Abstract
Multiple signaling systems and transcription factor cascades control pancreas development and endocrine cell fate determination. Epigenetic processes contribute to the control of this transcriptional hierarchy, involving both histone modifications and DNA methylation. Here, we summarize recent advances in the field that demonstrate the importance of epigenetic regulation in pancreas development, β-cell proliferation, and cell fate choice. These breakthroughs were made using the phenotypic analysis of mice with mutations in genes that encode histone modifying enzymes and related proteins; by application of activators or inhibitors of the enzymes that acetylate or methylate histones to fetal pancreatic explants in culture; and by genomic approaches that determined the patterns of histone modifications and chromatin state genome-wide.
1. Introduction
Pancreas development and endocrine cell fate determination are controlled by precisely timed signaling events, discussed in detail in (Serup, in this issue), which determine the chronology of activation and repression of transcriptional networks [1, 2]. The transcriptional hierarchy that regulates gene expression during development and disease is in part regulated by epigenetic process, involving both histone and DNA modifications, which in turn facilitate or prevent recruitment of effectors protein complexes.
Epigenetic events were originally defined as those heritable changes to phenotype that occurred without altering the DNA sequence itself. This definition has been loosened in recent years to include control of gene expression by DNA methylation and histone modification, even when this is not heritable through mitosis. One example of the striking functional significance of epigenetic alterations is the silencing of tumor suppressor genes that can occur in cancer and which is mediated through DNA methylation and silencing of promoters. In mammalian cells, DNA methylation occurs on cytosines in the context of CpG dinucleotides at the 5 position to create 5-methylcytosine, and is mediated by methyltransferase enzymes encoded by three genes, Dnmt1, Dnmt3a and Dnmt3b. Over the past two decades, dozens of modifications to histones, including lysine (K) acetylation, lysine and arginine (R) methylation, serine (S) and threonine (T) phosphorylation, and lysine sumoylation and ubiquitination have been shown to affect gene expression in multiple ways.
Here we describe recent findings demonstrating the involvement of epigenetic mechanisms in pancreas development, post-natal regeneration of the insulin-producing β-cell, and preservation of lineage identity through cell divisions. These findings were obtained using three general approaches: (1) the phenotypic analysis of model organisms, chiefly mice, with mutations in one or several of the genes that encode histone modifying enzymes and related proteins; (2) the use of more or less specific activators or inhibitors of the enzymes that acetylate or methylate histones; and (3) genomic approaches that determine the patterns of histone modifications and chromatin state genome-wide, and make inferences about gene regulation by comparison to similar maps from other cell types.
2. Genetic analysis of mutations for histone modification enzymes in pancreas development and β-cell replication
Genome-wide location analysis of embryonic stem (ES) cells has identified a unique histone modification pattern, termed "bivalent domain," in which repressive histone marks, i.e. H3K27me3, and activating marks, i.e. H3K4me3, are present at the same location [3]. In general, the repressive mark is dominant, meaning the corresponding gene is silent in ES cells. Many of these bivalent domains were found at important developmental determination genes, which are thus ‘poised’ for rapid activation during ES cell differentiation by simple removal of the repressive mark.
Xu et al [4] explored the possibility that such mechanisms exist also in multipotent cells of the developing endoderm, prior to the fate choice between liver and ventral pancreas identity. Chromatin immunoprecipitation (ChIP) analyses of multiple histone modifications of early liver and pancreas-specific genes from FACS-sorted endoderm cells (day E8.25) or hepatoblasts (day E9.5), showed that all hepatic lineage-specific genes are marked as ‘silent’ in endoderm cells as expected, while these liver-specific genes become marked as ‘activated’ in hepatoblasts. Two chromatin marks were found to be different in endoderm cells between liver- and pancreas-specific genes. Thus, H3K9acK14ac, associated with gene activation, was poorly represented in the regulatory elements of liver-specific genes such as Alb1, Afp and Ttr, but enriched in the regulatory elements of PDX1, an early pancreatic gene. Similarly, H3K27me3, associated with gene silencing, was also under-represented in ‘liver elements’, but enriched at the promoter of the PDX1 gene. In other words, in early endodermal cells, PDX1 was bivalently marked, while liver-specific genes carried no histone modification marks at all. However, following differentiation into hepatoblasts, H3K9acK14ac increased on liver-specific elements, while H3K27me3 remained low, whereas the PDX1 promoter remained hyperacetylated and enriched for H3K27me3, indicative of distinct chromatin states for the two types of genes.
Based on these findings, Xu and colleagues studied the enzymatic machinery underlying this differential state, by using gene-targeted mice heterozygous for P300 (P300+/−), a histone acetyltransferase. ChIP analysis showed increased levels of histone acetylation in promoters of the early liver marker genes Alb1, Afp and Ttr, in wild type hepatoblasts compared to P300+/− hepatoblasts. In parallel, the expression of these early liver-specific genes was diminished in P300+/− hepatoblasts, whereas PDX1 expression was upregulated. These findings suggest that P300 is necessary for acetylation, and thus activation, of liver-specific regulatory elements, and that P300 modulates the cell fate choice between liver progenitors and pancreas progenitors.
Next, the authors showed, again using ChIP analysis, enrichment of Ezh2, a methyltransferase for H3K27me3 and a member of PRC2 (polycomb-repressive complex 2) at upstream regulatory elements in the PDX1 gene. Binding of Ezh2 was overlapping with H3K27me3, but absent from liver-specific regulatory elements in wild type endoderm cells. Following deletion of the Ezh2 allele in foregut endoderm using an Ezh2 conditional allele and the FoxA3-Cre transgene, embryos at E10 exhibited an expanded PDX1-positive ventral pancreas domain, accompanied by multiple bud-like structures, leading to an enlarged ventral pancreas at E11.5. This expansion of the pancreas occurred at the expense of liver development. In conclusion, Ezh2 normally promotes the liver program by restraining pancreatic commitment (See Figure 1 for schematic summary).
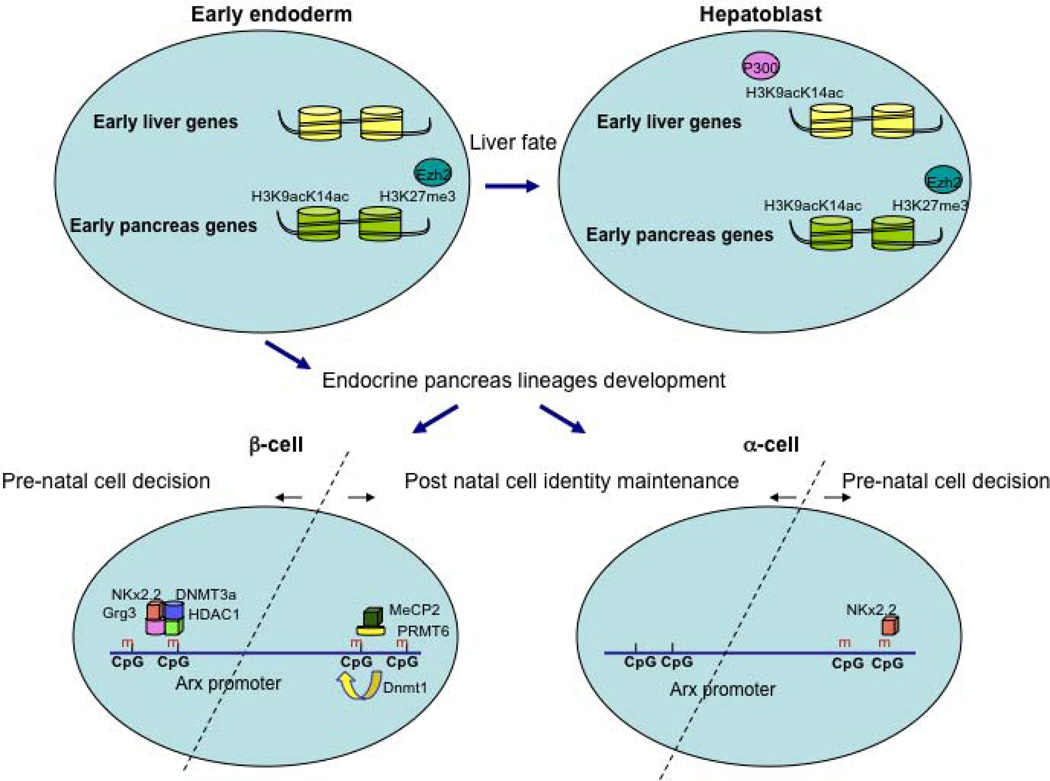
Figure 1.
