Abstract
The proteasome is the degradation machine at the center of the ubiquitin-proteasome system and controls the concentrations of many proteins in eukaryotes. It is highly processive so that substrates are degraded completely into small peptides, avoiding the formation of potentially toxic fragments. Nonetheless, some proteins are incompletely degraded, indicating the existence of factors that influence proteasomal processivity. We have quantified proteasomal processivity and determined the underlying rates of substrate degradation and release. We find that processivity increases with species complexity over a 5-fold range between yeast and mammalian proteasome, and the effect is due to slower but more persistent degradation by proteasomes from more complex organisms. A sequence stretch that has been implicated in causing incomplete degradation, the glycine-rich region of the NFκB subunit p105, reduces the proteasome’s ability to unfold its substrate, and polyglutamine repeats such as found in Huntington’s disease reduce the processivity of the proteasome in a length-dependent manner.
The proteasome is at the center of the ubiquitin-proteasome system (UPS), which controls the concentrations of many proteins in eukaryotes. The proteasome is a molecular machine of around 2.5 MDa and consists of approximately 40 proteins, organized into a large barrel-shaped structure (1-3). The proteolytic sites are buried deep within the particle and are accessible only through a narrow channel such that proteins have to be unfolded before they can be degraded (1-3). A ring of six ATP-dependent motor proteins, called Rpt1-6 in yeast, is located at the entrance of the degradation channel and controls access (1, 4). The degradation signal or degron in most proteasome substrates contains a proteasome-binding tag such as a poly-ubiquitin modification and an unstructured initiation region (5, 6). The binding tags can be recognized by several proteasome subunits and the initiation region is probably recognized by the motor proteins (1, 7). The motors engage the initiation region and then translocate the substrate into the degradation chamber, unfolding any structures that the proteasome encounters along the way (8-10).
The proteasome is thought to be highly processive so that substrate proteins are degraded completely into small peptides (8, 11, 12). This processivity is important biologically because it avoids the formation of potentially toxic protein fragments. ATP-dependent proteases similar to the proteasome control protein concentrations in bacteria and archaea (13). These proteases vary in their unfolding ability over several orders of magnitude, and these differences may contribute to substrate selection (12, 14-16). The eukaryotic proteasome is the most processive of the ATP-dependent proteases analyzed so far (12). Nonetheless, it degrades a handful of proteins only partially in vivo: several transcription factors are converted into fragments that have biological activities distinct from those of the full-length proteins (17-23). The molecular mechanism that causes this partial degradation is poorly understood. Presumably, the proteasome’s processivity depends on the resistance of a given domain to unfolding, the mechanical capability of the motors, and how well the proteasome holds on to the substrate during degradation (12, 15, 19). Indeed, some evidence suggests that domains that are able to resist unfolding stall the proteasome’s progression along a substrate (19, 24). The sequence stretch just preceding a folded domain also seems to play a role in inducing processing or reducing processivity (18, 19, 25, 26).
The first characterized example of processing was the conversion of p105 into the NFκB subunit p50 as part of its activation in mammalian cells (17). In p105, a Rel homology domain is followed first by a glycine-rich region (GRR) and then the degron responsible for degradation. Presumably, processing occurs when the proteasome encounters the folded Rel homology domain as the Rpt motors are interacting with the GRR (18, 27). The NFκB signaling pathway is not found in yeast (28) but it can be reconstituted by expressing the various components from plasmids (29, 30). Surprisingly, the GRR is not required for processing in yeast (30), suggesting there might be differences in proteasomal processivity across eukaryotic species.
Polyglutamine diseases may also provide an example of a protein that changes the proteasome’s processivity. The most common of this group of disorders is Huntington’s disease (HD). HD is associated with the formation of an N-terminal fragment of the Huntingtin protein (Htt), which contains a polyglutamine stretch (31, 32). Expansion of this repetitive amino acid region to more than 35 glutamines correlates with earlier onset of the disease(31). The Htt fragment accumulates (32) despite being ubiquitinated (32, 33) and this leads to cytotoxicity. There is some evidence that protein fragments containing these polyglutamine (polyQ) repeats resist degradation and may even impede the UPS, perhaps by clogging up the proteasome (34-36).
Here we define the processivity of proteasomal degradation quantitatively by measuring the rate constants with which the proteasome progresses along a substrate and with which partially degraded substrates are released. We find that the processivity varies between species over an approximately 5-fold range, with the mammalian and frog proteasome being the most processive, worm and yeast proteasome the least processive, and fly proteasome in between. The higher processivity of the mammalian proteasome is due to a roughly fifteen-fold slower substrate release rate than for the yeast proteasome, which is partially offset by a slower unfolding rate. Thus, the mammalian proteasome has a slower, more careful, motor than the yeast proteasome. The GRR increases fragment formation primarily by decreasing the ability of the proteasome to move forward to unfold its substrate and not by increasing substrate dissociation rates. Similarly, increasing the length of a polyglutamine stretch progressively decreases the processivity of the proteasome, which may at least in part explain why Htt fragments accumulate in cells.
RESULTS and DISCUSSION
Measuring proteasome processivity
To investigate proteasomal processivity, we followed the degradation of a two-domain protein by proteasome purified from different organisms. The substrate consisted of two domains fused to each other and to a degron: an E. coli dihydrofolate reductase (DHFR) domain at the N terminus, followed by a barnase domain, and then a degron derived from yeast Sic1 (37) at the C terminus (N-DHFR-barnase-degron-C; Figure 1) (12). The barnase domain is easily unfolded by the proteasome, whereas the DHFR domain is considerably harder to degrade (8, 12) and can be further stabilized by the addition of ligands. The substrate analog methotrexate (MTX) binds DHFR tightly (Kd ~ 1 nM) and prevents its degradation by the proteasome, whereas the cofactor NADPH binds less tightly (Kd ~ 1 μM) and should stabilize DHFR to a lesser extent (38-41).
