Abstract
Hydrogen sulfide (H2S) has long been known for its toxic properties; however, in recent years, evidence has emerged that this small, gaseous molecule may serve as an endogenous cell-signaling agent. Though perhaps surprising in light of its potential role as an endogenous signaling agent, a number of studies have provided evidence that H2S is a DNA-damaging mutagen. In the work reported here, the chemical mechanisms of DNA damage by H2S were examined. Using a plasmid-based DNA strand cleavage assay, it was found that micromolar concentrations of H2S generated single-strand DNA cleavage. Mechanistic studies indicate that this process involved autoxidation of H2S to generate superoxide, hydrogen peroxide and, ultimately, the well-known DNA-damaging agent hydroxyl radical via a trace metal-mediated Fenton-type reaction. Strand cleavage by H2S proceeded in the presence of physiological thiol concentrations and the known byproducts of H2S oxidation such as thiosulfate, sulfite, and sulfate do not contribute to the strand cleavage process. On the other hand, initially-generated oxidation products such as persulfide (S22−) likely undergo rapid autoxidation reactions that contribute to the generation of superoxide. The potential relevance of autoxidation processes to the genotoxic and cell signaling properties of H2S is discussed.
Introduction
Hydrogen sulfide (H2S) has long been known for its toxic properties1–5; however, in recent years, evidence has emerged that this small molecule may serve as a cell signaling agent in mammals.6–13 H2S has been implicated in the modulation of diverse processes including inflammation,14 angiogenesis,15 cytoprotection,16 nociception,17 stimulation of ATP-sensitive potassium ion channels,18 myocardial contractility,19 and vascular tone and blood pressure.20,21 Some sulfur-containing small molecules including leinamycin,22 1,2-dithiolan-3-ones,23 polysulfides,22,24–26 varacin,22,24–26 lissoclinotoxin A,22,24–26 3H-1,2-dithiole-3-thiones,27,28 and garlic-derived phytochemicals such as S-allylcysteine, 29,30 allicin,29,30 diallyl disulfide,21,31 and diallyl trisulfide 21 may gain at least a portion of their bioactivities via the release of H2S.
Though perhaps surprising in light of its potential role as an endogenous signaling agent, there is evidence that H2S is a DNA-damaging mutagen. For example, H2S showed genotoxicity in a modified comet assay where DNA repair was inhibited32 and in nontransformed human intestinal epithelial cells.33 In naked nuclei of Chinese hamster ovary cells, H2S caused nucleobase damage that was excised by the repair enzyme formamidopyrimidine glycosylase (FPG).34 Evidence for the oxidative nature of this base damage was inferred from the observation that the radical scavenger t-butylhydroxyanisole inhibited its formation. In a separate study, H2S was found to cause increased levels of 8-oxo-7,8-dihydro-2′-deoxyguanosine in coelomocytes and in Glycera dibranchiata.35 In cultured human lung fibroblasts, H2S induced a concentration-dependent increase in micronuclei, a finding suggestive of DNA damage.36 Finally, H2S was shown to be weakly mutagenic in the Salmonella typhimurium strain 1535.37 Transition metal-dependent autoxidation of H2S in the environment has been characterized,38,39 but the generation of DNA-damaging reactive oxygen species by these processes under physiologically-relevant conditions has not been well studied. In the work reported here, the chemical mechanisms of DNA damage initiated by the autoxidation of H2S under physiological conditions were examined.
Experimental
Materials
Reagents were purchased from the following suppliers and were of the highest purity available: sodium phosphate, ethidium bromide, mannitol, 2-mercaptoethanol, L-cysteine, dithiothreitol, and sodium bisulfite from Aldrich Chemical Co.; sodium sulfide nonahydrate, NaSH•xH2O, and H2S gas, superoxide dismutase (SOD), catalase, glutathione, sodium thiosulfate pentahydrate, Tris-HCl, EDTA from Sigma Chemical Co.; water (HPLC grade), sodium sulfite, sodium sulfate from Fisher Scientific; absolute ethanol from Decon Labs; diethylenetriaminepentaacetic acid (DETAPAC) from Fluka; and agarose from Lonza. Before use, the sodium sulfide was rinsed with distilled, deionized water to remove oxide impurities from the surface and then dried as described previously (weighing of freshly washed, damp material may lead to a slight underestimation of stock concentrations).40
Cleavage of plasmid DNA by H2S
CAUTION: H2S is highly toxic. Exposure to this gas can be fatal. Appropriate precautions must be taken when working with H2S gas and aqueous solutions containing the salts Na2S and NaSH.41 In a typical assay, supercoiled double-stranded plasmid DNA (pGL2-Basic, 1 μL of a 1 mg/mL solution in 10 mM Tris-HCl, 1 mM EDTA, pH 7.5) was added to water (17 μL), followed by addition of sodium sulfide nonahydrate (Na2S, 2 μL in sodium phosphate, pH 7, 500 mM). The solution was gently mixed, spun for 2 s in a tabletop centrifuge, and incubated at 37 °C for 12–14 h. Solutions of sodium sulfide were prepared immediately before use and used within 5 min of preparation. In mechanistic experiments where various additives were present in the assays, these agents were placed in the reaction mixture prior to the addition of DNA. After incubation, loading buffer (5 μL of a 50% glycerol loading buffer42) was added and the reaction mixtures were loaded onto a 0.9% agarose gel. The gel was electrophoresed for 2–3 h at 80 V in 1x TAE buffer and then stained in a dilute solution of aqueous ethidium bromide. The gel was placed on a UV transilluminator and the amount of DNA in each band quantified using an Alpha Innotech IS-1000 digital imaging system. Solutions of NaSH were calculated using a molecular weight of 56.1 g/mol. Because the NaSH salt contains waters of hydration (usually about 2–3), the reported concentrations are somewhat higher than the actual concentrations.