Epigenetic control in early pancreas development and endocrine cell fate allocation.
During early endoderm development into liver and pancreas fates, liver-specific genes and pancreas-specific genes are differentially marked in multipotent cells. While PDX1, and early pancreatic gene, is bivalently marked with both the activation-associated H3K9acK14ac histone modification and H3K27me3, associated with gene silencing, liver-specific genes such as Alb1, Afp and Ttr carry none of these histone marks. Upon differentiation into hepatoblasts, the H3K9acK14ac mark increases on liver-specific promoters, while H3K27me3 remains low, whereas the PDX1 promoter remains hyperacetylated and enriched for H3K27me3. These cell fate programs are modulated by the histone acetyltransferase P300, and by Ezh2, a methyltransferase for H3K27me3. During pancreatic islet development, the α and β-cells fate decision is epigenetically regulated by the differentially recruitment of a repression complex (DNMT3a, Grg3 and HDAC1) by Nkx2.2, to the Arx gene promoter in β-cells, but not α–cells. This differential binding is influenced by the methylation state of the Arx promoter.
In post-natal life, the maintenance of α and β-cell identity is epigenetically regulated by a differentially methylated state of part of the Arx promoter. In β-cells, this region is hypermethylated, and is occupied by a repressive complex of MeCP2 and PRMT6, to repress the Arx locus. In α-cells, this promoter region is kept hypomethylated.
During embryonic development, cell specification is achieved by activation and repression of transcription factors in response to inductive developmental signals. Transcriptional programs are somewhat plastic, and thus cellular fates can be “re-programmed” in extreme conditions. For example, misexpression of the α-cell-specific transcription factor Aristaless homeobox gene (Arx) in fetal β-cells causes β-cell to α-cell conversion [5]. Furthermore, Thorel and colleagues have shown that mice with over 90% reduction in β-cell mass are able to replenish some of the lost cells through up-regulation of β-cells transcription factors in α-cells, causing α to β-cell transdifferentiation [6]. However, the normal complement of functional β-cells was not restored in this model. Nevertheless, these examples of transdifferentiation point to a close developmental relationship between α and β-cells, and suggest that they exist in a similar epigenetic state.
Nkx2.2 is a homeodomain transcription factor required for pancreatic islet cell fate decisions [7]. In a recent study, Papizan and colleagues showed that in β-cells, Nkx2.2 is part of a repression complex, together with DNMT3a - a de novo DNA methyltransferase important for establishing methylation patterns during development [8], the groucho-related repressor Grg3, and the histone deacetylase HDAC1. To investigate the role of this complex in pancreatic islet development, they derived mice homozygous for a specific point mutation in the tinman (TN) domain of Nkx2.2 (Nkx2.2TNmut/TNmut), which disrupts the interaction between NKx2.2 and Grg3. These mice develop hyperglycemia and do not survive beyond eight weeks of age. Mutant islets are smaller and contain fewer β-cells and more α–cells, presumed to have formed at the expense of the β-cells population. Interestingly, by the end of the gestation, the mutant mice present a distinct population of β-cells expressing the α-cell specific transcription factor Arx.
Based on the hypothesis that the mutation in the Nkx2.2 TN domain is causing derepression of the Arx gene in β-cells, Papizan and colleagues permanently marked β-cells by genetic lineage tracing. This involved transgenic mice with an insulin promoter driven Cre recombinase (Ins:Cre) transgene combined with the Rosa26:LacZ reporter allele, which leads to permanent expression of β-galactosidase, the product of the bacterial LacZ gene, in all cells that express Cre recombinase and their descendants. Indeed, the authors found that a fraction of the β-galactosidase marked cells expressed Arx, demonstrating that β-cells had been reprogrammed towards an α-cell fate in the absence of fully functional Nkx2.2. Bisulfite analyses to determine the CpG methylation status at the Arx promoter showed that Nkx2.2 occupies the hypermethylated Arx promoter in both α and β cells. However, the other members of the repressive complex, i.e. Grg3, HDAC1 and Dnmt3a, were found preferentially at the Arx promoter in β-cells. Taken together, this study demonstrates the role of Nkx2.2 and Dnmt3a in recruiting a repressor complex to the Arx promoter in β-cells to maintain their identity. Although Nkx2.2 is expressed and functions in both α and β-cells, its binding is epigenetically regulated to preferentially occupy the Arx promoter in β-cells. This differential binding is influenced by both the methylation state of the Arx promoter and by the DNA modifications induced by Dnmt3a (see Figure 1).
In the post-natal pancreas, terminally differential β-cells preserve the potential to proliferate during maturation and in response to injury, in order to maintain glucose homeostasis [9–11]. Therefore, a tight regulatory system is needed to limit proliferation and to maintain cell identity after cell division. DNA methylation is one of the mechanisms that can ensure stable inheritance of repressed genes. Recently, Dhawan and colleagues [12] used β-cell specific ablation of the DNMT1 gene, a DNA methyltransferase that restores CpG methylation pattern after DNA replication in S-phase of the cell cycle (for review see [13]), to elucidate a possible role for DNA methylation in assuring β-cell identity. They focused on two transcription factor genes; the aforementioned Arx and the paired box factor Pax4, found to be crucial for controlling endocrine lineage cell fate, and expressed exclusively in α and β cell lineages, respectively [14–17]. The authors derived a mouse model in which β-cells are selectively deficient for Dnmt1, by crossing mice transgenic for Cre recombinase under the control of rat insulin promoter (Rip-Cre) with Dnmt1fl/fl mice [18, 19]. The absence of Dnmt1 resulted in a passive loss of DNA methylation with age. This was accompanied by an increase in the number of glucagon-expressing α-cells, reduction in the number of insulin-expressing β–cells, and abnormal glucose homeostasis. Using the genetic lineage-tracing technique described above to heritably label insulin-expressing cells, they demonstrated that a subset of glucagon-expressing cells had originated from insulin-expressing ancestral cells.
Analysis of the methylation profiles of endocrine cell fate determination genes identified CpG-rich areas in the regulatory region of Arx, a key player in α cell fate [14, 20, 21]. Bisulfite-sequencing analysis of FACS-purified α– and β-cells revealed that one of the CpG-rich areas in the Arx promoter is hypermethylated in β-cells, but hypomethylated in α-cells. Using the same analysis on heritably labeled β-cells isolated from Dnmt1-ablated mice, they found two subpopulations. These two populations were differently methylated at the Arx regulatory region, indicating clonal heterogeneity. In addition, the subpopulation of β-cells that had reduced methylation at the Arx promoter was expressing high levels of the α-cell transcription factors Arx and MafB, and significant lower levels of the β-cell regulators Pdx1 and Pax4. ChIP analyses revealed further that MeCP2, a methyl DNA-binding protein [22] and PRMT6, an H3R2 methyltransferase associated with repression of transcription [23, 24] form a repressive complex that is recruited to the methylated regions of the Arx promoter in β-cells, but not in α-cells, to propagate the repressive chromatin structure at the Arx locus (see Figure 1).