Figure 1. Measuring the unfolding ability of the proteasome.
a) Schematic depiction of the substrate, which consists of an N-terminal DHFR domain, which is followed by a barnase domain and finally a degron derived from the yeast Sic1 protein at the C terminus. Ubiquitinated substrate binds to the proteasome and either becomes deubiquitinated with the rate constant kdeubi, or degraded. During degradation, the barnase domain disappears with the rate constant kdegfull-length but the proteasome stalls at the DHFR domain. At this point the proteasome can either proceed degrade DHFR (kdegfrag) or release a DHFR fragment (krelfrag). The partitioning between the pathways is determined by the ratio of the rate constants for the two processes. b) Degradation of radiolabeled ubiquitinated full-length substrate (N-DHFR-barnase-degron-C; circles) by 40 nM yeast proteasome. No detectable DHFR fragment (N-DHFR---C’; triangles) is formed. The amounts of full-length protein and fragment are shown as a percentage of the ubiquitinated substrate presented to the proteasome at the beginning of the reaction. Error bars are the SEM from 3 experiments. c) Degradation assay as in b but in the presence of 500 μM NADPH. Approximately 35% of full-length protein that is degraded is converted to fragment. The lower fragment band most likely arises from trimming of the initial fragment that is formed, which contains an unstructured tail in addition to the DHFR domain. Error bars are the SEM of 15 experiments.
The substrate was synthesized by coupled in vitro transcription and translation, ubiquitinated by the ubiquitin-protein ligase (E3) Rsp5 (37), and highly ubiquitinated forms were purified. The ubiquitinated substrate was then presented to the proteasome under single-turnover conditions, which means that proteasome was present in great excess over substrate and each proteasome complex likely degraded at most one substrate molecule during the reaction. When the substrate binds to the proteasome it can either become degraded or it can become deubiquitinated before the unfolding motors are fully engaged, leading to dissociation (42). Degradation occurs sequentially from the C-terminal degron through the proximal barnase domain and then the N-terminal DHFR domain (Figure 1). When the proteasome encounters the DHFR domain, the reaction partitions between two branches. In the first branch, the DHFR domain becomes unfolded and the remainder of the substrate is translocated into the proteasome and proteolysed with the rate constant kdegfrag. The process reflected by this rate constant in principle includes unfolding, translocation and proteolysis steps, but it is dominated by its slowest step, the unfolding reaction (see supplementary information). In the second branch, the DHFR domain together with the remainder of the substrate dissociates from the proteasome irreversibly with the rate constant krelfrag, so that a partially degraded fragment is released (Figure 1a). We define the unfolding ability or processivity of the proteasome as the ratio of the two branches of the reaction (U = kdegfrag/krelfrag) (12). This ratio can be determined most easily from the endpoint of a degradation reaction by comparing the amount of the full-length protein that disappears (i.e., all the protein for which at least the barnase domain is degraded) to the amount of DHFR fragment formed (U = (Fraction Barnase degraded/Fraction DHFR fragment released) – 1) (Figure 1) (12).
In our degradation assays, the yeast proteasome was highly processive, as we had expected, and it degraded the N-DHFR-barnase-degron-C substrate completely without the formation of any detectable fragments, except for a small fraction of full-length protein that was deubiquitinated and therefore escaped degradation (Figure 1b). Stabilizing the DHFR domain with 500 μM NADPH stalled the proteasome’s progression such that a third of the time the DHFR domain, together with a small remnant of preceding sequence that had not yet been degraded, dissociated and escaped degradation; the other two thirds of the time the substrate was degraded completely (Figure 1c). This ratio of partitioning between the partial and complete degradation corresponds to an unfolding ability U of 1.9 ± 0.2 and is a direct measure of proteasomal processivity.
The mammalian proteasome is more processive than the yeast proteasome
We next carried out this partitioning assay with mammalian proteasome that had been affinity-purified from rabbit reticulocytes (Figure 2). We found the mammalian proteasome to be ~5-fold more processive than the yeast proteasome with an unfolding ability of U = 11 ± 2 compared to U = 1.9 ± 0.2 for the yeast proteasome (Table 1). This difference in unfolding ability is surprisingly large considering that the machines have been optimized for ostensibly the same purpose in both organisms, and the amino acid sequences of the motor subunits are approximately 60-70% identical between yeast and mammals. To establish that this difference is not simply an idiosyncrasy of the particular substrate chosen, we reversed the order of the sequence elements within the substrate, such that DHFR is degraded from the N-terminus rather than the C-terminus. As the local structure first encountered by the proteasome determines a folded domain’s ability to resist degradation (8), this reversed substrate orientation effectively presents the proteasome with a different structure to unfold. The mammalian proteasome was much more processive (U = 29 ± 7) than the yeast proteasome (U = 2.0 ± 0.2) from this direction as well (Supplementary Figure 1).
Figure 2. The unfolding ability of proteasome from rabbit as well as worms, flies, and frogs.

a,b) Degradation of radiolabeled ubiquitinated substrate (N-DHFR-barnase-degron-C; circles) by 10 nM rabbit proteasome and the accumulation of partially degraded DHFR fragment (N-DHFR---C’; triangles) in the absence (a) and presence of 500 μM NADPH (b). The amounts of full-length protein and fragment are shown as a percentage of the ubiquitinated substrate presented to the proteasome at the beginning of the reaction. Error bars are SEM of 3-4 experiments. c) Unfolding ability for proteasome prepared from different organisms as determined from degradation reactions with radiolabeled ubiquitinated substrate.
Table 1.
Unfolding abilities and kinetic parameters of proteasomes from different species.
| UNADPHa | UNADPHb | Ufreeb |
krelfrag (min−1) |
kdegfrag (min−1) |
kdegfragNADPH (min−1) |
ATPasec (min−1) |
|
|---|---|---|---|---|---|---|---|
| Yeast | 1.9±0.2 | 2.3±0.4 | 42±6 | 0.35±0.05 | 14.5±0.7 | 0.80±0.08 | 110±30 |
| Worms | 1.9±0.1 | ND | ND | ND | ND | ND | ND |
| Fly | 7.6±2 | 6.0±0.5 | 56±5 | 0.12±0.01 | 6.6±0.3 | 0.71±0.03 | 27 ± 5 |
| Frog | ND | 9±1 | 110±10 | 0.028±0.002 | 3.1±0.1 | 0.26±0.01 | 16±3 |
| Rabbit | 11±1 | 9±1 | 140±20 | 0.021±0.002 | 3.0±0.2 | 0.19±0.01 | 27±5 |
From endpoints of single-exponential fits at low proteasome concentration.
From global fitting kinetic parameters to the model of Figure 1b.
Measured in the absence of substrate. For rabbit proteasome, no difference in ATPase rate was detected in the presence of 1 μM ubiquitinated substrate.