Results and Discussion
DNA strand cleavage by H2S
H2S can be introduced into aqueous solutions as the gaseous form or as the salts Na2S or NaSH.41 Regardless of the form in which it is introduced into solution, at pH 7.4 and 25 °C, an equilibrium mixture of composed of H2S (~30%) and the monoanion HS− (~70%) are established, with the dianion S2− present in a very small amount (for H2S, pKa1 = 6.98, pKa2 ~ 19).41 Here, we refer to this collective equilibrium mixture as “H2S”. Sulfur anions readily undergo trace metal-mediated oxidation in aerobic solution to generate superoxide radical (O2•−, Eqns 1–3).26,38,43–47 Superoxide radical disproportionates to yield hydrogen peroxide (H2O2) that, in turn, can undergo a Fenton-type reaction involving adventitious traces of transition metals to yield the well known DNA strand cleaving agent hydroxyl radical (HO•, Eqn 4 and 5).48 Accordingly, we examined the activity of H2S in a plasmid-based assay that readily measures strand cleavage by reactive oxygen species (ROS).22,24,26 In this assay, single-strand cleavage converts supercoiled plasmid DNA (form I) to the open-circular form (form II).49–51 The two forms of plasmid DNA are then separated using agarose gel electrophoresis, the gel stained with a DNA-binding dye such as ethidium bromide, and the relative amounts of cleaved and intact plasmid quantitatively determined by digital image analysis. Direct strand breaks (not requiring thermal or basic workup) monitored in this type of experiment typically arise via the reaction of radicals with hydrogen atoms on 2′-deoxyribose residues in the backbone of DNA.52–55
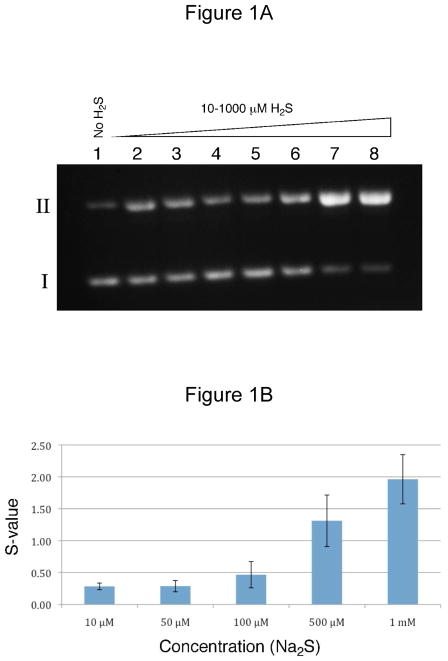
Using Na2S as a source of H2S we observed concentration-dependent cleavage of duplex DNA (Figure 1). At Na2S concentrations in the range of 10–1000 μM, nicked (form II) plasmid resulting from single-strand cleavage was observed, while no linearized (form III) plasmid arising from double-strand breakage was seen. Significant strand cleavage was observed at Na2S concentrations as low as 10 μM. The data suggests a non-linear increase in the yields of DNA strand breaks with increasing Na2S concentration in the range 10–1000 μM (Figure 1B). Previous reports indicates that high H2S:O2 ratios favor the formation of elemental sulfur as an oxidation product.44,56 Elemental sulfur may react with HS− to generate polysulfides Sn2− that, in turn, react with O2 to generate additional superoxide radical.44,56–58 To probe this possibility directly, we investigated the effect of added elemental sulfur (S8, added as a suspension in water) on DNA cleavage by Na2S. These reactions wre carried out under our standard reaction conditions reported in the Legend of Figure 1, except at 24 °C. S8 alone (16 nM) generated 0.44 ± 0.20 strand breaks above background, comparable to the 0.41 ± 0.11 strand breaks generated by Na2S (250 μM) alone. The combination of S8 (16 nM) and Na2S gave a synergistic increase to yield 1.98 ± 0.10 strand breaks above background.
Figure 1. DNA strand cleavage by H2S.
Strand cleavage assays were performed as described in the Experimental Section. Briefly, supercoiled double-stranded plasmid DNA (pGL2-Basic, 1 μL of a 1 mg/mL solution in 10 mM Tris-HCl, 1 mM EDTA, pH 7.5) was added to water (17 μL), followed by addition of sodium sulfide nonahydrate (2 μL in sodium phosphate, pH 7, 500 mM). After12 h incubation, Form I and II plasmid DNA were resolved on an agarose gel and stained with ethidium bromide. The DNA was visualized by UV-transillumination and the amounts measured by digital image analysis. The number of single strand breaks per plasmid DNA molecule (S) was calculated using the equation S = −ln f1 where f1 is the fraction of plasmid present as form I.97 Panel A. Treatment of plasmid DNA with H2S led to an increase in the amount of cleaved, form II DNA. Lane 1 contained plasmid with no H2S while lanes 2–8 contained 10, 25, 50, 100, 250, 500, and 1000 μM H2S, respectively. Panel B. A plot of strand cleavage yields (S) versus H2S concentration. Here the yields of strand cleavage were corrected for the amount of form II plasmid present in untreated plasmid DNA (lane 1).
As part of this work, we compared strand cleavage by Na2S to that by NaSH. Interestingly, at higher concentrations (50–1000 μM) the yields of strand cleavage engendered by NaSH were significantly greater than that observed for Na2S. For example, at concentrations of 500 μM, Na2S and NaSH gave 1.3 ± 0.4 and 2.1 ± 0.4 strand breaks per plasmid, respectively, under the standard reaction conditions described in the Legend of Figure 1. This may reflect the action of polysulfide contaminants such as S32− that are commonly present in the (NaSH•xH2O) reagent.41 Differences in the properties of Na2S and NaSH may be noteworthy in light of the widespread use of NaSH as a source of H2S in biological studies.
Mechanism of strand cleavage by H2S
As noted above, trace metal-mediated autoxidation of HS− may generate O2•−.26,38,43–47 To probe the involvement of O2•−, H2O2, HO•, and adventitious transition metals in strand cleavage by H2S, we performed a series of cleavage assays in the presence of additives that interact with various species shown in Eqns 2–5.48 For example, we found that strand cleavage was inhibited by the classical hydroxyl radical scavengers48 methanol, ethanol, and DMSO (Table 1). The hydrogen peroxide-destroying enzyme catalase also inhibited strand cleavage.
Table 1. Effect of additives on DNA cleavage by Na2S.