To summarize, this study showed that propagation of DNA methylation pattern forms the backbone for transmitting a repressive chromatin structure at the Arx locus, which is stably inherited through cell divisions to preserve β-cell identity. By reprogramming Dnmt1-deficient β-cells towards the α-cell fate, this study also supports the notion that adult endocrine cells can be manipulated to change identity or regain plasticity, which is an exciting prospect for the future development of cell-based therapies for diabetes.
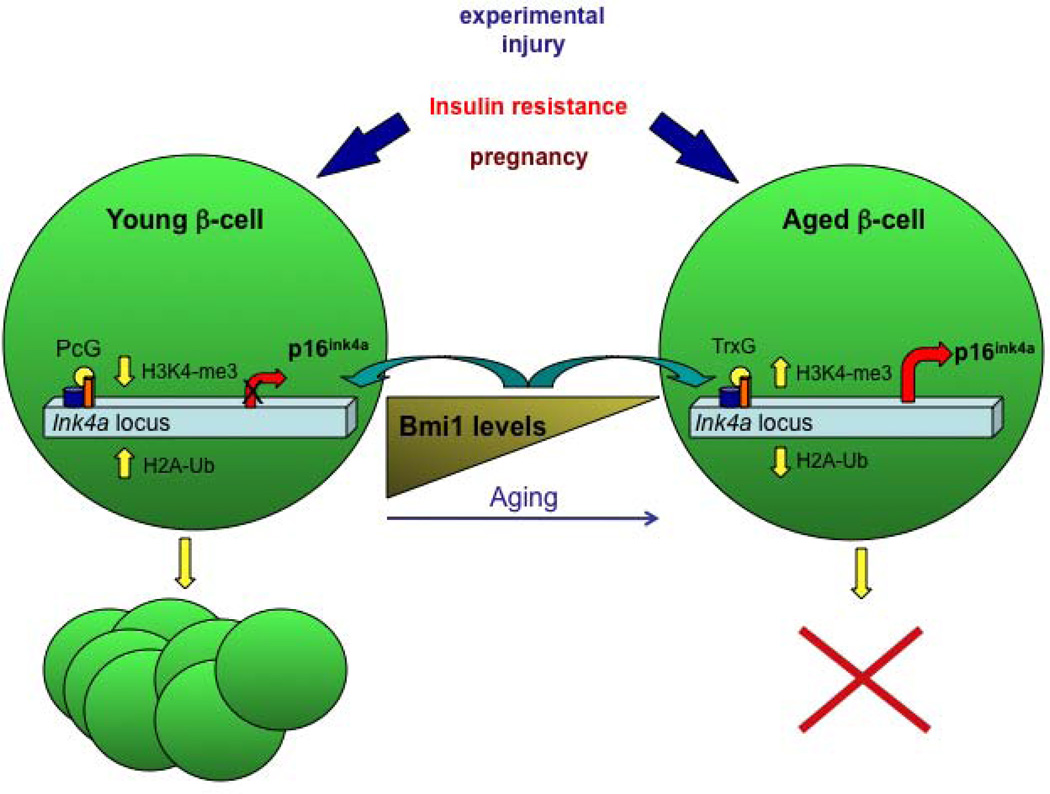
The endocrine pancreas responses to physiological changes, such as peripheral insulin resistance and pregnancy [25] by adaptive changes of β-cell mass, which was shown to be mainly due to proliferation of developmentally matured β-cells, at least in rodents [26]. Nevertheless, the capacity of β-cells to proliferate in response to changes in metabolic demand decreases dramatically with age [27, 28], possibly contributing to the pathogenesis of diabetes. Studying the mechanism that controls the regeneration capacity of β-cells will greatly contribute to the development of therapeutic approaches aimed to renew and restore the number of functioning β-cells in diabetic patients. Tschen and colleagues [29] investigated the role of the aging process in the regeneration and mass expansion capacity of β-cells in response to increased metabolic demand in several hyperglycemic mice models. To assess the influence of age on adaptive β-cell mass expansion associated with insulin resistance, one and seven to eight months old mice were fed a high fat diet. Glucose tolerance tests taken after eight weeks on the diet found old mice to be glucose intolerant, while young mice remained normal glucose homeostasis on the high fat diet. Fasting blood glucose levels was also above normal in the old mice, suggesting that these mice are unable to maintain glucose homeostasis. Indeed, the β-cell mass of the young mice had expanded five-fold after eight weeks on the high fat diet, whereas there was no change in the β-cell mass of old mice. The authors further analyzed the protein levels of p16ink4a, a CDK4-D type cyclin inhibitor and a product of the Ink/Arf locus [30, 31]. Immunohistochemistry confirmed that the protein expression levels of p16Ink4a are low in young mice and increase with age. Western blot analysis of p16Ink4a protein levels in islets isolated from old and young mice after eight weeks of high-fat diet showed that the low levels in young mice where yet lower after high fat diet feeding, while the levels remained high in old mice regardless of the diet. The levels of Bmi1, a polycomb group protein known to regulate the Ink4a locus, increased in young mice after high fat diet, but stayed low in old mice on the same diet. These observations suggest that p16Ink4a levels may play a role in the adaptive expansion of β-cell mass in response to high-fat diet. Similar to the poor adaptive expansion response of β-cells of old mice, following insulin resistance, treatment with exendin-4, a glucagon-like peptide 1 (GLP-1) analog that can induce β-cell replication in some conditions, failed to induce β-cell expansion in old mice. Using Bmi1 null mice [32], the authors showed further that Bmi1 specifically represses p16Ink4a expression by modulation of chromatin structure. Treatment of Bmi1 null mice with exendin-4 did not increase β-cell mass in these mice, supporting that Bmi1 expression and regulation of p16Ink4a levels is important in the age-dependant rise of p16Ink4a and in the decline in β-cell proliferation capacity with age. Finally, using low-doses of the β-cell toxin Streptozocin (STZ) treatment as a model of β-cell regeneration, the authors demonstrated that β-cell did not proliferate in old mice, compared with a four-fold increase in proliferating β-cells in young mice under these condition. Together, their data showed clearly that young but not old mice have the capacity to regenerate β-cell mass (see Figure 2).
Figure 2.
Epigenetically-based mechanisms control the regeneration capacity of β-cells:
The capacity of β-cells to proliferate in response to changes in metabolic demand declines with age. In several models of beta-cell replication, such as insulin resistance, treatment with exendin-4, or following Streptozocin (STZ) treatment, only β-cells from young mice have the capacity to regenerate β-cell mass. The cell cycle inhibitor p16Ink4a, a product of the Ink/Arf locus, increases with age. Conversely, the levels of Bmi1, a polycomb-repressive complex 1 (PRC1) protein known to regulate the Ink4a locus, decline with age. In young and regenerating β-cells, increased binding of Bmi-1 and Ezh2 (a polycomb histone methyltransferase) coincides with decreased H3K4 trimethylation, increased H2A ubiquitylation and H3K27 trimethylation and repression of the Ink/Arf locus, and thus high β-cell proliferative capacity. With aging, reduced Bmi-1 binding facilitates H3K4 trimethylation by MLL1, a trithorax group (TrxG) protein and an H3K4 methyltransferase, reduced H2A ubiquitylation and increased Ink4a/Arf expression, therefore reduced β-cell replication.