ND: Not Determined
Several observations suggest that this difference in processivity reflects intrinsic properties of the proteasomes and not the presence of processivity factors co-purified with one proteasome but not the other, or other defects in one of the proteasome preparations. First, both yeast and mammalian proteasome preparations contained comparable proportions of singly- and doubly-capped core particles, with little if any free core particle (Supplementary Figure 2a). Second, mass spectroscopy analysis of the purified proteasomes showed that both yeast and mammalian proteasomes contained similar sets of known proteasome subunits and proteasome-associated proteins such as Rad23 and Cdc48/p97 (Supplementary Information). Indeed, the yeast proteasome preparation contained more additional associated proteins than the rabbit proteasome, including members of the hsp20, hsp70, and hsp104 chaperone families, which are not found associated with the rabbit proteasome, suggesting that the yeast proteasome has not lost any factors that might aid in unfolding or processivity in the purification. Third, yeast proteasome preparation using different conditions such as the higher salt washes used for mammalian proteasome purification, or affinity purification via a different tag all consistently yielded yeast proteasome with similar processivities, all significantly lower than that of mammalian proteasome (Supplementary Figure 2b). Finally, we mixed yeast proteasome with rabbit proteasome treated with a proteasome inactivator (AdaAhx3L3-vinyl sulfone) to test whether the rabbit proteasome preparation might contain a processivity factor that could be transferred to the yeast proteasome preparation. The yeast proteasome’s ability to unfold the DHFR domain did not increase, although we cannot rule out very slowly exchanged processivity factors (Supplementary Figure 2c-g). Together, these observations suggest that the processivities we measured reflect the proteasome particles’ intrinsic abilities to unfold domains in their substrates.
Processivity of proteasome from other species
The observed difference in processivity between yeast and mammalian proteasomes could be merely an idiosyncratic difference between the two proteasomes, or it could reflect the result of an underlying evolutionary pressure for a more processive proteasome in more complex eukaryotes. To test this idea, we compared the unfolding abilities of affinity-purified proteasomes from three additional species: worm (C. elegans), fly (D. melanogaster) and frog (X. laevis). All proteasomes were characterized by mass spectrometry (Supplementary Information) and native gel analysis (Supplementary Figure 2a) and the preparations appeared to be of similar purity. The unfolding abilities of the proteasomes increased in the order yeast ~ worm < fly < frog ~ rabbit (Figure 2c and Table 1).
There are at least two potential driving forces for greater proteasome processivity. First, the complex regulatory network in metazoans might require a more processive degradation machine, simply because there are more pathways where an undegraded fragment would wreak havoc. Second, more complex eukaryotes have a higher proportion of multi-domain proteins than simpler eukaryotes (Supplementary Information). In multi-domain proteins the domains must be unfolded and degraded one after the other and each domain can potentially cause the dissociation of an undegraded fragment. Thus, the more complex protein architecture could lead to a need for a proteasome with greater processivity. Indeed, the ability of the proteasome to unfold a domain increased with the prevalence of multi-domain proteins in the proteome (Figure 3). In addition, proteomic studies have begun to identify the collection of all proteasome substrates in different organisms and it appears that mammalian proteasome substrates also include more multi-domain proteins than their yeast orthologs (Supplementary Table).
Figure 3. Correlation between proteasomal processivity and protein complexity in different species.

The processivity of proteasomes from different organisms (yeast, worm, fly, frog, and rabbit) was compared to the fraction of multi-domain proteins in the proteome of the different organisms. The fraction was calculated by dividing the number of proteins with more than one annotated domain by the number of proteins with at least one annotated domain to eliminate any species differences in either annotation quality or the number of unstructured proteins.
A transient intermediate allows determination of unfolding and release rates
The mammalian proteasome could be more processive than the yeast proteasome because it unfolds and proteolyzes DHFR more rapidly (larger rate constant kdegfrag) or because it releases the partially degraded DHFR fragment more slowly (smaller rate constant krelfrag) (Figure 1a). To measure these rate constants, we increased the proteasome concentration in the reaction to accelerate substrate encounter and thus the rate with which the full-length protein is degraded (larger rate constant kdegfull-length). This allowed us to observe the rapid formation of DHFR fragment as the proteasome first runs into the DHFR domain and stalls. The intermediate consisted of the DHFR domain and a tail of some 80 amino acids. Most likely, the DHFR domain was located at the entrance to the degradation channel and the undegraded tail reached down the degradation channel past the ATPase subunits to the proteolytic sites. Some of the fragment then decayed as the proteasome was able to unfold and degrade the DHFR domain. During this time, the undecayed fragment remained associated with the proteasome as a productive reaction intermediate, as it could not be competed away with an excess of unlabeled substrate added to the reaction at different times (Supplementary Figure 3). At the end of the reaction a certain amount of fragment that the proteasome failed to degrade remained as an inert end product (Figure 4). The final amount of fragment formed and thus the unfolding strength of the proteasome did not depend on the proteasome concentration in the reaction.
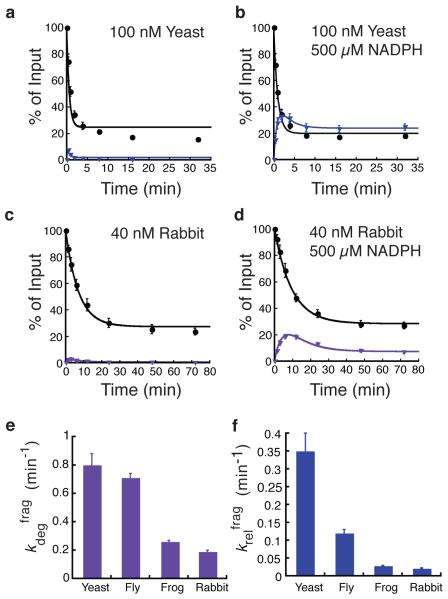
Figure 4. Kinetic mechanism of proteasomal processivity for different species.
a-d) Degradation of radiolabeled ubiquitinated full-length substrate (N-DHFR-barnase-degron-C; circles) and formation and decay of DHFR fragment (N-DHFR---C’; triangles) with distinct kinetics under the following conditions: 100 nM yeast proteasome (a), 100 nM yeast proteasome and 500 μM NADPH (b), 40 nM rabbit proteasome (c), and 40 nM rabbit proteasome and 500 μM NADPH (d). Solid lines are fits to the kinetic model in Figure 1a, in which a DHFR fragment is formed transiently and is then either released or degraded. The amounts of full-length protein and fragment are shown as a percentage of the ubiquitinated substrate presented to the proteasome at the beginning of the reaction. Errors bars are SEM of 5-10 experiments. e,f) Rate constant for fragment release krelfrag (e) and fragment degradation kdegfrag (f) as determined by global curve fitting of degradation reactions with N-DHFR-barnase-degron-C and proteasome from different organisms.