Strand cleavage assays were performed as described in the Experimental Section. Briefly, supercoiled double-stranded plasmid DNA (pGL2-Basic, 1 μL of a 1 mg/mL solution in 10 mM Tris-HCl, 1 mM EDTA, pH 7.5) was added to water (17 μL), followed by addition of sodium sulfide nonahydrate (2 μL in sodium phosphate, pH 7, 500 mM). Form I and II plasmid DNA were resolved on an agarose gel and stained with ethidium bromide. The DNA was visualized by UV-transillumination and the amounts measured by digital image analysis. The number of single strand breaks per plasmid DNA molecule (S) was calculated using the equation S = −ln f1 where f1 is the fraction of plasmid present as form I.97
| Reaction/Additive | % Nicked, Form II DNA | S-valueb |
|---|---|---|
| DNA Alone | 32.2 | 0.39±0.04 |
| 250 μM Na2S (Std.) | 49.8 | 0.70±0.09 |
| Std. + methanol (500 mM) | 33.1 | 0.40±0.06 |
| Std. + ethanol (500 mM) | 31.4 | 0.38±0.05 |
| Std. + mannitol (100 mM) | 32.8 | 0.40±0.04 |
| Std. + DETAPAC (10 mM) | 29.8 | 0.36±0.03 |
| Std. + SOD (100 μg/mL) | 88.2 | 2.18±0.23 |
| Std. + catalase (100 μg/mL) | 42.4 | 0.55±0.03 |
| (1) |
| (2) |
| (3) |
| (4) |
| (5) |
| (6) |
Interestingly, addition of superoxide dismutase (SOD) significantly increased the yield of DNA strand breaks. There are at least two possible reasons for this effect, both of which are consistent with the reaction cascade shown in Eqns 1–5. First, SOD catalyzes the disproportionation of O2•− to H2O2 and O2.48 Although spontaneous disproportionation of O2•− is fast,48,59 the ability of SOD to accelerate this reaction nonetheless has the potential to increase the yield of H2O2 formation, thus increasing the yields of strand cleavage stemming from the reactions shown in Eqns 1–5. Second, and likely more important, SOD acts as an HS−:O2 oxidoreductase that converts HS− and O2 into H2O2 and S° (Eqn 6, where S° is defined as elemental sulfur and related “sulfane” species in which sulfur is bonded only to sulfur).60 Under the reaction conditions employed here, the resulting elemental sulfur is expected25,61 to react with HS− to generate polysulfides (Sn2−) that can react with O2 to generate additional superoxide radical via reactions analogous to those shown in Eqns 2 and 3. Indeed, we presented evidence above showing that addition of elemental sulfur to a standard Na2S reaction significantly increased strand cleavage.
DNA cleavage also was effectively suppressed by the presence of diethylenetriaminepentaacetic acid (DETAPAC), a chelator of adventitious metals that inhibits transition metal-dependent Fenton-type reactions (Eqn 5).48 In analogy with the ability of chelators to inhibit metal-mediated oxidation of organic thiolates (RS−),45 it also was expected that DETAPAC could inhibit the initial metal-mediated oxidation of HS− (Eqn 2).38 To provide further evidence for the role of metals in the H2S-mediated strand cleavage process, we carried out a complimentary experiment in which we added small amounts of the transition metal Fe(III) to the reaction mixtures. We found that Fe(III) concentrations between 1 nM and 1 μM significantly increased strand cleavage induced by H2S (Table 2). Fe(III) at these low concentrations did not induce significant strand cleavage on its own. These results confirmed the metal-dependence of H2S-mediated strand cleavage process.
Table 2. Cleavage of plasmid DNA by H2S in the presence of various concentrations of iron(III).
Strand cleavage assays were performed as described in the Experimental Section. Briefly, supercoiled double-stranded plasmid DNA (pGL2-Basic, 1 μL of a 1 mg/mL solution in 10 mM Tris-HCl, 1 mM EDTA, pH 7.5) was added to water (17 μL), followed by addition of sodium sulfide nonahydrate (2 μL in sodium phosphate, pH 7, 500 mM). Form I and II plasmid DNA were resolved on an agarose gel and stained with ethidium bromide. The DNA was visualized by UV-transillumination and the amounts measured by digital image analysis. The number of single strand breaks per plasmid DNA molecule (S) was calculated using the equation S = −ln f1 where f1 is the fraction of plasmid present as form I.97
| Reaction/Additive | % Nicked, Form II DNA | S-valueb |
|---|---|---|
| DNA Alone | 33.9 | 0.40±0.05 |
| 250 μM Na2S alone (Std.) | 48.2 | 0.60±0.04 |
| Std. + FeSO4 (1 nM) | 53.7 | 0.80±0.14 |
| Std. + FeSO4 (100 nM) | 66.3 | 1.01±0.06 |
| Std. + FeSO4 (1 μM) | 83.9 | 1.83±0.04 |
| FeSO4 alone (1 μM) | 32.1 | 0.36±0.07 |
Investigating strand cleavage by the H2S decomposition products thiosulfate, sulfite, and sulfate
H2S readily undergoes oxidation both in aerobic solution and in cells.6,41,62–65 The products of this oxidation process include thiosulfate (S2O32−), sulfite (SO32−), and sulfate (SO42−).6,41,66 Given that sulfite and bisulfite (HSO3−, the protonated form of sulfite), at least, have been reported to undergo metal-mediated autoxidation reactions that generate reactive oxygen and sulfur species,67–69 we felt it was important to investigate the ability of various H2S oxidation products to cause strand cleavage under the conditions of our assay. In the event, we found that none of these H2S oxidation products generated significant levels of strand cleavage. Overall, the data indicates that the expected H2S oxidation products thiosulfate, sulfite, and sulfate do not contribute to the DNA strand cleavage observed under our reaction conditions.
Effect of thiols on DNA strand cleavage by H2S
Cells contain millimolar concentrations of thiols such as glutathione.70–72 Therefore, we examined the effects of added thiols on the ability of H2S to cause DNA strand cleavage. We find that Na2S (1 mM) in the presence of 2-mercaptoethanol (10 mM) yields 1.2 ± 0.3 strand breaks above background. Under the same conditions – except in the absence of thiol – Na2S generates 1.9 ± 0.4 strand breaks. The compound 2-mercaptoethanol alone (10 mM) generates 0.6 ± 0.5 strand breaks above the background levels of strand breaks present in the plasmid substrate, under the reaction conditions employed here. The result of this control reaction is consistent with previous reports indicating that thiols alone generate DNA strand breaks via autoxidation processes that produce ROS.73,74 Overall, our results provide evidence that H2S generates DNA strand cleavage in the presence of thiols, although the cleavage yields are somewhat diminished. Similar results were observed at lower concentrations of Na2S and thiol and with the biological thiol, glutathione. In general, thiols have the potential to act as either prooxidants or antioxidants. When incubated alone in the plasmid-based DNA-cleavage assay, the mild prooxidant properties of 2-mercaptoethanol are displayed. However, with respect to the strand cleavage caused by Na2S added thiol appears to serve as an antioxidant.
Discussion
Our results indicate that H2S undergoes trace metal-mediated autoxidation to generate superoxide, hydrogen peroxide and, ultimately, the well-known DNA-cleaving agent hydroxyl radical. The metal dependence of this process is consistent with previous reports regarding the role of transition metals in the environmental oxidation of sulfide.38,39 Though cells contain little or no free transition metals, it is clear that protein bound metals are capable of participating in such redox processes.48 Indeed hemeprotein-mediated autoxidation processes have been suggested to contribute to the cytotoxicity of H2S.4 It may be significant that, at physiological pH, significant amounts of H2S exist as the sulfur anion HS− that is the principal substrate for aerobic oxidation.41,43 In contrast, typical pKa values for thiols are substantially higher (e.g. the pKa of the thiol group in cysteine is 8.3) and relatively small amounts of the thiolate anions (RS−) are present.