In addition to the findings that the expression of p19Arf and p16Ink4a increases with age [30, 31] and is linked to the reduced proliferative capacity of β-cells [33], genome-wide association studies identified the INK/ARF locus as contributing to the genetic risk of type-2 diabetes [34–37]. Dhawan and colleagues studied the regulation of the Ink/Arf locus by comparing pancreatic β-cells from mice at different ages, and again demonstrated that the levels of Bmi-1 decline with age. Bmi-1 is part of the polycomb-repressive complex 1 (PRC1), which possesses histone H2A-K119 ubiquitin E3 ligase activity [38]. Bmi-1 repression of the Ink/Arf locus, previously demonstrated in cancer cell lines and embryonic fibroblasts, was found to be dependent on the presence of PRC2, which possesses H3K27 methyltransferase activity, mediated by Ezh2, already introduced above [39, 40]. Using ChIP analyses and comparing islets from wild type and Bmi-1 null mice [32], they showed that PcG and Trithorax-group (TrxG, associated with gene activation) complexes have opposite roles in regulating the Ink/Arf locus. Thus, decreased binding of Bmi-1 to the Ink/Arf locus in aged islets resulted in reduced H2A ubiquitylation, stimulating recruitment of MLL1 (a trithorax group (TrxG) protein and an H3K4 methyltransferase), and increased H3K4 trimethylation, causing increased transcription from this locus. However, in the regenerating β-cell, increased binding of Bmi-1 coincides with decreased H3K4 trimethylation and repression of the Ink/Arf locus. Using Bmi-1 null mice, Dhawan and colleagues established that the main mechanism by which Bmi-1 regulates β-cell proliferation is through repression of p16Ink4a (see Figure 2).
Accordingly, the epigenetically-based mechanism revealed in this study suggests that in young islets, high levels of Bmi-1 ensure increased H2A ubiquitylation, repression of the Ink4a/Arf locus and high β-cell proliferative capacity. With aging, reduced Bmi-1 binding would facilitate H3K4 trimethylation, increased Ink4a/Arf expression, and reduced β-cell replication. Finally, experiments with STZ-induced β-cell regeneration suggest that the modulation of Ink4a/Arf expression levels by Bmi-1 is critical to the β-cell regeneration process.
In a parallel study, Chen and colleagues examined the role of the polycomb histone methyltransferase Ezh2 in the regenerative capacity of β-cells [41]. ChIP analysis of the Ink4a/Arf locus in islets isolated from young and old mice showed a decrease in Ezh2 binding and H3K27 trimethylation in old mice, concomitant with increased H3K4 trimethylation and acetylation. Again, the authors employed the Cre/loxP technology for β-cell specific ablation of Ezh2. Loss of Ezh2 in β-cells led to derepression of established Ezh2 targets, premature reduction of H3K27 trimethylation levels, and increased H3K4 trimethylation and H3 acetylation at the Ink4a/Arf locus. Physiologic characterization revealed that Ezh2 deficient mice suffer from mild diabetes mellitus, and display reduced pancreatic insulin content. Incorporation of bromodeoxyuridine (BrdU) in Ezh2-deficient mice at one month of age showed that β-cell proliferation was reduced compared with controls, suggesting that Ezh2 is essential for β-cell proliferation and establishing the appropriate β-cell mass in juvenile mice.
To test the hypothesis that increased levels of Ink4a/Arf promote the pathogenesis of diabetes in Ezh2 deficient mice, the authors intercrossed mice to generate RIP-Cre; Ezh2f/f; Cdkn2a (p16Ink4A) null animals (double mutants). Unlike single Ezh2-deficient littermates, which developed fasting hyperglycemia and overt postprandial diabetes by four weeks of age, double mutant mice maintained normal glucose levels during fasting and random feeding, and showed no evidence of impaired glucose tolerance. Thus, Cdkn2a deficiency was sufficient to restore normoglycemia and insulin levels to mice lacking Ezh2 in β-cells. Moreover, pancreatic insulin content, β-cell mass, and β-cell proliferation in double mutant mice and littermate controls were indistinguishable. Taken together, these data indicate that increased Ink4a/Arf expression, resulting in reduced β-cell proliferation, is a principal cause of premature diabetes onset in mice lacking Ezh2 in β-cells. Finally, following STZ treatment to destroy most of the β-cells, Ezh2 mRNA and protein levels were increased in surviving β-cells, while Ink4a/Arf expression was reduced, suggesting that Ezh2 is required for repression of Ink4a/Arf expression during β-cell regeneration.
3. Interrogating the epigenetic control of pancreas development using small molecules
The development of the pancreas from the dorsal and the ventral region of the foregut endoderm is controlled by the hierarchical expression of transcription factors [42] beginning with the winged helix factors Foxa1 and Foxa2, which are required for the induction of the homeodomain factor PDX1 in the pancreas-committed endodermal region [43–45], followed by neurogenin 3 (NGN3) expression, which initiates the endocrine differentiation program [46, 47], and continues with the expression of transcription factors that are involved in determining endocrine cell fate, such as Arx in α and Pax4 in β and δ -cell fates [14, 15]. Haumaitre and colleagues examined the possible role of histone acetylation and deacetylation status in controlling the timing of pancreatic differentiation and embryonic pancreas cell fate decision. The acetylation state of histones, controlled by histone acetyltransferases (HATs) and histone deacetylases (HDACs) was previously shown to be involved in regulating proliferation and differentiation processes in many tissues [48–51]. Here, the authors used an in vitro model of culturing E13.5 rat pancreatic primordia as explants on floating filters at the air-medium interface for up to two weeks. Under these conditions, acinar and endocrine cells can develop in a similar way to in-vivo pancreatic development. At E13.5, the pancreatic cells expressed class I and class II HDACs; however, their expression levels declined towards E17.5, concomitant with an increase in histone acetylation levels (as detected using antibodies against histone acetyl-H3 and -H4). Manipulation of the histone-acetylation levels with the HDAC inhibitors (HDACi) valproic acid (VPA) and trichostatin A (TSA), which target class I HDACs and both class I and class II HDACs, respectively, resulted in abnormal morphology of the developed rat pancreas at E17.5. Examination of the expression levels of transcription factor that are involved in acinar cell differentiation (P48/Ptf1a and Mist1) and markers of mature acinar cells (amylase), using both quantitative real time PCR (qPCR) and immunostaining, showed decreased expression levels of these genes, and a reduced number of acinar cells in the HDACi-treated explants compared to controls. On the other hand, the expression levels of the ductal cell differentiation marker SPP1 were increased by HDACi. These results demonstrated that HDACi treatment caused a decrease in acinar linage differentiation in favor of the ductal linage. Next, the authors examined the effect of HDACi on the regulation of the endocrine lineage by examining the expression levels of Ngn3, a pro-endocrine transcription factor. In control cells the expression levels of Ngn3 peaked at day 3 of culture (similar to E16.5 in vivo) and decreased thereafter, whereas in HDACi-treated cells, Ngn3 levels increased to a peak at day 7, indicative of an increase in the pool of endocrine progenitor cells. Further examination of HDACi influence on the various endocrine cell types showed an increase in expression of Arx, the α and pancreatic polypeptide (PP) cell fate specific transcription factor, as well as increases in the expression of glucagon and PP, implying that HDACi treatment promotes α and PP cell lineage allocation. However, when treating the explants with VPA, which targets only class I HDACs, there was decreased expression of PAX4, a β and δ cell fate transcription factor, associated with a dramatic decrease in insulin and somatostatin expression, whereas treatment with TSA, which targets also class II HDACs, caused activation of Pax4 and increased β cell mass. Finally, HDACi treatment had no effect on proliferation and apoptosis rates, suggesting that the effect observed in this study was due to altered cell lineage allocation from precursors. Taken together, this study highlights the potent effect of HDACi treatment on pancreatic differentiation. Since treatment with the HDACi TSA decreased acinar lineage differentiation to the benefit of endocrine cells, and especially insulin-expressing β cells, this finding may have important therapeutic implications for cell therapy in diabetes.