To determine the rate constants kdegfrag and krelfrag from the degradation curves we used global curve fitting of the disappearance of full-length protein and the formation and degradation of fragment to the reaction model described above (Figure 4; Table 1) (see Supplementary Methods for details). The unfolding abilities calculated from the ratio of kdegfrag to krelfrag obtained by curve fitting were essentially identical to those determined directly from the final levels of fragment formed by yeast and rabbit proteasome in the presence of NADPH (Table 1), which validates the parameters obtained by curve fitting.
Mammalian proteasome releases substrate more slowly than yeast proteasome
Now that the individual rate constants could be determined, we could establish the mechanism for the greater processivity of the rabbit proteasome. The ~5-fold greater unfolding ability results from a ~16-fold slower fragment release rate (krelfrag), which is offset by a ~4-fold slower rate of unfolding and proteolyzing of DHFR (kdegfrag) (Figure 4 & Table 1). Thus, the mammalian proteasome is slower but also more persistent than the yeast proteasome. The slower degradation rate of the rabbit proteasome may be a reflection of a slower overall catalytic cycle for the proteasome, and consistent with this hypothesis the mammalian proteasome had a 4-fold slower ATPase rate than the yeast proteasome (Table 1). The situation is similar for proteasome from flies and frogs: the increase in processivity was due principally to a decrease in krelfrag for proteasome from more complex organisms (Figure 4e & Table 1), offset by smaller changes in kdegfrag (Figure 4f & Table 1).
Despite its lower unfolding rate, the mammalian proteasome is as powerful as the yeast proteasome. If each ATP hydrolysis corresponds to one cycle of pulling, we can determine the number of pulls required to degrade DHFR by dividing the ATP hydrolysis rate by the forward degradation rate kdegfrag (43). According to this calculation, the mammalian proteasome can degrade DHFR in 9 ± 2 ATPs or pulls, while the yeast proteasome requires 8 ± 2 ATPs or pulls on average. In the presence of saturating NADPH, ~140 pulls are required to degrade the stabilized DHFR for both yeast and mammalian proteasome. Thus, approximately the same number of ATP-hydrolysis events are required for any of the proteasomes to unfold DFHR. This result suggests that unfolding and translocation are coupled tightly to the ATP-hydrolysis cycle – regardless of the species of origin each ATP-dependent powerstroke has the same probability of unfolding the domain, and therefore likely has the same translocation step-size and exerts the same amount of force on the substrate. However, yeast proteasome releases its substrate once every ~300 ATP hydrolysis events, while the mammalian proteasome only releases once every ~1300 cycles (ATPase rate divided by krelfrag).
A processing element decreases the unfolding rate
Although the proteasome is quite processive, certain amino acid sequences can reduce this processivity and lead to the release of undigested protein fragments. The partial degradation seems to be part of some regulatory processes in the cell and to control the activities of several transcription factors including p105, Spt23, Mga2, Ci and the Gli proteins (17-23). The conversion of p105 to p50 in mammalian cells requires a specific amino acid sequence, the 35 amino acid long GRR (18, 27). The GRR also directs fragment formation in model proteins when placed adjacent to folded domains such as DHFR (18, 19). To determine how the GRR reduces proteasome processivity, we inserted the GRR or a 35 amino acid long control sequence between barnase and DHFR. The GRR reduced the processivity of the yeast proteasome by roughly an order of magnitude relative to the control sequence both in the presence and absence of NADPH (Figure 5). In the presence of NADPH less than half of the protein containing the GRR is degraded completely. Global fitting of the degradation data again allowed us to determine the rate constants for partitioning between fragment degradation and release (Figure 1a; Table 2). These showed that the GRR reduced the effective unfolding ability of the proteasome entirely by reducing the degradation rate constant kdegfrag ~17-fold while leaving the release rate constant krelfrag largely unaffected. Thus, the GRR promotes fragment formation by preventing the proteasome from unfolding DHFR rather than by accelerating the release of the fragment. The GRR is dispensable for processing of p105 when it is expressed artificially in yeast (30), and this could be due to the lower intrinsic unfolding ability of the yeast proteasome.
Figure 5. The GRR sequence leads to a decrease in proteasomal processivity.
a) Degradation of N-DHFR-GRR-barnase-degron-C (circles) and formation of DHFR fragment (N-DHFR---C’; triangles) by 100 nM yeast proteasome. b) Degradation of DHFR-35ctrl-barnase-degron, where 35ctrl is a control sequence derived from the pre-sequence to yeast cytochrome b2, by 100 nM yeast proteasome. For the control sequence, no fragment is formed in the absence of NADPH. c,d) As in a,b but in the presence of 500 μM NADPH. The GRR sequence leads to significantly more fragment than the control sequence. Solid lines are fits from kinetic modeling. The amounts of full-length protein and fragment are shown as a percentage of the ubiquitinated substrate presented to the proteasome at the beginning of the reaction. Error bars are SEM of 4-9 experiments.
Table 2.
Unfolding abilities and kinetic parameters for GRR and control inserts calculated from kinetic modeling.
| UNADPH | U |
krelfrag (min−1) |
kdegfrag (min−1) |
kdegfragNADPH (min−1) |
|
|---|---|---|---|---|---|
| GRR | 0.9±0.3a | 18±8 | 0.08±0.03 | 1.5±0.3 | 0.072±0.007 |
| 35ctrl | 6±1 | 56±5 | 0.18±0.01 | 10.2±0.2 | 1.05±0.05 |
Calculating the unfolding ability from the endpoints of single-exponential fits yields U = 0.47±0.05
This reduction in kdegfrag suggests that the Rpt motors exert less unfolding force than usual on the folded domain while interacting with the GRR. ClpXP and the other bacterial AAA+ proteases use aromatic paddles to move the substrate through the degradation channel and the proteasome likely uses a similar mechanism (7, 9, 10, 44, 45). Perhaps the GRR contains fewer chemical features that the paddles can interact with to pull on the substrate. The polypeptide chain may also interact transiently with additional binding sites in the motor or translocation channel as it moves through the protease, for example to prevent back-slipping of the substrate. Perhaps the GRR sequence can bind to these sites only weakly, such that the substrate slips back more frequently allowing partially unfolded domains to snap back more frequently. Similar effects of the GRR on unfolding ability were seen with mammalian proteasome, but the kinetics were such that the underlying rate constants could not be determined (Supplementary Figure 4).