Our results showed that the H2S oxidation products thiosulfate, sulfite, and sulfate do not contribute to the strand cleavage processes examined here. On the other hand, initially-generated oxidation products such as persulfide (S22−) likely undergo rapid autoxidation reactions that contribute to the generation of superoxide under our reaction conditions.44,56–58 The non-linear increase in DNA strand cleavage with increasing H2S concentrations may mesh with previous reports indicating that high H2S:O2 ratios favor the formation of elemental sulfur as an oxidation product.44,56 Elemental sulfur may react with HS− to generate polysulfides Sn2− that, in turn, react with O2 to generate additional superoxide radical.44,56–58 In this manner, polysulfides act as catalysts for sulfide oxidation and the concomitant production of ROS.22,75 Indeed, we provided evidence here that addition of even small amounts of elemental sulfur dramatically increased the DNA-cleaving properties of Na2S. Finally, we showed that DNA strand cleavage by H2S proceeded in the presence of physiological thiol concentrations.
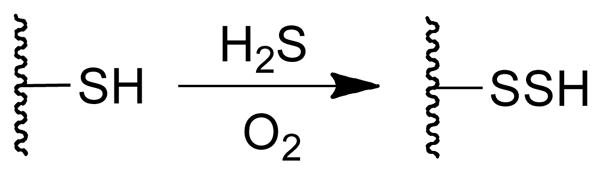
Redox chemistry of the type described here could underlie much of the biological activity associated with H2S. For example, the ability of H2S to generate superoxide, hydrogen peroxide, and hydroxyl radical under physiological conditions may explain the mutagenic properties of H2S.32,34–37 Hydroxyl radical is a well characterized mutagen.52,76 Furthermore, H2O2 produced in the autoxidation of H2S may be relevant to the putative cell signaling properties of H2S, given that H2O2 has recently become established as a cell signaling agent in its own right.77–83 Thus, it is possible that, under some circumstances, H2S serves as a means for generating H2O2 in cell signaling processes. Our results further highlight a potential role for the enzyme superoxide dismutase in catalyzing the generation of H2O2 from H2S.60 The production of ROS (specifically H2O2) may explain the ability of H2S to activate the transcription factors such as Nrf2.16,84 Likewise, H2S could contribute to the activation of Nrf2 by agents such as 3H-1,2-dithiole-3-thiones and diallyl sulfides.21,85–90 The oxidation of H2S in the presence of protein thiols has the potential to generate protein persulfides (Scheme 1). It has been suggested that such protein sulfhydration reactions are involved in the cell signaling properties of H2S.7,91,92 Finally, it has been proposed that sulfane byproducts of H2S oxidation (e.g. Sn2−, S8) may be the actual regulatory agents generated by H2S.93,94
Scheme 1.
Our work highlights some similarities between the cell signaling agents nitric oxide, H2O2, and H2S. Each of these species can mediate controlled biological responses via the selective reactions with specific target proteins, yet also have the potential to cause toxicity via “off target” reactions with bystander proteins and nucleic acids.81,95,96 The DNA-damaging properties of H2S discussed here and elsewhere32–37 emphasize the importance of spatial control in the generation of H2S if this agent does indeed serve as a cell signaling agent.
Table 3. Cleavage of plasmid DNA by H2S oxidation products.
Strand cleavage assays were performed as described in the Experimental Section. Briefly, supercoiled double-stranded plasmid DNA (pGL2-Basic, 1 μL of a 1 mg/mL solution in 10 mM Tris-HCl, 1 mM EDTA, pH 7.5) was added to water (17 μL), followed by addition of sodium sulfide nonahydrate (2 μL in sodium phosphate, pH 7, 500 mM). Form I and II plasmid DNA were resolved on an agarose gel and stained with ethidium bromide. The DNA was visualized by UV-transillumination and the amounts measured by digital image analysis. The number of single strand breaks per plasmid DNA molecule (S) was calculated using the equation S = −ln f1 where f1 is the fraction of plasmid present as form I.97
| Reaction/Additive | % Nicked, Form II DNA | S-valueb |
|---|---|---|
| DNA Alone | 31.8 | 0.33±0.02 |
| 500 μM Na2S | 43.2 | 0.56±0.08 |
| 1 mM Na2S | 46.3 | 0.63±0.12 |
| 500 μM NaS2O3 | 27.8 | 0.30±0.02 |
| 1 mM NaS2O3 | 28.1 | 0.30±0.02 |
| 500 μM Na2SO3 | 28.3 | 0.31±0.04 |
| 1 mM Na2SO3 | 27.8 | 0.29±0.03 |
| 500 μM Na2SO4 | 23.7 | 0.23±0.02 |
| 1 mM Na2SO4 | 24.7 | 0.23±0.04 |
| 500 μM NaHSO3 | 29.8 | 0.28±0.00 |
| 1 mM NaHSO3 | 31.4 | 0.31±0.03 |
Acknowledgments
Funding Support. We are grateful to the National Institutes of Health for support of this work (CA83925 and 119131). In addition, this work was made possible in part by Grant Number P50AT006273 from the National Center for Complementary and Alternative Medicines (NCCAM), the Office of Dietary Supplements (ODS), and the National Cancer Institute (NCI). Its contents are solely the responsibility of the authors and do not necessarily represent the official views of the NCCAM, ODS, NCI, or the National Institutes of Health.
