2 score test (see methods).
Results after conditioning on the indicated HLA genes or alleles.
Results using only markers with p values < 5 × 10−8 in the initial association tests.
Conditioning on Specific HLA alleles
We next examined the effect of conditioning on specific DRB1, DQA1, DQB1 and DPB1 classical genes and AAs. A clear pattern emerged showing that the association of groups of SNPs was specifically controlled by different alleles. These results are highlighted in Table 4 and in a more complete version Supporting Table 3. Individually, the specific SNPs conditioned a part of the association signal largely corresponding to those SNPs in moderate or strong LD (r2 > 0.5) with the particular classical specificity or AA. Similar effects were observed for specific alleles in one gene in strong LD with another gene. This is particularly evident for the DRB1*14 and DQB1*05:03 in which the effects of controlling for either of these alleles was virtually indistinguishable (Supporting Table 3). Other pairs (e.g. DRB1*08 and DQB1*04:02) showed small but consistent differences in which the DRB1 allele diminished more signal than when the DQB1 allele was used in the conditioning analyses (Supporting Table 3). Notably the strongest AA association was at position 74 that is in the antigen binding pocket of DRB111, 12.
Table 4.
Conditioning on Specific HLA DRB1 and DPB1 Alleles
| Groupa | Marker/Amino Acid/HLA determinantb |
base pair position (HG19)c |
p-valued | Cond. p- value DRB1*08e |
Cond. p- value DRB1*11e |
Cond. p- value DRB1*14e |
Cond. p- value DPB1*03:01e |
|---|---|---|---|---|---|---|---|
| 1 | rs114991247 | 32583504 | 5.45E-13 | 2.41E-03 | 3.96E-13 | 2.15E-18 | 3.75E-14 |
| 1 | rs116240177 | 32561662 | 8.20E-13 | 4.82E-03 | 1.17E-12 | 3.66E-18 | 4.29E-14 |
| 1 | DRB1-AA74L | 32551948 | 1.33E-11 | 4.72E-01 | 2.26E-11 | 3.67E-16 | 1.48E-12 |
| 1 | HLA-DRB1*08 | 32552064 | 1.59E-11 | 1.00E+00 | 2.73E-11 | 4.55E-16 | 1.77E-12 |
| 1 | DQA1-AA69T | 32609279 | 2.75E-11 | 8.93E-01 | 3.55E-11 | 5.99E-16 | 2.35E-12 |
| 1 | HLA-DQB1*04:02 | 32631061 | 1.38E-10 | 5.79E-01 | 1.86E-10 | 4.02E-15 | 1.63E-11 |
| 1 | HLA-DQA1*04:01 | 32608306 | 1.90E-10 | 9.58E-01 | 1.73E-10 | 5.55E-15 | 2.01E-11 |
| 2 | rs9461776 | 32575735 | 3.02E-12 | 2.39E-14 | 3.35E-03 | 9.62E-15 | 1.53E-15 |
| 2 | rs115671039 | 32603212 | 1.98E-11 | 9.29E-13 | 4.62E-02 | 5.22E-13 | 2.91E-14 |
| 2 | HLA-DRB1*11 | 32552064 | 1.42E-10 | 1.34E-12 | 9.99E-01 | 4.34E-13 | 9.25E-14 |
| 2 | DRB1-AA58E | 32551996 | 1.45E-10 | 1.34E-12 | 9.98E-01 | 4.38E-13 | 9.30E-14 |
| 2 | DQB1-AA71T | 32632646 | 5.25E-10 | 3.65E-09 | 2.62E-04 | 4.68E-10 | 1.69E-12 |
| 2 | HLA-DQB1*03:01 | 32631061 | 6.10E-09 | 3.29E-10 | 9.76E-01 | 3.95E-10 | 3.25E-11 |
| 3 | rs380911 | 32209451 | 8.94E-09 | 1.36E-10 | 4.52E-06 | 3.71E-04 | 5.26E-09 |
| 3 | rs115240978 | 32672383 | 1.28E-08 | 1.30E-10 | 2.24E-06 | 6.75E-03 | 8.16E-10 |
| 3 | HLA-DQB1*05:03 | 32631061 | 6.21E-07 | 4.79E-09 | 5.13E-05 | 6.15E-01 | 2.31E-08 |
| 3 | HLA-DRB1*14 | 32552064 | 6.89E-07 | 5.29E-09 | 4.83E-05 | 9.99E-01 | 2.46E-08 |
| 3 | DRB1-AA57A | 32551999 | 6.90E-07 | 5.29E-09 | 4.82E-05 | 9.99E-01 | 2.46E-08 |
| 3 | DRB1-AA60H | 32551990 | 6.90E-07 | 5.29E-09 | 4.82E-05 | 9.99E-01 | 2.46E-08 |
| 4 | rs116751915 | 33052502 | 8.11E-09 | 9.21E-09 | 3.09E-06 | 1.76E-10 | 1.69E-04 |
| 4 | rs114761768 | 33050442 | 1.79E-08 | 3.09E-08 | 4.73E-06 | 4.90E-10 | 3.98E-04 |
| 4 | DPB1-AA76M | 33048662 | 6.13E-08 | 3.78E-07 | 2.11E-05 | 1.07E-08 | 8.71E-03 |
| 4 | DPB1-AA170I | 33052958 | 1.06E-07 | 2.61E-07 | 1.61E-05 | 3.21E-09 | 8.41E-03 |
| 4 | HLA-DPB1*03:01 | 33049368 | 9.18E-07 | 2.05E-06 | 1.01E-06 | 3.70E-09 | 9.98E-01 |
The group corresponds to those markers that are successfully controlled by specific HLA alleles (p value after conditioning is >1 × 10–4. The bold highlighted p-values indicate the p-values used in the “group” assignment. For each group the following are shown in this abbreviated summary: i) the two SNPs with the strongest association in the group; ii) the two HLA amino acids (AA) with the strongest association; and iii) all classical HLA alleles with p <10–6 prior to conditioning. The p values were determined using SNPTEST V2 (see methods) and used the genotype probabilities. A more complete summary of strongly association markers is provided in Supplementary Table 2.