Glutamine repeats reduce unfolding ability in a length-dependent manner
Next, we asked whether polyglutamine (polyQ) repeats such as those found in the Htt protein associated with HD might affect proteasomal processivity. We inserted stretches of different numbers of glutamine residues (11, 33 and 53) into the substrate proteins and found that the processivity of the rabbit and yeast proteasomes decreased with longer polyQ insertions up to 10-fold (Figure 6). We could not determine the unfolding and release rate constants for the fragment because the kinetics of its formation followed single exponential behavior at all tested proteasome concentrations. The fact that the polyQ sequence prevented the transient accumulation of intermediate observed in the parent construct by itself does not provide a clue for the mechanism. Its transient accumulation could be prevented either because the polyQ sequence caused the proteasome-bound fragment to disappear more rapidly through degradation or release, or because the polyQ sequence caused the fragment to be formed more slowly. The simplest explanation is that polyQ sequence acts like the GRR sequence and slows the forward movement of the proteasome along its substrates, however we cannot establish this mechanism directly. In the absence of NADPH, fragment formation was seen only for the longer 53 glutamine repeats (Figure 6 & Supplementary Figure 5). Thus, the proteasome is able to degrade polyQ sequences, but they appear to slow proteasome progression along a substrate, presumably allowing the release of partially degraded Htt fragments that can go on to form aggregates. PolyQ tracts might also prevent degradation due to their resistance to proteolytic cleavage (36) (i.e., due to sequence preferences of the proteolytic sites on the proteasome) although more recent studies indicate that the proteasome can cleave the polyQ tract at multiple places (46).
Figure 6. PolyQ insertions lead to a length-dependent decrease in processivity.
a-f) Degradation of N-DHFR-polyQ-barnase-degron-C (circles) by 40 nM rabbit proteasome and formation of partially degraded substrate (N-DHFR---C’; triangles) in the absence (a-c) and presence of 500 μM NADPH (d-f). In the absence of NADPH, only Q53 leads to substantial fragment formation (a-c), whereas in the presence of NADPH, increasing numbers of glutamines lead to increasing final levels of fragment (d-f). The amounts of full-length protein and fragment are shown as a percentage of the ubiquitinated substrate presented to the proteasome at the beginning of the reaction. g,h) Plots of unfolding ability as determined by the endpoints of degradation kinetics against the length of the polyQ inserts in N-DHFR-polyQ-barnase-degron-C substrates for rabbit proteasome (g) and for yeast proteasome (h). Error bars are SEM of 4-6 experiments.
PolyQ stretches appear to have a similar effect on proteasomal processivity as the GRR. PolyQ and GRR sequences are both characterized by a strongly biased amino acid composition and it has been proposed that these low complexity regions are part of a general processing signal (19). It is possible that low complexity regions in general slow the progression of the proteasome along a substrate because they lack sequence features that can be recognized by the ATPase motors. The likelihood of particular consensus motifs occurring by chance in an amino acid sequence becomes lower as the amino acid composition of that sequence becomes less complex. Interestingly, inserting the control sequence into the substrate increased the unfolding ability of the proteasome compared to constructs without the insert (essentially comparing the effect of the control sequence to that of the barnase sequence), suggesting that amino acid sequence in general, and not just GRR and polyQ sequences, can affect processivity and that sequence complexity is not the only factor.
In Huntington’s disease, ubiquitinated aggregates of Htt exon1 fragments persist in neurons and accumulate slowly over many years to eventually form visible inclusions (32). The accumulation of aggregates correlates with the disease phenotype, though soluble forms of Htt exon1 rather than the aggregates themselves may be the toxic species (47). The poor degradation of polyQ-containing proteins observed here might be a reason why Htt exon1 protein is not cleared from cells effectively.
In our assays, all of the polyQ insertions decreased the degradation of DHFR stabilized by NADPH, and did so the more effectively the longer they were. Only the longest polyglutamine stretch examined here (53 glutamines) began to prevent degradation of DHFR in the absence of NADPH. This observation is reminiscent of the finding that only repeats of more than 35 glutamines in Htt appear to lead to significant aggregation and HD clinically (31). The test substrates with the unstabilized DHFR domain may represent a model for pre-aggregation degradation, where longer glutamine expansions begin to hamper proteasomal degradation and allow the slow formation and accumulation of polyQ fragments that will eventually aggregate. Indeed, at least some htt aggregates contain polyQ stretches of different lengths as would be created when the proteasome is no longer fully processive (48). The test substrates in which the DHFR domain is stabilized by the NADPH ligand might represent a model for htt exon1 proteins where some of the polyQ stretch began to form aggregates, which act like stable folded domains so that the proteasome is unable to unfold and clear the proteins. If the polyQ sequences inhibit proteasome progress along its substrate just as the GRR does, these sequences may attenuate overall proteasome activity in the cell by increasing the time that partially degraded substrates remain associated with proteasomes. However, the evidence whether or not the UPS is attenuated in a clinically meaningful way in HD is contradictory (34, 49-52).
Conclusion
We have established an assay to describe the processivity of the proteasome quantitatively by measuring the forward degradation and substrate dissociation rates that determine whether the proteasome digests or releases a substrate as it proceeds along the polypeptide chain. The mammalian proteasome is the most processive proteasome amongst those we investigated. Its higher unfolding and degradation power is caused by a much slower release of partially degraded substrates than was observed for the yeast proteasome. This may allow the mammalian proteasome to handle complex proteins reliably. However, the mammalian proteasome’s slower release rate may also make it more likely to choke on hard-to-degrade substrates such as Htt aggregates, potentially leading to an increase in toxicity in any number of protein misfolding diseases.
METHODS
Proteasome purification
Proteasomes were purified as described previously or by adapting previous protocols (Supplementary Information).