References
- 1.Beauchamp RO, Bus JC, Popp JA, Boreiko CJ, Andjelkovich DA. A critical review of the literature on hydrogen sulfide toxicity. Crit Rev Toxicol. 1984;13:25–97. doi: 10.3109/10408448409029321. [DOI] [PubMed] [Google Scholar]
- 2.Truong DH, Eghbal MA, Hindmarsh W, Roth SH, O’Brien PJ. Molecular mechanisms of hydrogen sulfide toxicity. Drug Metab Rev. 2006;38:733–744. doi: 10.1080/03602530600959607. [DOI] [PubMed] [Google Scholar]
- 3.Dorman DC, Moulin FJM, McManus BF, Mahle KC, James AL, Struve M. Cytochrome oxidase inhibition induced by acute hydrogen sulfide inhalation: correlation with tissue sulfide concentrations in the rat brain, liver, lung, and nasal epithelium. Toxicol Sci. 2002;65:18–25. doi: 10.1093/toxsci/65.1.18. [DOI] [PubMed] [Google Scholar]
- 4.Eghbal MA, Pennefather PS, O’Brien PJ. H2S cytotoxicity mechanism involves reactive oxygen species formation and mitochodrial depolarisation. Toxicology. 2004;203:69–76. doi: 10.1016/j.tox.2004.05.020. [DOI] [PubMed] [Google Scholar]
- 5.Smith L, Kruszyna H, Smith RP. The effect of methemoglobin on the inhibition of cytochrome c oxidase by cyanide, sulfide, or azide. Biochem Pharmacol. 1977;26:2247–2250. doi: 10.1016/0006-2952(77)90287-8. [DOI] [PubMed] [Google Scholar]
- 6.Li L, Rose P, Moore PK. Hydrogen sulfide and cell signaling. Annu Rev Pharmacol Toxicol. 2011;51:169–187. doi: 10.1146/annurev-pharmtox-010510-100505. [DOI] [PubMed] [Google Scholar]
- 7.Mustafa AK, Gadalla MM, Snyder SH. Signaling by gasotransmitters. Sci Signal. 2009;2:re2. doi: 10.1126/scisignal.268re2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Olson KR. Is hydrogen sulfide a circulating “gasotransmitter” in vertebrate blood? Biochim Biophys Acta. 2009;1787:856–863. doi: 10.1016/j.bbabio.2009.03.019. [DOI] [PubMed] [Google Scholar]
- 9.Mancardi D, Penna C, Merlino A, Del Soldato P, Wink DA, Pagliaro P. Physiological and pharmacological features of the novel gasotransmitter: hydrogen sulfide. Biochim Biophys Acta. 2009;1787:864–872. doi: 10.1016/j.bbabio.2009.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pryor WA, Houk KN, Foote CS, Fukuto JM, Ingarro LJ, Squadrito GL, Davies KJA. Free radical biology and medicine: it’s a gas, man! Am J Physiol Regul Integr Comp Physiol. 2006;291:R491–R511. doi: 10.1152/ajpregu.00614.2005. [DOI] [PubMed] [Google Scholar]
- 11.Gadalla MM, Snyder SH. Hydrogen sulfide as a gasotransmitter. J Neurochem. 2010;113:14–26. doi: 10.1111/j.1471-4159.2010.06580.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Banerjee R, Kabil O. Redox biochemistry of hydrogen sulfide. J Biol Chem. 2010;285:21903–21907. doi: 10.1074/jbc.R110.128363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Olson KR. The therapeutic potential of hydrogen sulfide: separating hype from hope. Am J Physiol Regul Integr Comp Physiol. 2011;301:R297–R312. doi: 10.1152/ajpregu.00045.2011. [DOI] [PubMed] [Google Scholar]
- 14.Zhu L, Ang AD, Zhang H, Moore PK, Bhatia M. Hydrogen sulfide induces the synthesis of proinflammatory cytokines in human monocyte cell line U937 via the ERK-NF-kB pathway. J Leukocyte Biol. 2007;81:1322–1332. doi: 10.1189/jlb.1006599. [DOI] [PubMed] [Google Scholar]
- 15.Papapetropoulos A, Pyriochou A, Altaany Z, Yang G, Marazioti A, Zhou Z, Jeschke MG, Branski LK, Herndon DN, Wang R, Szabo C. Hydrogen sulfide is an endogenous stimulator of angiogenesis. Proc Nat Acad Sci USA. 2009;106:21972–21977. doi: 10.1073/pnas.0908047106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Calvert JW, Jha S, Gundewar S, Elrod JW, Ramachandran A, Patillo CB, Kevil CG, Lefer DJ. Hydrogen sulfide mediates cardioprotection through Nrf2 signaling. Circ Res. 2009;105:365–374. doi: 10.1161/CIRCRESAHA.109.199919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kawabata A, Ishiki T, Nagasawa K, Yoshida S, Maeda Y, Takahashi T, Sekiguchi F, Wada T, Ichida S, Nishikawa H. Hydrogen sulfide as a novel nociceptive messenger. Pain. 2007;132:74–81. doi: 10.1016/j.pain.2007.01.026. [DOI] [PubMed] [Google Scholar]
- 18.Zhao W, Zhang J, Lu Y, Wang R. The vasorelaxant effect of H2S as a novel endogenous gaseous K-ATP channel opener. EMBO J. 2001;20:6008–6016. doi: 10.1093/emboj/20.21.6008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Geng B, Yang J, Qi Y, Zhao J, Pang Y, Du T, Tang C. H2S generated by heart in rat and its effects on cardiac function. Biochem Biophys Res Commun. 2004;313:362–368. doi: 10.1016/j.bbrc.2003.11.130. [DOI] [PubMed] [Google Scholar]
- 20.Yang G, Wu L, Jiang B, Yang W, Qi J, Cao K, Meng Q, Mustafa AK, Mu W, Zhang S, Snyder SH, Wang R. H2S as a physiologic vasorelaxant: hypertension in mice with deletion of cystathione gamma-lyase. Science. 2008;322:587–590. doi: 10.1126/science.1162667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Benavides GA, Squadrito GL, Mills RW, Patel RP, Darley-Usmar VM, Doeller JE, Kraus DW. Hydrogen sulfide mediates vasoactivity of garlic. Proc Nat Acad Sci USA. 2007;104:17977–17982. doi: 10.1073/pnas.0705710104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mitra K, Kim W, Daniels JS, Gates KS. Oxidative DNA cleavage by the antitumor antibiotic leinamycin and simple 1,2-dithiolan-3-one 1-oxides: Evidence for thiol-dependent conversion of molecular oxygen to DNA-cleaving oxygen radicals mediated by polysulfides. J Am Chem Soc. 1997;119:11691–11692. [Google Scholar]