The SNP name, classical HLA determinant or AA is shown. For the AA the letter following the residue number provides the relevant AA.
The position corresponding to the marker is shown (chromosomal base pair positions from HG19).
p values after conditioning using population substructure and gender as covariates.
p value after conditioning (Cond) on the allele(s) listed in the heading line.
Similar to when we conditioned on genes (“all” alleles at a particular gene), the DPB1 region SNP signal was only substantially reduced when DPB1*03:01 (Table 4 and Supporting Table 3) or when specific DPB1 AA’s were used in conditioning. The strongest effects were observed for the lysine at AA position 11 and the methionine at AA position 76 that are both members of the 16 AA in the putative antigen binding pocket of this gene11.
Thus, the vast majority of the HLA region association signal can be accounted individually by conditioning on one of four specific alleles, three in DRB1 (*08, *11, and *14), and one in DPB1 (*03:01). Combinations of these specific alleles accounted for most of the remaining signal (Supporting Table 3 and Fig. 1F) and are also reflected in the strong reduction in beta estimates (Table 3). However, there are signals from several SNPs in the DRB1-DQB1 region that are not accounted for by these conditioning studies. None of these SNPs with signals p <10−5 after conditioning on DRB*08, *11 and *14, and DPB1*03:01 were among the stronger associated SNPs prior to conditioning (all with original association p values >10−6). In particular, the strongest associated SNP after conditioning (rs9268668, p = 1.67 × 10−7), showed no signal prior to conditioning (p = 0.40). Whether these residual or new signals are also due to other specific classical HLA genes is not clear, however, conditioning on “all” DRB1 and DPB1 alleles ablated all signals with resulting p-values > 10−5 (Fig. 1F) suggesting that additional sequence differences (e.g. putative regulatory SNPs) do not have to be postulated.
Most of the signal observed for specific AAs was also specifically eliminated when conditioning on the DRB1 or DPB1 classical alleles. However, there were several exceptions in which the association signal was not readily decreased by controlling for single classical HLA alleles. These AAs included DRB1-AA47F, DRB1 AA74A, DQB1-AA26G, and DQB1-AA74S. For these AAs the signal was ablated when conditioning on two DRB1 alleles (DRB1*08 and DRB1*11) (Supporting Table 3). Conversely, conditioning on these AA could not account for the association of the most of the other SNPs that were not ablated by single classical HLA alleles (Supporting Table 3). Therefore, it may be less likely that these particular AAs are critical to explaining the association patterns we observed. However, we cannot exclude a specific functional role for these AAs and it is notable that DRB1-AA47 and DRB1-AA74 are both in the antigen binding pocket of DRB111, 12 and that conditioning on DRB1-AA47F did ablate several of the association signals that were not controlled by individual DRB1 classical alleles.
Genotypic Associations
We also examined genotypic associations including combinations of susceptibility alleles and combinations of risk susceptibility and protective alleles. Examining individuals with combinations of risk alleles DRB1*08 combined with DPB1*03:01 or DRB1*14 combined with DPB1*03:01 we found higher odds ratios for disease association than when examining only individuals with single susceptibility alleles (Supporting Table 4). There were insufficient numbers of DRB1*08/DRB1*14 (frequency <1%) to evaluate these heterozygote genotypes. The increased risk of the combining DPB1*03:01 with DRB1*08 or DRB1*14 was observed whether DPB1*0301 was on the same or different haplotype as that of the DRB1 risk allele (Supporting Table 4). When risk alleles were combined with the DRB1*11 protective allele the odds ratios were near 1 and there was no significant association with disease.