Constructs
Constructs encoding substrate proteins were cloned into pGEM-3Zf+ (Promega). The main unfolding ability substrate consisted of an N-terminal His-tag followed by E. coli dihydrofolate reductase (DHFR) followed by a catalytically inactive (H102A) Bacillus amyloliquefaciens barnase with all lysines replaced by arginine, methionine or alanine followed by 60 amino acids from the Sic1 protein containing a PPXY Rsp5 ubiquitination motif. The complex insertion between DHFR and barnase corresponded to the first 35 amino acids of the S. cerevisiae cytochrome b2 mitochondrial targeting presequence with lysines changed to arginines or glutamines. The GRR insertion corresponded to residues 369-403 of human p105. The polyQ insertions consisted of 11, 33 or 53 glutamine repeats. For protein expression in E. coli, the unfolding ability construct was subcloned into pet44a just downstream of the T7-lac promoter. Constructs were assembled using a combination of traditional restriction enzyme cloning and In-Fusion (Clontech).
Substrates
Radioactive substrates were in vitro translated using the RTS 100 E. coli HY kit (5 PRIME), supplemented with 35S methionine. After high-speed ultracentrifugation, substrates were affinity purified using Talon magnetic beads (Clontech) as described previously (24). The substrates were then in vitro ubiquitinated and purified (Supplementary Information).
Proteasomal degradation assay
Assays were carried out under single turnover conditions (enzyme in vast excess of substrate) at 30 °C essentially as described previously (24). Degradation data were analyzed by curve fitting. Single exponential curve fits were performed using non-linear least-squares fitting in Igor Pro (Wavemetrics). Global fits were performed by kinetic modeling, as described in the Supplementary Information.
ATPase assays
ATPase assays were performed using a coupled pyruvate kinase/lactate dehydrogenase assay at 30 °C in the presence of saturating ATP. Briefly, proteasome (10-100 nM) was mixed with 0.4 mM NADH, 2 mM phosphoenolpyruvate, 1 mM DTT and 1 mM ATP in a buffer consisting of 50 mM HEPES*KOH pH 7.5, 50 mM KCl, 5 mM MgCl2, and the disappearance of NADH was monitored by absorbance in a Molecular Devices SpectraMaxPlus plate reader.
Mass Spectrometry and Proteomic Analysis
Supplementary Material
ACKNOWLEDGEMENTS
We thank R. Carthew, C. LaBonne, R. Morimoto, M. Saks, and S. Wignall all in the Department of Molecular Biosciences at Northwestern University for excellent advice and for generously providing materials. We are also grateful to the members of the Carthew, Morimoto and Matouschek labs for their help. This work was supported by the National Institutes of Health (grants R01GM63004 and U54 CA143869) and the American Cancer Society through a Postdoctoral Fellowship (PF-09-084-01-TBE) to D.A.K..
Footnotes
Supporting Information Available: This material is free via the Internet.
REFERENCES
- 1.Finley D. Recognition and processing of ubiquitin-protein conjugates by the proteasome. Annu Rev Biochem. 2009;78:477–513. doi: 10.1146/annurev.biochem.78.081507.101607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Voges D, Zwickl P, Baumeister W. The 26S proteasome: a molecular machine designed for controlled proteolysis. Annu Rev Biochem. 1999;68:1015–1068. doi: 10.1146/annurev.biochem.68.1.1015. [DOI] [PubMed] [Google Scholar]
- 3.Groll M, Bochtler M, Brandstetter H, Clausen T, Huber R. Molecular machines for protein degradation. Chembiochem. 2005;6:222–256. doi: 10.1002/cbic.200400313. [DOI] [PubMed] [Google Scholar]
- 4.Bar-Nun S, Glickman MH. Proteasomal AAA-ATPases: Structure and function. Biochim Biophys Acta. 2012;1823:67–82. doi: 10.1016/j.bbamcr.2011.07.009. [DOI] [PubMed] [Google Scholar]
- 5.Schrader EK, Harstad KG, Matouschek A. Targeting proteins for degradation. Nat Chem Biol. 2009;5:815–822. doi: 10.1038/nchembio.250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Prakash S, Tian L, Ratliff KS, Lehotzky RE, Matouschek A. An unstructured initiation site is required for efficient proteasome-mediated degradation. Nat Struct Mol Biol. 2004;11:830–837. doi: 10.1038/nsmb814. [DOI] [PubMed] [Google Scholar]
- 7.Martin A, Baker TA, Sauer RT. Pore loops of the AAA+ ClpX machine grip substrates to drive translocation and unfolding. Nat Struct Mol Biol. 2008;15:1147–1151. doi: 10.1038/nsmb.1503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lee C, Schwartz MP, Prakash S, Iwakura M, Matouschek A. ATP-dependent proteases degrade their substrates by processively unraveling them from the degradation signal. Mol Cell. 2001;7:627–637. doi: 10.1016/s1097-2765(01)00209-x. [DOI] [PubMed] [Google Scholar]
- 9.Aubin-Tam M-E, Olivares AO, Sauer RT, Baker TA, Lang MJ. Single-Molecule Protein Unfolding and Translocation by an ATP-Fueled Proteolytic Machine. Cell. 2011;145:257–267. doi: 10.1016/j.cell.2011.03.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Maillard RA, Chistol G, Sen M, Righini M, Tan J, Kaiser CM, Hodges C, Martin A, Bustamante C. ClpX(P) Generates Mechanical Force to Unfold and Translocate Its Protein Substrates. Cell. 2011;145:459–469. doi: 10.1016/j.cell.2011.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nussbaum AK, Dick TP, Keilholz W, Schirle M, Stevanović S, Dietz K, Heinemeyer W, Groll M, Wolf DH, Huber R, Rammensee HG, Schild H. Cleavage motifs of the yeast 20S proteasome beta subunits deduced from digests of enolase 1. Proc Natl Acad Sci USA. 1998;95:12504–12509. doi: 10.1073/pnas.95.21.12504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Koodathingal P, Jaffe NE, Kraut DA, Prakash S, Fishbain S, Herman C, Matouschek A. ATP-dependent proteases differ substantially in their ability to unfold globular proteins. J Biol Chem. 2009;284:18674–18684. doi: 10.1074/jbc.M900783200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sauer RT, Baker TA. AAA+ proteases: ATP-fueled machines of protein destruction. Annu Rev Biochem. 2011;80:587–612. doi: 10.1146/annurev-biochem-060408-172623. [DOI] [PubMed] [Google Scholar]