- 23.Sivaramakrishnan S, Gates KS. Possible mechanisms underlying the antitumor activity of S-deoxyleinamycin. Bioorg Med Chem Lett. 2008;18:3076–3080. doi: 10.1016/j.bmcl.2007.11.092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chatterji T, Gates KS. DNA cleavage by 7-methylbenzopentathiepin: a simple analog of the antitumor agent varacin. Bioorg Med Chem Lett. 1998;8:535–538. doi: 10.1016/s0960-894x(98)00066-3. [DOI] [PubMed] [Google Scholar]
- 25.Chatterji T, Gates KS. Reaction of thiols with 7-methylbenzopentathiepin. Bioorganic Med Chem Lett. 2003;13:1349–1352. doi: 10.1016/s0960-894x(03)00103-3. [DOI] [PubMed] [Google Scholar]
- 26.Chatterji T, Keerthi K, Gates KS. Generation of reactive oxygen species by a persulfide (BnSSH) Bioorg Med Chem Lett. 2005;15:3921–3924. doi: 10.1016/j.bmcl.2005.05.110. [DOI] [PubMed] [Google Scholar]
- 27.Fiorucci S, Orlandi S, Mencarelli A, Caliendo G, Santagada V, Distrutti E, Santucci L, Cirino G, Wallace JL. Enhanced activity of a hydrogen sulphide-releasing derivative of masalamine (ATB-429) in a mouse model of colitis. Br J Pharmacol. 2007;150:996–1002. doi: 10.1038/sj.bjp.0707193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wallace JL, Caliendo G, Santagada V, Cirino G, Fiorucci S. Gastrointestinal safety and anti-inflammatory effects of a hydrogen sulfide-releasing diclofenac derivative in the rat. Gastroenterology. 2007;132:261–271. doi: 10.1053/j.gastro.2006.11.042. [DOI] [PubMed] [Google Scholar]
- 29.Das DK. Hydrogen sulfide preconditioning by garlic when it starts to smell. Am J Physiol Heart Circ Physiol. 2007;293:H2629–H2630. doi: 10.1152/ajpheart.00996.2007. [DOI] [PubMed] [Google Scholar]
- 30.Chuah SC, Moore PK, Zhu YZ. S-allylcysteine mediates cardioprotection in an acute myocardial infarction rat model via a hydrogen sulfide-mediated pathway. Am J Physiol Heart Circ Physiol. 2007;293:H2693–H2701. doi: 10.1152/ajpheart.00853.2007. [DOI] [PubMed] [Google Scholar]
- 31.Lefer DJ. A new gaseous signaling molecule emerges: cardioprotective role of hydrogen sulfide. Proc Nat Acad Sci USA. 2007;104:17907–17908. doi: 10.1073/pnas.0709010104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Attene-Ramos MS, Wagner ED, Plewa MJ, Gaskins HR. Evidence that hydrogen sulfide is a genotoxic agent. Mol Cancer Res. 2006;4:9–14. doi: 10.1158/1541-7786.MCR-05-0126. [DOI] [PubMed] [Google Scholar]
- 33.Attene-Ramos MS, Nava GM, Muellner MG, Wagner ED, Plewa MJ, Gaskins HR. DNA damage and toxicogenomic analysis of hydrogen sulfide in human intestinal epithelial FHs 74 Int cells. Env Mol Mutagenesis. 2010;51:304–314. doi: 10.1002/em.20546. [DOI] [PubMed] [Google Scholar]
- 34.Attene-Ramos MS, Wagner ED, Gaskins HR, Plewa MJ. Hydrogen sulfide induces direct radical-associated DNA damage. Mol Cancer Res. 2007;5:455–459. doi: 10.1158/1541-7786.MCR-06-0439. [DOI] [PubMed] [Google Scholar]
- 35.Joyner-Matos J, Predmore BL, Stein JR, Christiaan L, Julian D. Hydrogen sulfide induces oxidative damage to RNA and DNA in a sulfide-tolerant marine invertebrate. Physiol Biochem Zoology. 2010;83:356–365. doi: 10.1086/597529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Baskar R, Li L, Moore PK. Hydrogen sulfide-induces DNA damage and changes in apoptotic gene expression in human lung fibroblast cells. FASEB J. 2007;21:247–255. doi: 10.1096/fj.06-6255com. [DOI] [PubMed] [Google Scholar]
- 37.Gocke E, King MT, Eckhardt K, Wild D. Mutagenicity of cosmetics ingredients licensed by the European Communities. Mutation Res. 1981;90:91–109. doi: 10.1016/0165-1218(81)90072-0. [DOI] [PubMed] [Google Scholar]
- 38.Luther GWI, Findlay AJ, MacDonald DJ, Owings SM, Hanson TE. Thermodynamics and kinetics of sulfide oxidation by oxygen: a look at inorganically controlled reactions and biologically mediated processes in the environment. Front Microbiol. 2011;2:1–9. doi: 10.3389/fmicb.2011.00062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhang JA, Millero FJ. The products from the oxidation of H2S in seawater. Geochimica Acta. 1993;57:1705–1718. [Google Scholar]
- 40.Perrin DD, Armarego WLF. Purification of Laboratory Chemicals. Butterworth Heinemann; Oxford: 1997. [Google Scholar]
- 41.Hughes MN, Centelles MN, Moore KP. Making and working with hydrogen sulfide. The chemistry and generation of hydrogen sulfide in vitro and its measurement in vivo: a review. Free Rad Biol Med. 2009;47:1346–1353. doi: 10.1016/j.freeradbiomed.2009.09.018. [DOI] [PubMed] [Google Scholar]
- 42.Sambrook J, Fritsch EF, Maniatis T. Molecular Cloning: A Lab Manual. Cold Spring Harbor Press; Cold Spring Harbor, NY: 1989. [Google Scholar]
- 43.Millero FJ, Hubinger S, Fernadez M, Garnett S. Oxidation of H2S in seawater as a function of temperature, pH, and ionic strength. Environ Sci Technol. 1987;21:439–443. doi: 10.1021/es00159a003. [DOI] [PubMed] [Google Scholar]
- 44.Chen KY, Morris JC. Kinetics of oxidation of aqueous sulfide by O2. Environ Sci Technol. 1972;6:529–537. [Google Scholar]
- 45.Misra HP. Generation of superoxide free radical during the autooxidation of thiols. J Biol Chem. 1974;249:2151–2155. [PubMed] [Google Scholar]
- 46.Albro PW, Corbett JT, Schroeder JL. Generation of hydrogen peroxide by incidental metal ion-catalyzed autooxidation of glutathione. J Inorg Biochem. 1986;27:191–203. doi: 10.1016/0162-0134(86)80060-5. [DOI] [PubMed] [Google Scholar]
- 47.Cullis CF, Trimm DL. Homogeneous catalysis of the oxidation of thiols by metal ions. Disc Faraday Soc. 1968;46:144–149. [Google Scholar]
- 48.Halliwell B, Gutteridge JMC. Role of free radicals and catalytic metal ions in human disease: an overview. Methods Enzymol. 1990;186:1–85. doi: 10.1016/0076-6879(90)86093-b. [DOI] [PubMed] [Google Scholar]