Finally, we also examined the cumulative combination of risk predisposing and protective alleles (Table 5 and Supporting Table 4). The count of predisposing alleles minus protective alleles showed a strong correspondence with the odds ratio for PBC. Individuals with an excess of one or two or more risk alleles showed an odds ratio (OR) of 3.05 and 5.25 between cases and controls, and conversely individuals with an excess of one or two or more protective alleles had OR of 0.5 or 0.38, respectively (Table 5). All results were compatible with an additive model of action between each of the alleles similar to our previous observations6.
Table 5.
Cumulative Effect of Risk and Protective Classical HLA Alleles for PBC Susceptibility
| Categorya | Controls Number (%)b |
Cases Number (%) |
Odds Ratio | p-value |
|---|---|---|---|---|
| RISK - PROTECTIVE =/< −2 | 177 (12.3) | 34 (5.0) | 0.38 | 5.87E-08 |
| RISK - PROTECTIVE =/< −1 | 682 (48.1) | 185 (27.4) | 0.50 | 2.03E-12 |
| RISK - PROTECTIVE = 0 | 522 (36.3) | 246 (36.4) | 1.00 | NS |
| RISK - PROTECTIVE =/> +1 | 226 (15.7) | 245 (36.2) | 3.05 | 6.94E-25 |
| RISK - PRPTECTIVE =/> 2 | 32 (2.2) | 72 (10.7) | 5.25 | 1.37E-15 |
This table shows the results of categorizing each participant based on the sum of the each risk allele (positive number) and each protective allele (negative number). The risk alleles are DRB1*08, DRB1*14 and DPB1*03:01. The protective alleles are DRB1*11 and DRB1*13. The alleles were determined from the most probable allele after imputation and haplotype analyses (see Methods).
The number of subjects that are in each category are shown together with the % of total controls (1440) or percent of cases (676).
The two tailed Fisher exact p value from contingency table analyses comparing cases and controls in each category.
Discussion
The current study of PBC association with HLA differs from previous investigations by providing the most comprehensive analysis of the entire HLA region while correcting for multiple confounding factors. Our results are consistent with a predominant role for class II genes and we believe exclude any substantial effect from either HLA class I or class III genes (there were no residual signals for these genes with p <0.0005 after accounting for class II genes). This contrasts other autoimmune diseases in which HLA class I or class III plays a predominant role (e.g. myasthenia gravis13) or strong class I gene effects are observed independent of class II associations (e.g. type 1 diabetes14 and multiple sclerosis15, 16).
Our study strongly suggests that the major gene in HLA that underlies susceptibility to PBC is DRB1. Overall DRB1 alleles show the strongest associations and conditioning studies show that DRB1 could account for almost all (except DPB1 region) of the association signal. HLA-DQB1 shows association that is only marginally less than that observed for DRB1. However, several points suggest that these associations are secondary to the strong LD between DRB1 and DQB1: 1) the overall strength of association of particular DRB1 alleles is stronger than the corresponding DQB1 allele; 2) conditioning on DRB1 could account for all DQB1 associations; and 3) residual beta-estimates after conditioning showed a substantially stronger DRB1 than DQB1 effect.
In addition, our study provides strong evidence for an independent effect of DPB1. Although previous studies have as indicated in the introduction suggested DPB1 associations in PBC, these were based on small subject sets and were difficult to evaluate. Our study demonstrates that the association of DPB1 cannot be accounted for by controlling for other HLA region genes. These results are also consistent with findings in some, but not other, autoimmune diseases in which an independent effect of DPB1 has been reported. These include juvenile idiopathic arthritis17, type 1 diabetes18, multiple sclerosis16, 19 and particular autoantibodies in systemic lupus erythematosus20.
We note that our study does not directly address whether DRB3, DRB4, DRB5 or structural variations might have additional independent associations. At present such studies are challenging due to absence of reference sets for imputation and/or difficulty in assessing these polymorphisms including whether missing genotypes (excluded SNPs with call rates <0. 95) may have excluded analysis or inclusion of SNPs within these genes in available arrays.
This study has also addressed the association of specific HLA-gene alleles. Most of the HLA association with PBC can be attributed to specific associations with DRB1*08, DRB1*11, DRB1*14 and DPB1*03:01. DRB1*08 has the strongest association, followed by DRB1*11 consistent with several previous studies6, 7. PBC associations with DRB1*14 has not been previously demonstrated, however, this weaker effect is supported by our conditioning studies that show that this classical allele can control for a set of associated SNPs and AAs that are not strongly influenced by other classical alleles (Table 3 and Supporting Table 3). The DPB1*0301 association is consistent with a previous study of a small German cohort7.