- 14.Herman C, Prakash S, Lu CZ, Matouschek A, Gross CA. Lack of a robust unfoldase activity confers a unique level of substrate specificity to the universal AAA protease FtsH. Mol Cell. 2003;11:659–669. doi: 10.1016/s1097-2765(03)00068-6. [DOI] [PubMed] [Google Scholar]
- 15.Gur E, Sauer RT. Degrons in protein substrates program the speed and operating efficiency of the AAA+ Lon proteolytic machine. Proc Natl Acad Sci USA. 2009;106:18503–18508. doi: 10.1073/pnas.0910392106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gur E, Vishkautsan M, Sauer RT. Protein unfolding and degradation by the AAA+ lon protease. Protein Sci. 2012;21:268–278. doi: 10.1002/pro.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Palombella VJ, Rando OJ, Goldberg AL, Maniatis T. The ubiquitin-proteasome pathway is required for processing the NF-kappa B1 precursor protein and the activation of NF-kappa B. Cell. 1994;78:773–785. doi: 10.1016/s0092-8674(94)90482-0. [DOI] [PubMed] [Google Scholar]
- 18.Lin L, Ghosh S. A glycine-rich region in NF-kappaB p105 functions as a processing signal for the generation of the p50 subunit. Mol Cell Biol. 1996;16:2248–2254. doi: 10.1128/mcb.16.5.2248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tian L, Holmgren RA, Matouschek A. A conserved processing mechanism regulates the activity of transcription factors Cubitus interruptus and NF-kappaB. Nat Struct Mol Biol. 2005;12:1045–1053. doi: 10.1038/nsmb1018. [DOI] [PubMed] [Google Scholar]
- 20.Aza-Blanc P, Ramírez-Weber FA, Laget MP, Schwartz C, Kornberg TB. Proteolysis that is inhibited by hedgehog targets Cubitus interruptus protein to the nucleus and converts it to a repressor. Cell. 1997;89:1043–1053. doi: 10.1016/s0092-8674(00)80292-5. [DOI] [PubMed] [Google Scholar]
- 21.Rape M, Jentsch S. Taking a bite: proteasomal protein processing. Nat Cell Biol. 2002;4:E113–116. doi: 10.1038/ncb0502-e113. [DOI] [PubMed] [Google Scholar]
- 22.Hoppe T, Matuschewski K, Rape M, Schlenker S, Ulrich HD, Jentsch S. Activation of a membrane-bound transcription factor by regulated ubiquitin/proteasome-dependent processing. Cell. 2000;102:577–586. doi: 10.1016/s0092-8674(00)00080-5. [DOI] [PubMed] [Google Scholar]
- 23.Wang B, Fallon JF, Beachy PA. Hedgehog-regulated processing of Gli3 produces an anterior/posterior repressor gradient in the developing vertebrate limb. Cell. 2000;100:423–434. doi: 10.1016/s0092-8674(00)80678-9. [DOI] [PubMed] [Google Scholar]
- 24.Kraut DA, Matouschek A. Proteasomal Degradation from Internal Sites Favors Partial Proteolysis via Remote Domain Stabilization. ACS Chem Biol. 2011;6:1087–1095. doi: 10.1021/cb2002285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hoyt MA, Zich J, Takeuchi J, Zhang M, Govaerts C, Coffino P. Glycine-alanine repeats impair proper substrate unfolding by the proteasome. EMBO J. 2006;25:1720–1729. doi: 10.1038/sj.emboj.7601058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Daskalogianni C, Apcher S, Candeias MM, Naski N, Calvo F, Fåhraeus R. Gly-Ala repeats induce position- and substrate-specific regulation of 26 S proteasome-dependent partial processing. J Biol Chem. 2008;283:30090–30100. doi: 10.1074/jbc.M803290200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Orian A, Schwartz AL, Israël A, Whiteside S, Kahana C, Ciechanover A. Structural motifs involved in ubiquitin-mediated processing of the NF-kappaB precursor p105: roles of the glycine-rich region and a downstream ubiquitination domain. Mol Cell Biol. 1999;19:3664–3673. doi: 10.1128/mcb.19.5.3664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gilmore TD. Introduction to NF-kappaB: players, pathways, perspectives. Oncogene. 2006;25:6680–6684. doi: 10.1038/sj.onc.1209954. [DOI] [PubMed] [Google Scholar]
- 29.Epinat JC, Whiteside ST, Rice NR, Israël A. Reconstitution of the NF-kappa B system in Saccharomyces cerevisiae for isolation of effectors by phenotype modulation. Yeast. 1997;13:599–612. doi: 10.1002/(SICI)1097-0061(19970615)13:7<599::AID-YEA109>3.0.CO;2-H. [DOI] [PubMed] [Google Scholar]
- 30.Sears C, Olesen J, Rubin D, Finley D, Maniatis T. NF-kappa B p105 processing via the ubiquitin-proteasome pathway. J Biol Chem. 1998;273:1409–1419. doi: 10.1074/jbc.273.3.1409. [DOI] [PubMed] [Google Scholar]
- 31.Zuccato C, Valenza M, Cattaneo E. Molecular mechanisms and potential therapeutical targets in Huntington’s disease. Physiol Rev. 2010;90:905–981. doi: 10.1152/physrev.00041.2009. [DOI] [PubMed] [Google Scholar]
- 32.DiFiglia M, Sapp E, Chase KO, Davies SW, Bates GP, Vonsattel JP, Aronin N. Aggregation of huntingtin in neuronal intranuclear inclusions and dystrophic neurites in brain. Science. 1997;277:1990–1993. doi: 10.1126/science.277.5334.1990. [DOI] [PubMed] [Google Scholar]
- 33.Kalchman MA, Graham RK, Xia G, Koide HB, Hodgson JG, Graham KC, Goldberg YP, Gietz RD, Pickart CM, Hayden MR. Huntingtin is ubiquitinated and interacts with a specific ubiquitin-conjugating enzyme. J Biol Chem. 1996;271:19385–19394. doi: 10.1074/jbc.271.32.19385. [DOI] [PubMed] [Google Scholar]