Based on the findings described above, Lenoir and colleagues further investigated the involvement of class II HDACs in pancreatic cell differentiation. Using qPCR, they showed that the class IIa HDACs members HDAC4, -5, and -9 are specifically expressed in β and/or δ cells during mouse pancreas development at E15.5 and in adulthood. In contrast, these HDACs were not expressed in either glucagon-expressing cells or in pancreatic acinar tissue. Analysis of the pancreas of mutant mice lacking HDAC4 [52] on postnatal day 1 (P1) revealed an increased mass of δ-cells. Lentivirus-mediated overexpression of HDAC4 at E13.5 caused decreased expression of insulin and somatostatin as observed by both qPCR and immunostaining analyses, implying that HDAC4 is involved in the control of β and δ-cells development. However, the expression levels of glucagon or amylase, markers of differentiated endocrine α-cells and acinar cells, respectively were unaffected.
Next, Lenoir and colleagues examined the pancreas of mice lacking HDAC5 [53] during embryogenesis (E18.5) and after birth (P7), and similarly found that these mice have enhanced β and δ-cells mass. Overexpression of HDAC5 in E13.5 pancreas explants again caused a decrease in the number of β and δ-cells as well as reduced expression levels of β- and δ-cell-specific transcription factors such as NeuroD1, Pdx1, Nkx2.2, MafA, Znt8, and Ia1. A similar inhibitory effect on β-cell mass was found for HDAC9. Finally, using the in-vitro system described above, the authors treated E13.5 pancreas with MC1568, a class II HDAC inhibitor that modulates the stability of the HDAC-MEF2 complexes, blocking MEF2-mediated transactivation [54]. This treatment caused enhanced expression of Pax4, as well as insulin, somatostatin and additional β-and δ-cell-specific transcription factors. To summarize, this study highlights the epigenetic mechanisms underlying the regulation of endocrine cell development by suggesting that class IIa HDACs have an important role in controlling β and δ-cells development.
4. Epigenomic analysis of pancreas development and differentiation
The cascade of transcription regulators and signaling pathways that direct the differentiation of the foregut endoderm into the mature pancreas has been largely decoded in the last decade by developmental genetic approaches [55]. This knowledge was the foundation that enabled the development of in vitro protocols aimed to induce differentiation of embryonic stem cells (ESCs) and pluripotent stem cells (iPSCs) to engineered β-cells for therapy of diabetes [56, 57] (reviewed in Nostro and Keller, this issue). However, although these protocols can be used to generate β-like cells that produce insulin, they do not respond normally to an extracellular glucose stimulus [57]. Only when transplanted into mice for an ‘in vivo maturation’ period of about five month, these human ES-derived, β-like cells are capable of maintaining normal blood glucose levels [58]. It is likely that important epigenetic modifications, which define the fully matured state of α and β-cells, are missing from these ESCs and iPSCs-generated endocrine cells. Therefore, the epigenomic study of pancreatic islets cells has become a necessity in order to overcome the obstacles in cell replacement therapy for treating diabetes.
The first genome-wide epigenetic studies of human pancreatic islets, using chromatin immunoprecipitation followed by high-throughput sequencing (ChIP-Seq analysis), focused on characterizing histone modifications that are involved in gene activation and repression such as H3K4me1, H3K4me2, H3K4me3, H3K79me2 and H3K27me3, as well as mapping the binding sites for the transcriptional repressor and insulator CTCF [59, 60]. Surprisingly, these studies revealed that the promoters of the highly expressed insulin and glucagon genes were only scarcely occupied by H3K4-trimethylation, the typical mark of active promoters. The authors suggested that H3K4-trimethylation occupancy of promoters lacking CpG islands does not correlate well with gene expression levels, while promoters containing a CpG island are likely to be more H3K4-methylated with increasing expression level. Although previous studies suggested that tissue-specific chromatin structure occurs mainly at enhancers [61] rather than promoters, comparing the H3K4-trimethylation occupancy in human islets and CD4+ T-cells [62] demonstrated a tissue-specific pattern in CpG-less promoters. Finally, the two studies found the present of some bivalent marks in developmental regulatory genes of adult human islets.
A comparative study of the histone modification profile of murine neural tissue, pancreatic progenitor cells, and differentiated β and acinar cells demonstrated a similar profile of the active H3K4-trimethylation mark between β cells and neural tissue, whereas the signature of the repressive mark H3K27-trimethylation showed a similar profile between β-cells and acinar cells [63]. The similar activation mark may reflect a cell type signature representing the similar functional state of β-cells and neuronal cells, which includes stimulated secretions of hormone or neurotransmitter-containing vesicles, respectively. On the other hand, the H3K27-trimethylation signature may signify the developmental origin of the cell, where both β-cells and acinar cells are derived from foregut endoderm, emphasizing again the importance of cell-type specific analysis.
Further progress in the field will be aided by the recent development of a panel of antibodies to cell surface antigens that allows for the sorting of the individual human pancreatic cell populations. Using these novel tools, Dorrell and colleagues used microarray and qPCR analyses to determined the mRNA transcriptome of FACS-sorted human pancreatic endocrine and exocrine α, β, duct and acinar cells [64]. Importantly, computational analysis of the transcriptomes revealed a gene expression pattern in α, β, large duct, small duct and acinar cells that lacked some of the regulatory genes found in the same murine pancreatic cells. For example, MAFB, a member of the v-maf musculoaponeurotic fibrosarcoma oncogene homologue (avian) family of basic leucine-zipper transcription factors, expression of which is restricted to α cells in adult mice [65], was found to be expressed in both adult human β cells. This transcriptome database will be an important resource for the research of diabetes and β cell regeneration.
Transcription regulation involves the recruitment of DNA binding proteins to gene regulatory elements, a process that is often associated with the eviction of nucleosomes [66], and an “open chromatin” structure that can be identified by its hypersensitivity to nucleases [67]. One way to study the involvement of chromatin structure in gene regulation is to define these open and closes regions and identity active DNA regulatory regions using FAIRE technology (Formaldehyde-Assisted Isolation of Regulatory Elements) [68, 69], in which nucleosome-depleted DNA is isolated and analyzed. During this procedure, chromatin is crosslinked with formaldehyde in vivo, sheared by sonication, and phenol-chloroform extracted. The DNA that is recovered in the aqueous phase is either fluorescently labeled and hybridized to a DNA microarray, or amplified and sequenced. Gaulton and colleagues [70] used FAIRE technology coupled to high-throughput sequencing, to identify sites of open chromatin in human pancreatic islets. The sites of open chromatin showed an overall correlation between known transcription start sites and transcript levels [71]. Comparing the islet FAIRE-seq data with five non-islet cell lines demonstrated a high level of unique open chromatin sites to islets, among them several at islet-specific genes involved in human diabetes such as PDX1, ABCC8, SLC30A8, G6PC2, and GAD2, as well as islet developmental regulators (NEUROD1, NKX6-1, PAX6, and ISL1), suggesting that the FAIRE method was able to detect open chromatin regions that are linked to islet-cell biology and disease. Interestingly, this analysis detected, in addition to promoter-associated open chromatin regions also distal, intergenic open chromatin sites. These are likely to have regulatory function as enhancers or silencers, based on their enrichment for predicted transcription factor binding sites and regulatory modules, as well as CTCF binding sites. The authors also noticed that open chromatin sites are distributed unevenly, and defined the term ‘COREs’ (Clusters of Open Regulatory Elements) for open chromatin sites that are clustered. Frequently, these clusters were found in islet-selective open chromatin sites. All together, this study represents a high-resolution map of regulatory elements in pancreatic islets and reveals new insights into the organization of tissue-specific cis-regulatory elements, identifying a large number of islet-specific regulatory domains.