In a previous study we observed that DRB1*13 was a protective allele6. In the current study the association of DRB1*13 was weak (p = 4.9 × 10−3, OR= 0.69, 95% CL = 0.53 – 0.89) compared to the previous study (p = 3.6 × 10−6). This may be due to several factors: i) the previous study did not explicitly control for population substructure; ii) the overlap of subjects with the previous study is < 25% and the difference may reflect statistical noise; and iii) the previous study used DNA typing rather than the imputation used in the current study. It may be worth noting that DRB1*13 like DRB1*11 has an alanine at AA position 74 and thus contributes to the protective effect observed for this AA (p = 1.33 × 10−11, Supporting Table 3). Similarly, our previous study6 showed only a marginal association of DRB1*14 (uncorrected p value = 0.004) compared with a strong association (p = 6.9 × 10−7) observed in our current study. Here, the conditioning study results including the effect of controlling SNPs with very strong associations (see group 3, Table 4) provide additional support for the role of DRB1*14.
Finally, we have also considered specific HLA AAs. Most of the associated AAs are both nearly unique to the specific HLA classical alleles discussed above and also correspond to critical residues for the antigen binding pocket. Thus, associated AAs in DRB1 at AA positions 37, 47, 57, 60, 67, 70 and 74; and DPB1 at AA positions 9, 11, 76, 84, 87 are antigen pocket AAs11, 12. Consistent with our results strong associations have been recently observed with serine at position 57 and leucine at position 74 in a Japanese PBC cohort21. We also note that many of the associated DQB1 AAs are also in critical residues for antigen binding (DQB1-AA13, 26, 70, 71, 74)11, 12. Of the DRB1 associated only AA58 is not among the AAs in this functional class, whereas for DQB1 several are not in this functional class (DQB1-AA45, 56, 75, 167, 185).
In conclusion, the most parsimonious explanation consistent with the current study is that classical HLA genes and the coding variations within these genes are responsible for the HLA associations with PBC. Although we cannot exclude the possibility that other sequence variations affecting for example gene regulation could be important, our data indicates that a limited set of classical DRB1 and DPB1 alleles are sufficient to explain HLA associations with this disease. We believe the current data provides cogent information for understanding HLA-associations in PBC. Studies in other ethnic groups both within Europe and in other continental groups will also be important in further definition of the role of particular HLA genes and alleles. Lastly, our results provide additional rational for functional studies examining specific HLA genes and their relative binding to the putative disease associated epitopes of the PDC-E2 the immunodominant autoantigen epitopes of PBC22 23.
Materials and Methods
Study Population and design
The Italian PBC cases were obtained through a multi-center study and met internationally accepted criteria for the diagnosis of PBC as detailed in a previous study6. Each of the included cases also met ancestry criteria as defined below (see Ancestry). Controls were derived from several sources and this sample set information is detailed in Supporting Table 1. After data filtering and ancestry analyses contained 676 Italian PBC cases and 1440 Italian controls. All subjects enrolled in the study provided written informed consent and the study followed ethical guidelines of the most recent revision of the Declaration of Helsinki (Edinburgh, 2000).
All samples were genotyped with either Illumina (San Diego, CA 92121) genome-wide and/or Immunochip SNP platforms and the participants included the dataset from our previous GWAS as well as new samples (see Supporting Table 1). With the exception of ancestry information and assessment of relatedness the current study was restricted to genotypes in an ~4 megabase segment of human chromosome 6 (bps 28911802 – 33813043, HG19 map). This dataset comprised a minimum of 1548 and a maximum of 5489 genotyped SNPs in each individual (Supporting Table 1) and was used for the SNP and HLA imputations (see Imputation).
Data Filtering
We used stringent quality control criteria to ensure that high-quality data were included in the analyses. We excluded individuals who had >5% missing data and all individuals with cryptic relatedness and duplicate samples based on identity-by-descent status for genome-wide SNPs (PI^ > 0.15) using PLINK24.
We included only SNPs with <5% missing data, Hardy-Weinberg (H-W) equilibrium p values >10−4 in controls and >10−5 in combined cases and controls (to exclude most genotyping errors) applying these procedures in a stepwise approach separately for each dataset. For each of the separately derived control genotyping sets (Supporting Table 1), SNPs were excluded if they failed the above criteria within the individual control set or in combination with any of the other control groups, or in the complete data set. The H-W criteria were applied after exclusion of non-European individuals (see Ancestry). Finally, SNPs were excluded if allele frequency differed by > 10% between different control subject groups.
Ancestry
European ancestry was determined using 883 genome-wide SNPs with minimal or no linkage disequilibrium (LD) (r2 < 0.1). SNPs analyzed using the STRUCTURE v2.1 program25 and subjects of known European, Amerindian, East Asian and West African origin as previously described26. We used STRUCTURE to exclude non-European and admixed study participants since this method allows exclusion/inclusion criteria to be set using reference populations. Subjects with >15% non-European ancestry were excluded from further analysis.