- 34.Bence NF, Sampat RM, Kopito RR. Impairment of the ubiquitin-proteasome system by protein aggregation. Science. 2001;292:1552–1555. doi: 10.1126/science.292.5521.1552. [DOI] [PubMed] [Google Scholar]
- 35.Verhoef LGGC, Lindsten K, Masucci MG, Dantuma NP. Aggregate formation inhibits proteasomal degradation of polyglutamine proteins. Hum Mol Genet. 2002;11:2689–2700. doi: 10.1093/hmg/11.22.2689. [DOI] [PubMed] [Google Scholar]
- 36.Venkatraman P, Wetzel R, Tanaka M, Nukina N, Goldberg AL. Eukaryotic proteasomes cannot digest polyglutamine sequences and release them during degradation of polyglutamine-containing proteins. Mol Cell. 2004;14:95–104. doi: 10.1016/s1097-2765(04)00151-0. [DOI] [PubMed] [Google Scholar]
- 37.Saeki Y, Isono E, Toh-E A. Preparation of ubiquitinated substrates by the PY motif-insertion method for monitoring 26S proteasome activity. Meth Enzymol. 2005;399:215–227. doi: 10.1016/S0076-6879(05)99014-9. [DOI] [PubMed] [Google Scholar]
- 38.Cayley PJ, Dunn SM, King RW. Kinetics of substrate, coenzyme, and inhibitor binding to Escherichia coli dihydrofolate reductase. Biochemistry. 1981;20:874–879. doi: 10.1021/bi00507a034. [DOI] [PubMed] [Google Scholar]
- 39.Johnston JA, Johnson ES, Waller PR, Varshavsky A. Methotrexate inhibits proteolysis of dihydrofolate reductase by the N-end rule pathway. J Biol Chem. 1995;270:8172–8178. doi: 10.1074/jbc.270.14.8172. [DOI] [PubMed] [Google Scholar]
- 40.Ainavarapu SRK, Li L, Badilla CL, Fernandez JM. Ligand binding modulates the mechanical stability of dihydrofolate reductase. Biophys J. 2005;89:3337–3344. doi: 10.1529/biophysj.105.062034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Touchette NA, Perry KM, Matthews CR. Folding of dihydrofolate reductase from Escherichia coli. Biochemistry. 1986;25:5445–5452. doi: 10.1021/bi00367a015. [DOI] [PubMed] [Google Scholar]
- 42.Crosas B, Hanna J, Kirkpatrick DS, Zhang DP, Tone Y, Hathaway NA, Buecker C, Leggett DS, Schmidt M, King RW, Gygi SP, Finley D. Ubiquitin chains are remodeled at the proteasome by opposing ubiquitin ligase and deubiquitinating activities. Cell. 2006;127:1401–1413. doi: 10.1016/j.cell.2006.09.051. [DOI] [PubMed] [Google Scholar]
- 43.Kenniston JA, Baker TA, Fernandez JM, Sauer RT. Linkage between ATP consumption and mechanical unfolding during the protein processing reactions of an AAA+ degradation machine. Cell. 2003;114:511–520. doi: 10.1016/s0092-8674(03)00612-3. [DOI] [PubMed] [Google Scholar]
- 44.Park E, Rho YM, Koh O-J, Ahn SW, Seong IS, Song J-J, Bang O, Seol JH, Wang J, Eom SH, Chung CH. Role of the GYVG pore motif of HslU ATPase in protein unfolding and translocation for degradation by HslV peptidase. J Biol Chem. 2005;280:22892–22898. doi: 10.1074/jbc.M500035200. [DOI] [PubMed] [Google Scholar]
- 45.Hinnerwisch J, Fenton WA, Furtak KJ, Farr GW, Horwich AL. Loops in the central channel of ClpA chaperone mediate protein binding, unfolding, and translocation. Cell. 2005;121:1029–1041. doi: 10.1016/j.cell.2005.04.012. [DOI] [PubMed] [Google Scholar]
- 46.Pratt G, Rechsteiner M. Proteasomes cleave at multiple sites within polyglutamine tracts: activation by PA28gamma(K188E) J Biol Chem. 2008;283:12919–12925. doi: 10.1074/jbc.M709347200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Slow EJ, Graham RK, Osmand AP, Devon RS, Lu G, Deng Y, Pearson J, Vaid K, Bissada N, Wetzel R, Leavitt BR, Hayden MR. Absence of behavioral abnormalities and neurodegeneration in vivo despite widespread neuronal huntingtin inclusions. Proc Natl Acad Sci USA. 2005;102:11402–11407. doi: 10.1073/pnas.0503634102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Mitsui K, Nakayama H, Akagi T, Nekooki M, Ohtawa K, Takio K, Hashikawa T, Nukina N. Purification of polyglutamine aggregates and identification of elongation factor-1alpha and heat shock protein 84 as aggregate-interacting proteins. J Neurosci. 2002;22:9267–9277. doi: 10.1523/JNEUROSCI.22-21-09267.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Maynard CJ, Böttcher C, Ortega Z, Smith R, Florea BI, Díaz-Hernández M, Brundin P, Overkleeft HS, Li J-Y, Lucas JJ, Dantuma NP. Accumulation of ubiquitin conjugates in a polyglutamine disease model occurs without global ubiquitin/proteasome system impairment. Proc Natl Acad Sci USA. 2009;106:13986–13991. doi: 10.1073/pnas.0906463106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Bowman AB, Yoo S-Y, Dantuma NP, Zoghbi HY. Neuronal dysfunction in a polyglutamine disease model occurs in the absence of ubiquitin-proteasome system impairment and inversely correlates with the degree of nuclear inclusion formation. Hum Mol Genet. 2005;14:679–691. doi: 10.1093/hmg/ddi064. [DOI] [PubMed] [Google Scholar]
- 51.Tokui K, Adachi H, Waza M, Katsuno M, Minamiyama M, Doi H, Tanaka K, Hamazaki J, Murata S, Tanaka F, Sobue G. 17-DMAG ameliorates polyglutamine-mediated motor neuron degeneration through well-preserved proteasome function in an SBMA model mouse. Hum Mol Genet. 2009;18:898–910. doi: 10.1093/hmg/ddn419. [DOI] [PubMed] [Google Scholar]
- 52.Ortega Z, Díaz-Hernández M, Maynard CJ, Hernández F, Dantuma NP, Lucas JJ. Acute polyglutamine expression in inducible mouse model unravels ubiquitin/proteasome system impairment and permanent recovery attributable to aggregate formation. J Neurosci. 2010;30:3675–3688. doi: 10.1523/JNEUROSCI.5673-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.