5. Concluding remarks
The findings summarized above clearly demonstrate that epigenetic mechanisms contribute to important cell fate decisions during endocrine pancreas development, and to the decline in replication ability that occurs as β–cell age. Important questions that remain include the analysis of chromatin states in the individual endocrine cell types, and the comparison of epigenetic marks in β-cells from diabetic individuals with those of healthy controls. In addition, it is likely that additional histone modifying enzymes are involved in fine-tuning development and function of pancreatic cell types. Finally, big challenges lie ahead in developing drugs that could target specific epigenetic modifications in pancreatic β-cells, both in vitro and in vivo, to improve the in vitro differentiation of β-cells from embryonic stem cells or induced pluripotent cells, or to enable the treatment of the epigenetically impaired β-cell in type 2 diabetics.
Highlights.
-
-
Epigenetic marks contribute to developmental choice between pancreas and liver
-
-
Transcription factors and epigenetic enzymes cooperate in cell fate maintenance
-
-
Declining proliferation of beta-cells with age is epigenetically controlled
-
-
Epigenetic drugs can be employed to alter fetal pancreas development
Acknowledgements
Related work in the Kaestner lab was supported by NIH grants R01-DK088383 and U01 DK089529.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Gittes GK. Developmental biology of the pancreas: a comprehensive review. Dev Biol. 2009;326:4–35. doi: 10.1016/j.ydbio.2008.10.024. [DOI] [PubMed] [Google Scholar]
- 2.Pan FC, Wright C. Pancreas organogenesis: from bud to plexus to gland. Dev Dyn. 2011;240:530–565. doi: 10.1002/dvdy.22584. [DOI] [PubMed] [Google Scholar]
- 3.Bernstein BE, Mikkelsen TS, Xie X, Kamal M, Huebert DJ, Cuff J, et al. A bivalent chromatin structure marks key developmental genes in embryonic stem cells. Cell. 2006;125:315–326. doi: 10.1016/j.cell.2006.02.041. [DOI] [PubMed] [Google Scholar]
- 4.Xu CR, Cole PA, Meyers DJ, Kormish J, Dent S, Zaret KS. Chromatin "prepattern" and histone modifiers in a fate choice for liver and pancreas. Science. 2011;332:963–966. doi: 10.1126/science.1202845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Collombat P, Hecksher-Sorensen J, Krull J, Berger J, Riedel D, Herrera PL, et al. Embryonic endocrine pancreas and mature beta cells acquire alpha and PP cell phenotypes upon Arx misexpression. J Clin Invest. 2007;117:961–970. doi: 10.1172/JCI29115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Thorel F, Nepote V, Avril I, Kohno K, Desgraz R, Chera S, et al. Conversion of adult pancreatic alpha-cells to beta-cells after extreme beta-cell loss. Nature. 2010;464:1149–1154. doi: 10.1038/nature08894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sussel L, Kalamaras J, Hartigan-O'Connor DJ, Meneses JJ, Pedersen RA, Rubenstein JL, et al. Mice lacking the homeodomain transcription factor Nkx2.2 have diabetes due to arrested differentiation of pancreatic beta cells. Development. 1998;125:2213–2221. doi: 10.1242/dev.125.12.2213. [DOI] [PubMed] [Google Scholar]
- 8.Hervouet E, Vallette FM, Cartron PF. Dnmt3/transcription factor interactions as crucial players in targeted DNA methylation. Epigenetics. 2009;4:487–499. doi: 10.4161/epi.4.7.9883. [DOI] [PubMed] [Google Scholar]
- 9.Dor Y, Brown J, Martinez OI, Melton DA. Adult pancreatic beta-cells are formed by self-duplication rather than stem-cell differentiation. Nature. 2004;429:41–46. doi: 10.1038/nature02520. [DOI] [PubMed] [Google Scholar]
- 10.Georgia S, Bhushan A. Beta cell replication is the primary mechanism for maintaining postnatal beta cell mass. J Clin Invest. 2004;114:963–968. doi: 10.1172/JCI22098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Teta M, Rankin MM, Long SY, Stein GM, Kushner JA. Growth and regeneration of adult Beta cells does not involve specialized progenitors. Dev Cell. 2007;12:817–826. doi: 10.1016/j.devcel.2007.04.011. [DOI] [PubMed] [Google Scholar]
- 12.Dhawan S, Georgia S, Tschen SI, Fan G, Bhushan A. Pancreatic beta cell identity is maintained by DNA methylation-mediated repression of Arx. Dev Cell. 2011;20:419–429. doi: 10.1016/j.devcel.2011.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Klose RJ, Bird AP. Genomic DNA methylation: the mark and its mediators. Trends Biochem Sci. 2006;31:89–97. doi: 10.1016/j.tibs.2005.12.008. [DOI] [PubMed] [Google Scholar]
- 14.Collombat P, Mansouri A, Hecksher-Sorensen J, Serup P, Krull J, Gradwohl G, et al. Opposing actions of Arx and Pax4 in endocrine pancreas development. Genes Dev. 2003;17:2591–2603. doi: 10.1101/gad.269003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sosa-Pineda B, Chowdhury K, Torres M, Oliver G, Gruss P. The Pax4 gene is essential for differentiation of insulin-producing beta cells in the mammalian pancreas. Nature. 1997;386:399–402. doi: 10.1038/386399a0. [DOI] [PubMed] [Google Scholar]
- 16.Colombo E, Collombat P, Colasante G, Bianchi M, Long J, Mansouri A, et al. Inactivation of Arx, the murine ortholog of the X-linked lissencephaly with ambiguous genitalia gene, leads to severe disorganization of the ventral telencephalon with impaired neuronal migration and differentiation. J Neurosci. 2007;27:4786–4798. doi: 10.1523/JNEUROSCI.0417-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Collombat P, Xu XB, Ravassard P, Sosa-Pineda B, Dussaud S, Billestrup N, et al. The Ectopic Expression of Pax4 in the Mouse Pancreas Converts Progenitor Cells into alpha and Subsequently beta Cells. Cell. 2009;138:449–462. doi: 10.1016/j.cell.2009.05.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Herrera PL. Adult insulin- and glucagon-producing cells differentiate from two independent cell lineages. Development. 2000;127:2317–2322. doi: 10.1242/dev.127.11.2317. [DOI] [PubMed] [Google Scholar]
- 19.Jackson-Grusby L, Beard C, Possemato R, Tudor M, Fambrough D, Csankovszki G, et al. Loss of genomic methylation causes p53-dependent apoptosis and epigenetic deregulation. Nat Genet. 2001;27:31–39. doi: 10.1038/83730. [DOI] [PubMed] [Google Scholar]
- 20.Collombat P, Hecksher-Sorensen J, Broccoli V, Krull J, Ponte I, Mundiger T, et al. The simultaneous loss of Arx and Pax4 genes promotes a somatostatin-producing cell fate specification at the expense of the alpha- and beta-cell lineages in the mouse endocrine pancreas. Development. 2005;132:2969–2980. doi: 10.1242/dev.01870. [DOI] [PubMed] [Google Scholar]