Italian ancestry was defined using principal components analyses (PCA). For subjects with GWAS data, we used the same methods and criteria applied in a previous study with largely the same dataset. Briefly, PCA was performed using the EIGENSOFT statistical package27 utilizing 34 thousand SNPs distributed throughout the genome (r2 < 0.1) that we have previously used to define population genetic substructure2. These analyses used an independent set of Italian subjects for establishing membership [+/− 2 standard deviations (sd) in first 4 principal components (PCs)]. In the current study, a substantial portion of the samples did not have GWAS data (Supporting Table 1). For these samples we used a set of 12,579 SNPs from the Immunochip for which our empiric analyses demonstrated the ability of this set to discern Italian ancestry and exclude both other European ethnicities including Sardinian Italians (Supporting Fig. 1). Using the GWAS defined individuals, the Immunochip only genotyped samples were included using 2 sd in the first 4 PCs. In addition to the subject selection, we used the eigenvalues from the first four PCs (only the first four PCs were significant based on Tracy-Widom statistics) as covariates in our association analyses.
Imputation
We imputed SNPs, HLA classical alleles, and HLA gene amino acids (AAs) using phased reference genotypes from both the 1000 genome sequencing project (interim release June 2011) (http://www.1000genomes.org/) and an HLA defined reference set8. For the 1000 genome imputation we used IMPUTE version 228 under default parameters. The reference haplotypes for this imputation were from 1094 subjects including 381 European subjects and 98 Tuscan Italians. The number of genotyped (inference) SNPs that overlapped with the 1000 genome reference set ranged from 1435 SNPs (samples typed by GWAS), 4386 (samples typed by Immunochip) to 4981 SNPs (samples typed by GWAS plus Immunochip) (Supporting Table 1). For subsequent data analyses we utilized only imputed genotypes with maximum posterior probability scores of > 0.90. Using this parameter our empiric testing (leave one-out analyses) indicated that the maximum error rate for genotype assignment was < 0.05 and the mean error rate was < 0.01. To impute classical HLA alleles and corresponding amino acids determinants we utilized a reference separate dataset of collected by the Type 1 Diabetes Genetics Consortium (T1DGC). This reference data contains genotype data for 2,537 SNPs, selected to tag the entire MHC, and classical types for HLA-A, B, C, DRB1, DQA1, DQB1, DPA1 and DPB1 at 4-digit resolution in 2767 unrelated individuals of European descent29. The Beagle software package30 was used for this imputation under default parameters. The number of inference SNPs that overlapped with this reference dataset ranged from 648 (samples typed by GWAS), 1444 SNPs (samples typed by Immunochip) to 1610 SNPs (GWAS plus Immunochip) (Supporting Table 1). Similar to the imputation using 1000 Genome data, only SNPs with posterior probabilities of >0.90 were included in our final analyses. For imputed SNPs that overlapped between the two imputation sets and algorithms used (Impute V2.0 and Beagle) there was a nearly complete concordance of the association testing results indicating similar performance of these algorithms for this dataset. After imputation and selecting only those markers meeting posterior probability criterion this region contained a total of 49,885 markers including the genotyped SNPs that were included in association test analyses.
Association and Conditional Association Tests of Imputed SNPs and HLA determinants
SNPTEST V2.028 (web) was used for the primary association analyses for the imputed genotypes. This software uses the genotype probabilities for the imputed SNPs or determinants and accounts for genotype uncertainty. The first four PC eigenvalue scores were used as continuous variables in the association test together with the gender covariate. Analyses were performed using the SNPTEST v2 Score test algorithm that enabled both the inclusion of the covariates and conditioning tests and all of our reported results used an additive model. To minimize potential spurious results we limited our main and conditioning analyses to markers with information scores (Inf) > 0.85. This parameter is a measure of the observed statistical information for the estimate of SNP allele frequency (for additional information see https://mathgen.stats.ox.ac.uk/genetics_software/snptest/snptest.v2.pdf).
Conditioning on multiple markers either separately or together was performed using an additive model. For the HLA region, over 150 conditional analyses were performed using the SNPs and HLA determinants including all HLA determinants with p values < 10−6.
Nominal p values after correction for covariates and conditioning are provided throughout the manuscript. The p-values <10−6 would remain significant after conservative (Bonferroni) correction for the number of markers (<50,000) tested after imputation.
Supplementary Material
Acknowledgements
This study was supported by NIH grants: R01 DK056839, R01DK091823, and K08AR055688, and HYPERGENES (European Network for Genetic-Epidemiological Studies HEALTH-F4-2007-201550).