- 49.Hintermann G, Fischer HM, Crameri R, Hutter R. Simple procedure for distinguishing CCC, OC, and L forms of plasmid DNA by agarose gel electrophoresis. Plasmid. 1981;5:371–373. doi: 10.1016/0147-619x(81)90012-3. [DOI] [PubMed] [Google Scholar]
- 50.Jonson PH, Grossman LI. Electrophoresis of DNA in agarose gels. Optimizing separations of conformational isomers of double and single-stranded DNAs. Biochemistry. 1977;16:4217–4224. doi: 10.1021/bi00638a014. [DOI] [PubMed] [Google Scholar]
- 51.Mirabelli CK, Huang CH, Fenwick RG, Crooke ST. Quantitative measurement of single- and double-strand breakage of DNA in Escherichia coli by the antitumor antibiotics bleomycin and talisomycin. Antimicrobial Agents Chemother. 1985;27:460–467. doi: 10.1128/aac.27.4.460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Greenberg MM. Elucidating DNA damage and repair processes by independently generating reactive and metastable intermediates. Org Biomol Chem. 2007;5:18–30. doi: 10.1039/b612729k. [DOI] [PubMed] [Google Scholar]
- 53.Pogozelski WK, Tullius TD. Oxidative strand scission of nucleic acids: routes initiated by hydrogen atom abstraction from the sugar moiety. Chem Rev. 1998;98:1089–1107. doi: 10.1021/cr960437i. [DOI] [PubMed] [Google Scholar]
- 54.Gates KS. Chemical reactions of DNA damage and degradation. In: Platz MS, Moss RA, Jones MJ, editors. Reviews of Reactive Intermediates. John Wiley and Sons, Inc; Hoboken: 2007. pp. 333–378. [Google Scholar]
- 55.Gates KS. An overview of chemical processes that damage cellular DNA: spontaneous hydrolysis, alkylation, and reactions with radicals. Chem Res Toxicol. 2009;22:1747–1760. doi: 10.1021/tx900242k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.O’Brien DJ, Birkner FB. Kinetics of oxygenation of reduced sulfur species in aqueous solution. Environ Sci Technol. 1977;11:1114–1120. [Google Scholar]
- 57.Kleinjan WE, de Keizer A, Janssen AJH. Kinetics of chemical oxidation of polysulfide anions in aqueous solution. Water Res. 2005;39:4093–4100. doi: 10.1016/j.watres.2005.08.006. [DOI] [PubMed] [Google Scholar]
- 58.Zhang JZ, Millero FJ. The products from the oxidation of H2S in seawater. Geochim Cosmochim Acta. 1993;57:1705–1718. [Google Scholar]
- 59.Fridovich I. Superoxide radical and superoxide dismutase. Acct Chem Res. 1972;5:321–326. [Google Scholar]
- 60.Searcy DG. HS–:O2 Oxidoreductase activity of Cu,Zn superoxide dismutase. Arch Biochem Biophys. 1996;334:50–58. doi: 10.1006/abbi.1996.0428. [DOI] [PubMed] [Google Scholar]
- 61.Vineyard BD. The versatility and mechanism of the n-butylamine-catalyzed reaction of thiols with sulfur. J Org Chem. 1967;32:3833–3836. [Google Scholar]
- 62.Hildebrandt TM, Grieshaber MK. Three enzymatic activities catalyze the oxidation of sulfide to thiosulfate in mammalian and invertebrate mitochondria. FEBS J. 2008;275:3352–3361. doi: 10.1111/j.1742-4658.2008.06482.x. [DOI] [PubMed] [Google Scholar]
- 63.Mitsuhashi H, Yamashita S, Ikeuchi H, Kuroiwa T, Keneko Y, Hiromura K, Ueki K, Nojima Y. Oxidative stress-dependent conversion of huydrogen sulfide to sulfite by activated neutrophils. Shock. 2005;24:529–534. doi: 10.1097/01.shk.0000183393.83272.de. [DOI] [PubMed] [Google Scholar]
- 64.Durand M, Weinstein P. Thiosulfate in human urine following minor exposure to hydrogen sulfide: implications for forensic analysis of poisoning. Forensic Toxicol. 2007;25:92–95. [Google Scholar]
- 65.Lagoutte E, Mimoun S, Andriamihaja M, Cheumontet C, Blachier F, Bouillaud F. Oxidation of hydrogen sulfide remains a priority in mammalian cells and causes reverse electron transfer in colonocytes. Biochim Biophys Acta. 2010;1797:1500–1511. doi: 10.1016/j.bbabio.2010.04.004. [DOI] [PubMed] [Google Scholar]
- 66.Avrahami M, Golding RM. The oxidation of sulphide ion at very low concentrations in aqueous solutions. J Chem Soc A. 1968:647–651. [Google Scholar]
- 67.Jameton RA, Muller JG, Burrows CJ. Oxidative DNA damage from sulfite autoxidation catalyzed by manganese(III) Comptes Rendus. 2002;5:461–466. [Google Scholar]
- 68.Hayatsu H, Miller RCJ. The cleavage of DNA by the oxygen-dependent reaction of disulfite. Biochem Biophys Res Commun. 1972;46:120–124. doi: 10.1016/0006-291x(72)90638-9. [DOI] [PubMed] [Google Scholar]
- 69.Muller JG, Hickerson RP, Perez RJ, Burrows CJ. DNA damage from sulfite autoxidation catalyzed by a nickel(II) peptide. J Am Chem Soc. 1997;119:1501–1506. [Google Scholar]
- 70.Meister A, Anderson ME. Glutathione. Ann Rev Biochem. 1983;52:711–760. doi: 10.1146/annurev.bi.52.070183.003431. [DOI] [PubMed] [Google Scholar]
- 71.Soboll S, Grundel S, Harris J, Kolb-Bachofen V, Ketterer B, Sies H. The content of glutathione and glutathione S-transferases and the glutathione peroxidase activity in rat liver nuclei determined by a non-aqueous technique of cell fractionation. Biochem J. 1995;311:889–894. doi: 10.1042/bj3110889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Hwang C, Sinskey AJ, Lodish HF. Oxidized redox state of glutathione in the endoplasmic reticulum. Science. 1992;257:1496–1502. doi: 10.1126/science.1523409. [DOI] [PubMed] [Google Scholar]
- 73.Bode VC. Single-strand scissions induced in circular and linear lambda DNA by the presence of dithiothreitol and other reducing agents. J Mol Biol. 1967;26:125–129. doi: 10.1016/0022-2836(67)90266-5. [DOI] [PubMed] [Google Scholar]