- 21.Collombat P, Hecksher-Sorensen J, Krull J, Berger J, Riedel D, Herrera PL, et al. Embryonic endocrine pancreas and mature beta cells acquire alpha and PP cell phenotypes upon Arx misexpression. Journal of Clinical Investigation. 2007;117:961–970. doi: 10.1172/JCI29115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bird A. DNA methylation patterns and epigenetic memory. Genes Dev. 2002;16:6–21. doi: 10.1101/gad.947102. [DOI] [PubMed] [Google Scholar]
- 23.Guccione E, Bassi C, Casadio F, Martinato F, Cesaroni M, Schuchlautz H, et al. Methylation of histone H3R2 by PRMT6 and H3K4 by an MLL complex are mutually exclusive. Nature. 2007;449:933–937. doi: 10.1038/nature06166. [DOI] [PubMed] [Google Scholar]
- 24.Hyllus D, Stein C, Schnabel K, Schiltz E, Imhof A, Dou Y, et al. PRMT6-mediated methylation of R2 in histone H3 antagonizes H3 K4 trimethylation. Genes Dev. 2007;21:3369–3380. doi: 10.1101/gad.447007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bouwens L, Rooman I. Regulation of pancreatic beta-cell mass. Physiol Rev. 2005;85:1255–1270. doi: 10.1152/physrev.00025.2004. [DOI] [PubMed] [Google Scholar]
- 26.Zhong L, Georgia S, Tschen SI, Nakayama K, Bhushan A. Essential role of Skp2-mediated p27 degradation in growth and adaptive expansion of pancreatic beta cells. J Clin Invest. 2007;117:2869–2876. doi: 10.1172/JCI32198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Meier JJ, Butler AE, Saisho Y, Monchamp T, Galasso R, Bhushan A, et al. Beta-cell replication is the primary mechanism subserving the postnatal expansion of beta-cell mass in humans. Diabetes. 2008;57:1584–1594. doi: 10.2337/db07-1369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Teta M, Long SY, Wartschow LM, Rankin MM, Kushner JA. Very slow turnover of beta-cells in aged adult mice. Diabetes. 2005;54:2557–2567. doi: 10.2337/diabetes.54.9.2557. [DOI] [PubMed] [Google Scholar]
- 29.Tschen SI, Dhawan S, Gurlo T, Bhushan A. Age-dependent decline in beta-cell proliferation restricts the capacity of beta-cell regeneration in mice. Diabetes. 2009;58:1312–1320. doi: 10.2337/db08-1651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nielsen GP, Stemmer-Rachamimov AO, Ino Y, Moller MB, Rosenberg AE, Louis DN. Malignant transformation of neurofibromas in neurofibromatosis 1 is associated with CDKN2A/p16 inactivation. Am J Pathol. 1999;155:1879–1884. doi: 10.1016/S0002-9440(10)65507-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Krishnamurthy J, Torrice C, Ramsey MR, Kovalev GI, Al-Regaiey K, Su L, et al. Ink4a/Arf expression is a biomarker of aging. J Clin Invest. 2004;114:1299–12307. doi: 10.1172/JCI22475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.van der Lugt NM, Domen J, Linders K, van Roon M, Robanus-Maandag E, te Riele H, et al. Posterior transformation, neurological abnormalities, and severe hematopoietic defects in mice with a targeted deletion of the bmi-1 proto-oncogene. Genes Dev. 1994;8:757–769. doi: 10.1101/gad.8.7.757. [DOI] [PubMed] [Google Scholar]
- 33.Krishnamurthy J, Ramsey MR, Ligon KL, Torrice C, Koh A, Bonner-Weir S, et al. p16INK4a induces an age-dependent decline in islet regenerative potential. Nature. 2006;443:453–457. doi: 10.1038/nature05092. [DOI] [PubMed] [Google Scholar]
- 34.Saxena R, Voight BF, Lyssenko V, Burtt NP, de Bakker PI, Chen H, et al. Genome-wide association analysis identifies loci for type 2 diabetes and triglyceride levels. Science. 2007;316:1331–1336. doi: 10.1126/science.1142358. [DOI] [PubMed] [Google Scholar]
- 35.Scott LJ, Mohlke KL, Bonnycastle LL, Willer CJ, Li Y, Duren WL, et al. A genome-wide association study of type 2 diabetes in Finns detects multiple susceptibility variants. Science. 2007;316:1341–1345. doi: 10.1126/science.1142382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sladek R, Rocheleau G, Rung J, Dina C, Shen L, Serre D, et al. A genome-wide association study identifies novel risk loci for type 2 diabetes. Nature. 2007;445:881–885. doi: 10.1038/nature05616. [DOI] [PubMed] [Google Scholar]
- 37.Zeggini E, Weedon MN, Lindgren CM, Frayling TM, Elliott KS, Lango H, et al. Replication of genome-wide association signals in UK samples reveals risk loci for type 2 diabetes. Science. 2007;316:1336–1341. doi: 10.1126/science.1142364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wang H, Wang L, Erdjument-Bromage H, Vidal M, Tempst P, Jones RS, et al. Role of histone H2A ubiquitination in Polycomb silencing. Nature. 2004;431:873–878. doi: 10.1038/nature02985. [DOI] [PubMed] [Google Scholar]
- 39.Bracken AP, Kleine-Kohlbrecher D, Dietrich N, Pasini D, Gargiulo G, Beekman C, et al. The Polycomb group proteins bind throughout the INK4A-ARF locus and are disassociated in senescent cells. Genes Dev. 2007;21:525–530. doi: 10.1101/gad.415507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kotake Y, Cao R, Viatour P, Sage J, Zhang Y, Xiong Y. pRB family proteins are required for H3K27 trimethylation and Polycomb repression complexes binding to and silencing p16INK4alpha tumor suppressor gene. Genes Dev. 2007;21:49–54. doi: 10.1101/gad.1499407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chen H, Gu X, Su IH, Bottino R, Contreras JL, Tarakhovsky A, et al. Polycomb protein Ezh2 regulates pancreatic beta-cell Ink4a/Arf expression and regeneration in diabetes mellitus. Genes Dev. 2009;23:975–985. doi: 10.1101/gad.1742509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Collombat P, Hecksher-Sorensen J, Serup P, Mansouri A. Specifying pancreatic endocrine cell fates. Mech Dev. 2006;123:501–512. doi: 10.1016/j.mod.2006.05.006. [DOI] [PubMed] [Google Scholar]
- 43.Gao N, LeLay J, Vatamaniuk MZ, Rieck S, Friedman JR, Kaestner KH. Dynamic regulation of Pdx1 enhancers by Foxa1 and Foxa2 is essential for pancreas development. Genes Dev. 2008;22:3435–3448. doi: 10.1101/gad.1752608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Jonsson J, Carlsson L, Edlund T, Edlund H. Insulin-promoter-factor 1 is required for pancreas development in mice. Nature. 1994;371:606–609. doi: 10.1038/371606a0. [DOI] [PubMed] [Google Scholar]
- 45.Offield MF, Jetton TL, Labosky PA, Ray M, Stein RW, Magnuson MA, et al. PDX-1 is required for pancreatic outgrowth and differentiation of the rostral duodenum. Development. 1996;122:983–995. doi: 10.1242/dev.122.3.983. [DOI] [PubMed] [Google Scholar]