Abbreviations
- AA
Amino Acids AA
- GWAS
genome-wide association study
- H-W
Hardy-Weinberg
- LD
linkage disequilibrium
- OR
odds ratio
- PBC
primary biliary cirrhosis
- PC
principal component
- PCA
principal components analysis
- SNP
single nucleotide polymorphism
Footnotes
Disclosures: The authors declare no potential conflicts of interest.
Supplementary Information is available at the Genes and Immunity Web site.
The Italian PBC Genetic Study Group
Piero L. Almasio (Gastroenterology & Hepatology Unit, Di.Bi.M.I.S., University of Palermo, Palermo), Domenico Alvaro (Department of Medico-Surgical Sciences and Biotechnologies, Fondazione Eleonora Lorillard Spencer Cenci, University Sapienza of Rome, Rome), Pietro Andreone (Dipartimento di Medicina Clinica, Università di Bologna, Bologna), Angelo Andriulli (IRCCS Casa Sollievo della Sofferenza Hospital, San Giovanni Rotondo), Cristina Barlassina (Department of Medicine, Surgery, & Dentistry, Università degli Studi di Milano, Milan), Antonio Benedetti (Università Politecnica delle Marche, Ancona), Francesca Bernuzzi (Center for Autoimmune Liver Diseases, IRCCS Istituto Clinico Humanitas, Rozzano), Ilaria Bianchi (Center for Autoimmune Liver Diseases, IRCCS Istituto Clinico Humanitas, Rozzano), MariaConsiglia Bragazzi (Department of Medico-Surgical Sciences and Biotechnologies, Fondazione Eleonora Lorillard Spencer Cenci, University Sapienza of Rome, Rome), Maurizia Brunetto (Azienda Ospedaliera Universitaria Pisana, Pisa), Savino Bruno (Department of Internal Medicine, Ospedale Fatebene Fratelli e Oftalmico, Milan), Lisa Caliari (Center for Autoimmune Liver Diseases, IRCCS Istituto Clinico Humanitas, Rozzano), Giovanni Casella (Medical Department, Desio Hospital, Desio), Fabiola Civardi (Center for Autoimmune Liver Diseases, IRCCS Istituto Clinico Humanitas, Rozzano IRCCS Istituto Clinico Humanitas, Rozzano), Barbara Coco (Azienda Ospedaliera Universitaria Pisana, Pisa), Agostino Colli (Department of Internal Medicine, AO Provincia di Lecco, Lecco), Massimo Colombo (Fondazione IRCCS Ca’ Granda, Ospedale Maggiore Policlinico, Milan), Silvia Colombo (Treviglio Hospital, Treviglio), Carmela Cursaro (Dipartimento di Medicina Clinica, Università di Bologna, Bologna), Lory Saveria Croce (University of Trieste, & Fondazione Italiana Fegato (FIF), Trieste), Andrea Crosignani (San Paolo Hospital Medical School, Università di Milano, Milan), Francesca Donato (Fondazione IRCCS Ca’ Granda, Ospedale Maggiore Policlinico, Milan), Luca Fabris (University of Padova, Padova), Carlo Ferrari (Azienda Ospedaliero-Universitaria di Parma, Parma), Annarosa Floreani (Dept. of Surgical, Oncological and Gastroenterological Sciences, University of Padova, Padova), Andrea Galli (University of Florence, Florence), Ignazio Grattagliano (Italian College of General Practicioners, ASL Bari), Roberta Lazzari (Department of Surgical, Oncological and Gastroenterological Sciences University of Padova, Padova), Fabio Macaluso (Gastroenterology & Hepatology Unit, Di.Bi.M.I.S., University of Palermo, Palermo), Fabio Marra (University of Florence, Florence), Marco Marzioni (Università Politecnica delle Marche, Ancona), Alberto Mattalia (Santa Croce Carle Hospital, Cuneo), Renzo Montanari (Ospedale di Negrar, Verona), Lorenzo Morini (Magenta Hospital, Magenta), Filomena Morisco (University of Naples, Federico II, Naples), Luca Moroni (Center for Autoimmune Liver Diseases, IRCCS Istituto Clinico Humanitas, Rozzano), Luigi Muratori (Department of Clinical Medicine, University of Bologna, Bologna), Paolo Muratori (Department of Clinical Medicine, University of Bologna, Bologna), Grazia Niro (IRCCS Casa Sollievo della Sofferenza Hospital, San Giovanni Rotondo), Antonio Picciotto (University of Genoa, Genoa), Piero Portincasa (Department of Interdisciplinary Medicine, University Medical School, Bari), Daniele Prati (Ospedale Alessandro Manzoni, Lecco, Fondazione IRCCS Ca’ Granda, Ospedale Maggiore Policlinico, Milan), Cleofe Prisco (Ospedale Niguarda, Milan), Floriano Rosina (Division of Gastroenterology & Hepatology, Center for Predictive Medicine, Gradenigo Hospital, Turin), Sonia Rossi (Department of Internal Medicine, Ospedale Fatebene Fratelli e Oftalmico, Milan), Carlo Selmi (IRCCS Istituto Clinico Humanitas, Rozzano), Giancarlo Spinzi (Azienda Ospedaliera Valduce, Como), Mario Strazzabosco (Yale University, New Haven, Connecticut 06511, USA and University of Milan-Bicocca, Monza), Sonia Tarallo (Division of Gastroenterology & Hepatology, Center for Predictive Medicine, Gradenigo Hospital, Turin), Claudio Tiribelli (University of Trieste, & Fondazione Italiana Fegato (FIF), Trieste), Pierluigi Toniutto (University of Udine, Udine), Maria Vinci (Ospedale Niguarda, Milan), Massimo Zuin (San Paolo Hospital Medical School, Università di Milano, Milan).