- 74.Rosenkrantz HS, Rosenkrantz S. Degradation of DNA by cysteine. Arch Biochem Biophys. 1971;146:483–487. doi: 10.1016/0003-9861(71)90152-4. [DOI] [PubMed] [Google Scholar]
- 75.Bowers JW, Fuller MJA, Packer JE. Autoxidation of aqueous sulfide solutions. Chem Ind (London) 1966:65–66. [Google Scholar]
- 76.Reid TM, Loeb LA. Mutagenic specificity of oxygen radicals produced by human leukemia cells. Cancer Res. 1992;52:1082–1086. [PubMed] [Google Scholar]
- 77.LaButti JN, Chowdhury G, Reilly TJ, Gates KS. Redox regulation of protein tyrosine phosphatase 1B by peroxymonophosphate. J Am Chem Soc. 2007;129:5320–5321. doi: 10.1021/ja070194j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.LaButti JN, Gates KS. Biologically relevant properties of peroxymonophosphate (=O3POOH) Bioorg Med Chem Lett. 2009;19:218–221. doi: 10.1016/j.bmcl.2008.10.133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Zhou H, Singh H, Parsons ZD, Lewis SM, Bhattacharya S, Seiner DR, LaButti JN, Reilly TJ, Tanner JJ, Gates KS. The biological buffer, bicarbonate/CO2, potentiates H2O2-mediated inactivation of protein tyrosine phosphatases. J Am Chem Soc. 2011;132:15803–15805. doi: 10.1021/ja2077137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Tanner JJ, Parson ZD, Cummings AH, Zhou H, Gates KS. Redox Regulation of Protein Tyrosine Phosphatases: Structural and Chemical Aspects. Antioxid Redox Signal. 2011;15:77–97. doi: 10.1089/ars.2010.3611. [DOI] [PubMed] [Google Scholar]
- 81.Rhee SG. H2O2, a necessary evil for cell signaling. Science. 2006;312:1882–1883. doi: 10.1126/science.1130481. [DOI] [PubMed] [Google Scholar]
- 82.Paulson CE, Carroll KS. Orchestrating redox signaling networks through regulatory cysteine switches. ACS Chem Biol. 2010;5:47–62. doi: 10.1021/cb900258z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Dickinson BC, Chang CJ. Chemistry and biology of reactive oxygen species in signaling and stress responses. Nat Chem Biol. 2011;7:504–511. doi: 10.1038/nchembio.607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Kimura Y, Kimura H. Hydrogen sulfide protects neurons from oxidative stress. FASEB J. 2004;18:1165–1167. doi: 10.1096/fj.04-1815fje. [DOI] [PubMed] [Google Scholar]
- 85.Kim W, Gates KS. Evidence for thiol-dependent production of oxygen radicals by 4-methyl-5-pyrazinyl-3H-1,2-dithiole-3-thione (oltipraz) and 3H-1,2-dithiole-3-thione: Possible relevance to the anticarcinogenic properties of 1,2-dithiole-3-thiones. Chem Res Toxicol. 1997;10:296–301. doi: 10.1021/tx9601667. [DOI] [PubMed] [Google Scholar]
- 86.Chen C, Pung D, Leong V, Hebbar V, Shen G, Nair S, Li W, Kong T. Induction of detoxifying enzymes by garlic organosulfur compounds through transcription factor Nrf2: effect of chemical structure and stress signals. Free Rad Biol Med. 2004;37:1578–1590. doi: 10.1016/j.freeradbiomed.2004.07.021. [DOI] [PubMed] [Google Scholar]
- 87.Munchberg U, Anwar A, Mecklenberg S, Jacob C. Polysulfides as biologically active ingredients of garlic. Org Biomol Chem. 2007;5:1505–1518. doi: 10.1039/b703832a. [DOI] [PubMed] [Google Scholar]
- 88.Munday R, Munday JS, Munday CM. Comparative effects of mono-, di-, tri-, and tetrasulfides derived from plants of the Allium family: redox cycling in vitro and hemolytic activity and phase 2 enzyme induction in vivo. Free Rad Biol Med. 2003;34:1200–1211. doi: 10.1016/s0891-5849(03)00144-8. [DOI] [PubMed] [Google Scholar]
- 89.Munday R, Munday CM. Induction of phase II enzymes by aliphatic sulfides derived fromo garlic and onions: an overview. Methods Enzymol. 2004;382:449–456. doi: 10.1016/S0076-6879(04)82024-X. [DOI] [PubMed] [Google Scholar]
- 90.Holland R, Navamal M, Velayutham M, Zweier JL, Kensler TW, Fishbein JC. Hydrogen peroxide is a second messenger in phase 2 enzyme induction by cancer chemopreventive dithiolethiones. Chem Res Toxicol. 2009;22:1427–1434. doi: 10.1021/tx900110n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Mustafa AK, Gadalla MM, Sen N, Kim S, Mu W, Gazi SK, Barrow RK, Yang G, Wang R, Snyder SH. H2S signals through protein S-sulfhydration. Sci Signal. 2009;2:1–8. doi: 10.1126/scisignal.2000464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Krishnan N, Fu C, Pappin DJ, Tonks NK. Hydrogen Sulfide-Induced Sulfhydration of the Phosphatase PTP1B and Its Role in the Endoplasmic Reticulum Stress Response. Sci Signal. 2011;4:ra86. doi: 10.1126/scisignal.2002329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Toohey JI. Sulfur signaling: Is the agent sulfide or sulfane? Anal Biochem. 2011;413:1–7. doi: 10.1016/j.ab.2011.01.044. [DOI] [PubMed] [Google Scholar]
- 94.Toohey JI. Sulfane sulfur in biological systems: a possible regulatory role. Biochem J. 1989;264:625–632. doi: 10.1042/bj2640625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Thomas DD, Ridnour LA, Isenberg JS, Flores-Santana W, Switzer CH, Donzelli S, Hussain P, Vecoli C, Paolocci N, Ambs S, Colton CA, Harris CC, Roberts DD, Wink DA. The chemical biology of nitric oxide: implications in cellular signaling. Free Rad Biol Med. 2008;45:18–31. doi: 10.1016/j.freeradbiomed.2008.03.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Ferrer-Suelta G, Radi R. Chemical biology of peroxynitrite: kinetics, diffusion, and radicals. ACS Chem Biol. 2009;4:161–177. doi: 10.1021/cb800279q. [DOI] [PubMed] [Google Scholar]
- 97.Hertzberg RP, Dervan PB. Cleavage of DNA with methidiumpropyl-EDTA-Iron(II): Reaction conditions and product analyses. Biochemistry. 1984;23:3934–3945. doi: 10.1021/bi00312a022. [DOI] [PubMed] [Google Scholar]