- 46.Gradwohl G, Dierich A, LeMeur M, Guillemot F. neurogenin3 is required for the development of the four endocrine cell lineages of the pancreas. Proc Natl Acad Sci U S A. 2000;97:1607–1611. doi: 10.1073/pnas.97.4.1607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Gu G, Dubauskaite J, Melton DA. Direct evidence for the pancreatic lineage: NGN3+ cells are islet progenitors and are distinct from duct progenitors. Development. 2002;129:2447–2457. doi: 10.1242/dev.129.10.2447. [DOI] [PubMed] [Google Scholar]
- 48.Chang S, Young BD, Li S, Qi X, Richardson JA, Olson EN. Histone deacetylase 7 maintains vascular integrity by repressing matrix metalloproteinase 10. Cell. 2006;126:321–334. doi: 10.1016/j.cell.2006.05.040. [DOI] [PubMed] [Google Scholar]
- 49.Lagger G, O'Carroll D, Rembold M, Khier H, Tischler J, Weitzer G, et al. Essential function of histone deacetylase 1 in proliferation control and CDK inhibitor repression. EMBO J. 2002;21:2672–2681. doi: 10.1093/emboj/21.11.2672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.McKinsey TA, Zhang CL, Olson EN. Control of muscle development by dueling HATs and HDACs. Curr Opin Genet Dev. 2001;11:497–504. doi: 10.1016/s0959-437x(00)00224-0. [DOI] [PubMed] [Google Scholar]
- 51.Montgomery RL, Davis CA, Potthoff MJ, Haberland M, Fielitz J, Qi X, et al. Histone deacetylases 1 and 2 redundantly regulate cardiac morphogenesis, growth, and contractility. Genes Dev. 2007;21:1790–1802. doi: 10.1101/gad.1563807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Vega RB, Matsuda K, Oh J, Barbosa AC, Yang X, Meadows E, et al. Histone deacetylase 4 controls chondrocyte hypertrophy during skeletogenesis. Cell. 2004;119:555–566. doi: 10.1016/j.cell.2004.10.024. [DOI] [PubMed] [Google Scholar]
- 53.Chang S, McKinsey TA, Zhang CL, Richardson JA, Hill JA, Olson EN. Histone deacetylases 5 and 9 govern responsiveness of the heart to a subset of stress signals and play redundant roles in heart development. Mol Cell Biol. 2004;24:8467–8476. doi: 10.1128/MCB.24.19.8467-8476.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Nebbioso A, Manzo F, Miceli M, Conte M, Manente L, Baldi A, et al. Selective class II HDAC inhibitors impair myogenesis by modulating the stability and activity of HDAC-MEF2 complexes. EMBO Rep. 2009;10:776–782. doi: 10.1038/embor.2009.88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Pan FC, Wright C. Pancreas Organogenesis: From Bud to Plexus to Gland. Developmental Dynamics. 2011;240:530–565. doi: 10.1002/dvdy.22584. [DOI] [PubMed] [Google Scholar]
- 56.D'Amour KA, Bang AG, Eliazer S, Kelly OG, Agulnick AD, Smart NG, et al. Production of pancreatic hormone-expressing endocrine cells from human embryonic stem cells. Nat Biotechnol. 2006;24:1392–1401. doi: 10.1038/nbt1259. [DOI] [PubMed] [Google Scholar]
- 57.Furth ME, Atala A. Stem cell sources to treat diabetes. J Cell Biochem. 2009;106:507–511. doi: 10.1002/jcb.22000. [DOI] [PubMed] [Google Scholar]
- 58.Kroon E, Martinson LA, Kadoya K, Bang AG, Kelly OG, Eliazer S, et al. Pancreatic endoderm derived from human embryonic stem cells generates glucose-responsive insulin-secreting cells in vivo. Nat Biotechnol. 2008;26:443–452. doi: 10.1038/nbt1393. [DOI] [PubMed] [Google Scholar]
- 59.Bhandare R, Schug J, Le Lay J, Fox A, Smirnova O, Liu C, et al. Genome-wide analysis of histone modifications in human pancreatic islets. Genome Res. 2010;20:428–433. doi: 10.1101/gr.102038.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Stitzel ML, Sethupathy P, Pearson DS, Chines PS, Song L, Erdos MR, et al. Global epigenomic analysis of primary human pancreatic islets provides insights into type 2 diabetes susceptibility loci. Cell Metab. 2010;12:443–455. doi: 10.1016/j.cmet.2010.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Heintzman ND, Hon GC, Hawkins RD, Kheradpour P, Stark A, Harp LF, et al. Histone modifications at human enhancers reflect global cell-type-specific gene expression. Nature. 2009;459:108–112. doi: 10.1038/nature07829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Barski A, Cuddapah S, Cui K, Roh TY, Schones DE, Wang Z, et al. High-resolution profiling of histone methylations in the human genome. Cell. 2007;129:823–837. doi: 10.1016/j.cell.2007.05.009. [DOI] [PubMed] [Google Scholar]
- 63.van Arensbergen J, Garcia-Hurtado J, Moran I, Maestro MA, Xu X, Van de Casteele M, et al. Derepression of Polycomb targets during pancreatic organogenesis allows insulin-producing beta-cells to adopt a neural gene activity program. Genome Res. 2010;20:722–732. doi: 10.1101/gr.101709.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Dorrell C, Schug J, Lin CF, Canaday PS, Fox AJ, Smirnova O, et al. Transcriptomes of the major human pancreatic cell types. Diabetologia. 2011;54:2832–2844. doi: 10.1007/s00125-011-2283-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Artner I, Le Lay J, Hang Y, Elghazi L, Schisler JC, Henderson E, et al. MafB: an activator of the glucagon gene expressed in developing islet alpha- and beta-cells. Diabetes. 2006;55:297–304. doi: 10.2337/diabetes.55.02.06.db05-0946. [DOI] [PubMed] [Google Scholar]
- 66.Henikoff S. Nucleosome destabilization in the epigenetic regulation of gene expression. Nat Rev Genet. 2008;9:15–26. doi: 10.1038/nrg2206. [DOI] [PubMed] [Google Scholar]
- 67.Wallrath LL, Lu Q, Granok H, Elgin SC. Architectural variations of inducible eukaryotic promoters: preset and remodeling chromatin structures. Bioessays. 1994;16:165–170. doi: 10.1002/bies.950160306. [DOI] [PubMed] [Google Scholar]
- 68.Giresi PG, Kim J, McDaniell RM, Iyer VR, Lieb JD. FAIRE (Formaldehyde-Assisted Isolation of Regulatory Elements) isolates active regulatory elements from human chromatin. Genome Res. 2007;17:877–885. doi: 10.1101/gr.5533506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Nagy PL, Cleary ML, Brown PO, Lieb JD. Genomewide demarcation of RNA polymerase II transcription units revealed by physical fractionation of chromatin. Proc Natl Acad Sci U S A. 2003;100:6364–6369. doi: 10.1073/pnas.1131966100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Gaulton KJ, Nammo T, Pasquali L, Simon JM, Giresi PG, Fogarty MP, et al. A map of open chromatin in human pancreatic islets. Nat Genet. 2010;42:255–259. doi: 10.1038/ng.530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Gunton JE, Kulkarni RN, Yim S, Okada T, Hawthorne WJ, Tseng YH, et al. Loss of ARNT/HIF1beta mediates altered gene expression and pancreatic-islet dysfunction in human type 2 diabetes. Cell. 2005;122:337–349. doi: 10.1016/j.cell.2005.05.027. [DOI] [PubMed] [Google Scholar]