References
- 1.Hirschfield GM, Liu X, Xu C, Lu Y, Xie G, Lu Y, et al. Primary biliary cirrhosis associated with HLA IL12A, and IL12RB2 variants. N Engl J Med. 2009;360(24):2544–2555. doi: 10.1056/NEJMoa0810440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Liu X, Invernizzi P, Lu Y, Kosoy R, Lu Y, Bianchi I, et al. Genome-wide meta-analyses identify three loci associated with primary biliary cirrhosis. Nat Genet. 2010;42(8):658–660. doi: 10.1038/ng.627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mells GF, Floyd JA, Morley KI, Cordell HJ, Franklin CS, Shin SY, et al. Genome-wide association study identifies 12 new susceptibility loci for primary biliary cirrhosis. Nat Genet. 2011;43(4):329–332. doi: 10.1038/ng.789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Invernizzi P. Human leukocyte antigen in primary biliary cirrhosis: an old story now reviving. Hepatology. 2011;54(2):714–723. doi: 10.1002/hep.24414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Donaldson PT, Baragiotta A, Heneghan MA, Floreani A, Venturi C, Underhill JA, et al. HLA class II alleles, genotypes, haplotypes, and amino acids in primary biliary cirrhosis: a large-scale study. Hepatology. 2006;44(3):667–674. doi: 10.1002/hep.21316. [DOI] [PubMed] [Google Scholar]
- 6.Invernizzi P, Selmi C, Poli F, Frison S, Floreani A, Alvaro D, et al. Human leukocyte antigen polymorphisms in italian primary biliary cirrhosis: A multicenter study of 664 patients and 1992 healthy controls. Hepatology. 2008;48:1906–1912. doi: 10.1002/hep.22567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mella JG, Roschmann E, Maier KP, Volk BA. Association of primary biliary cirrhosis with the allele HLA-DPB1*0301 in a German population. Hepatology. 1995;21(2):398–402. doi: 10.1002/hep.1840210221. [DOI] [PubMed] [Google Scholar]
- 8.Pereyra F, Jia X, McLaren PJ, Telenti A, de Bakker PI, Walker BD, et al. The major genetic determinants of HIV-1 control affect HLA class I peptide presentation. Science. 2010;330(6010):1551–1557. doi: 10.1126/science.1195271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cortes A, Brown MA. Promise and pitfalls of the Immunochip. Arthritis Res Ther. 2011;13(1):101. doi: 10.1186/ar3204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.de Bakker PI, McVean G, Sabeti PC, Miretti MM, Green T, Marchini J, et al. A high-resolution HLA and SNP haplotype map for disease association studies in the extended human MHC. Nat Genet. 2006;38(10):1166–1172. doi: 10.1038/ng1885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Salamon H, Klitz W, Easteal S, Gao X, Erlich HA, Fernandez-Vina M, et al. Evolution of HLA class II molecules: Allelic and amino acid site variability across populations. Genetics. 1999;152(1):393–400. doi: 10.1093/genetics/152.1.393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Karp DR, Marthandan N, Marsh SG, Ahn C, Arnett FC, Deluca DS, et al. Novel sequence feature variant type analysis of the HLA genetic association in systemic sclerosis. Hum Mol Genet. 2010;19(4):707–719. doi: 10.1093/hmg/ddp521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Vandiedonck C, Beaurain G, Giraud M, Hue-Beauvais C, Eymard B, Tranchant C, et al. Pleiotropic effects of the 8.1 HLA haplotype in patients with autoimmune myasthenia gravis and thymus hyperplasia. Proc Natl Acad Sci U S A. 2004;101(43):15464–15469. doi: 10.1073/pnas.0406756101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Noble JA, Valdes AM, Varney MD, Carlson JA, Moonsamy P, Fear AL, et al. HLA class I and genetic susceptibility to type 1 diabetes: results from the Type 1 Diabetes Genetics Consortium. Diabetes. 2010;59(11):2972–2979. doi: 10.2337/db10-0699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wu XM, Wang C, Zhang KN, Lin AY, Kira J, Hu GZ, et al. Association of susceptibility to multiple sclerosis in Southern Han Chinese with HLA-DRB1, - DPB1 alleles and DRB1-DPB1 haplotypes: distinct from other populations. Mult Scler. 2009;15(12):1422–1430. doi: 10.1177/1352458509345905. [DOI] [PubMed] [Google Scholar]
- 16.Field J, Browning SR, Johnson LJ, Danoy P, Varney MD, Tait BD, et al. A polymorphism in the HLA-DPB1 gene is associated with susceptibility to multiple sclerosis. PLoS One. 2010;5(10):e13454. doi: 10.1371/journal.pone.0013454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Runstadler JA, Saila H, Savolainen A, Leirisalo-Repo M, Aho K, Tuomilehto-Wolf E, et al. HLA-DRB1, TAP2/TAP1, and HLA-DPB1 haplotypes in Finnish juvenile idiopathic arthritis: more complexity within the MHC. Genes Immun. 2004;5(7):562–571. doi: 10.1038/sj.gene.6364129. [DOI] [PubMed] [Google Scholar]
- 18.Varney MD, Valdes AM, Carlson JA, Noble JA, Tait BD, Bonella P, et al. HLA DPA1, DPB1 alleles and haplotypes contribute to the risk associated with type 1 diabetes: analysis of the type 1 diabetes genetics consortium families. Diabetes. 2010;59(8):2055–2062. doi: 10.2337/db09-0680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bergamaschi L, Leone MA, Fasano ME, Guerini FR, Ferrante D, Bolognesi E, et al. HLA-class I markers and multiple sclerosis susceptibility in the Italian population. Genes Immun. 2010;11(2):173–180. doi: 10.1038/gene.2009.101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sebastiani GD, Galeazzi M, Tincani A, Scorza R, Mathieu A, Passiu G, et al. HLA-DPB1 alleles association of anticardiolipin and anti-beta2GPI antibodies in a large series of European patients with systemic lupus erythematosus. Lupus. 2003;12(7):560–563. doi: 10.1191/0961203303lu402oa. [DOI] [PubMed] [Google Scholar]
- 21.Umemura T, Joshita S, Ichijo T, Yoshizawa K, Katsuyama Y, Tanaka E, et al. Human leukocyte antigen class II molecules confer both susceptibility and progression in Japanese patients with primary biliary cirrhosis. Hepatology. 2012;55(2):506–511. doi: 10.1002/hep.24705. [DOI] [PubMed] [Google Scholar]
- 22.Gershwin ME, Mackay IR, Sturgess A, Coppel RL. Identification and specificity of a cDNA encoding the 70 kd mitochondrial antigen recognized in primary biliary cirrhosis. J Immunol. 1987;138(10):3525–3531. [PubMed] [Google Scholar]
- 23.Lleo A, Bowlus CL, Yang GX, Invernizzi P, Podda M, Van de Water J, et al. Biliary apotopes and anti-mitochondrial antibodies activate innate immune responses in primary biliary cirrhosis. Hepatology. 2010;52(3):987–998. doi: 10.1002/hep.23783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MA, Bender D, et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007;81(3):559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Falush D, Stephens M, Pritchard JK. Inference of population structure using multilocus genotype data: linked loci and correlated allele frequencies. Genetics. 2003;164(4):1567–1587. doi: 10.1093/genetics/164.4.1567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kosoy R, Nassir R, Tian C, White PA, Butler LM, Silva G, et al. Ancestry informative marker sets for determining continental origin and admixture proportions in common populations in America. Hum Mutat. 2009;30(1):69–78. doi: 10.1002/humu.20822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Price AL, Patterson NJ, Plenge RM, Weinblatt ME, Shadick NA, Reich D. Principal components analysis corrects for stratification in genome-wide association studies. Nat Genet. 2006;38(8):904–909. doi: 10.1038/ng1847. [DOI] [PubMed] [Google Scholar]
- 28.Marchini J, Howie B. Genotype imputation for genome-wide association studies. Nat Rev Genet. 2010;11(7):499–511. doi: 10.1038/nrg2796. [DOI] [PubMed] [Google Scholar]
- 29.Brown WM, Pierce J, Hilner JE, Perdue LH, Lohman K, Li L, et al. Overview of the MHC fine mapping data. Diabetes, obesity & metabolism. 2009;11(Suppl 1):2–7. doi: 10.1111/j.1463-1326.2008.00997.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Browning BL, Browning SR. A unified approach to genotype imputation and haplotype-phase inference for large data sets of trios and unrelated individuals. Am J Hum Genet. 2009;84(2):210–223. doi: 10.1016/j.ajhg.2009.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.