Abstract
Adenoviral infection induces nucleoplasmic redistribution of a nucleolar nucleophosmin 1/NPM1/B23.1. NPM1 is preferentially localized in the nucleoli of normal cells, whereas it is also present at the nuclear matrix in cancer cells. However, the biological roles of NPM1 during infection are unknown. Here, by analyzing a pV-deletion mutant, Ad5-dV/TSB, we demonstrate that pV promotes the NPM1 translocation from the nucleoli to the nucleoplasm in normal cells, and the NPM1 translocation is correlated with adenoviral replication. Lack of pV causes a dramatic reduction of adenoviral replication in normal cells, but not cancer cells, and Ad5-dV/TSB was defective in viral assembly in normal cells. NPM1 knockdown inhibits adenoviral replication, suggesting an involvement of NPM1 in adenoviral biology. Further, we show that NPM1 interacts with empty adenovirus particles which are an intermediate during virion maturation by immunoelectron microscopy. Collectively, these data implicate that pV participates in a process of viral assembly through NPM1.
Keywords: adenovirus, adenoviral assembly, cancer gene therapy, nucleophosmin 1, protein V
Introduction
Human adenoviruses are a member of Mastadenovirus, and a non-enveloped, icosahedral virus containing a double-stranded DNA genome (Davison, Benko, and Harrach, 2003), and contain a few genus-specific genes missing in the other three viral taxons. One of the genes is a minor core protein V (pV) (Davison, Benko, and Harrach, 2003), and an adenovirus virion contains approximately 157 copies of pV (van Oostrum and Burnett, 1985). Protein V is speculated to function as a bridge between the core and the capsid proteins (Chatterjee, Vayda, and Flint, 1985; Matthews and Russell, 1998; Vayda, Rogers, and Flint, 1983) and non-specifically associated with the double stranded viral genomic DNA (Anderson, Young, and Flint, 1989; Chatterjee, Vayda, and Flint, 1986; Lehmberg et al., 1999). Initially, pV localizes in the nucleoli during infection, then translocates from the nucleoli to the nucleoplasm as the infection progresses and is incorporated in infectious viral particles (Ugai et al., 2010). On the other hand, the deletion of the pV gene from the human adenovirus type 5 (Ad5) genome reveals that pV is not essential for adenoviral replication and assembly of infectious viral particles in cancerous cells (Ugai et al., 2007). Interestingly, transient expression of pV induces the redistribution of nucleophosmin 1/NPM1/B23.1 from the nucleoli to the nucleoplasm and the cytoplasm in vitro (Matthews, 2001). Also, adenoviral infection induces NPM1 nucleolar delocalization (Walton et al., 1989).
NPM1 is an abundant nucleolar phosphoprotein (Grisendi et al., 2006), but is expressed at fairly modest levels in human normal cells (Nozawa et al., 1996). In contrast to the expression of NPM1 in human normal cells, NPM1 is isolated as one of the most abundant nuclear matrix proteins in cancer cells (Mattern et al., 1996; Zink, Fischer, and Nickerson, 2004), and detected in the nucleoplasm as well as in the nucleoli of tumors (Subong et al., 1999) and overexpressed in various types of tumors (Grisendi et al., 2006). Thus, the localization and expression of NPM1 are altered in human cancers (Grisendi et al., 2006). On the other hand, NPM1.2 which lacks the 35 amino acids at the C-terminus of NPM1, encodes the RNA binding site, RNase domain, and nucleolar localization signal of NPM1 (Herrera, Savkur, and Olson, 1995; Hingorani, Szebeni, and Olson, 2000; Wang, Umekawa, and Olson, 1993) expresses in rat tissue and cells at a low level in the cytoplasm and nucleoplasm (Wang, Umekawa, and Olson, 1993). However, the NPM1.2 expression is not found in human normal cells. Although NPM1.2 expression in human is only detected in HeLa cells which stably express human papillomavirus (HPV) E6 and E7 proteins (Dalenc et al., 2002; Sautkina et al., 2008), the function is largely unclear (Herrera, Savkur, and Olson, 1995; Savkur and Olson, 1998). NPM1 directly interacts with tumor suppressor p14ARF (ARF) which is an alternative reading frame product of the p16INK4a locus and antagonizes its function in the nucleoli of nonstressed cells (Itahana et al., 2003). In response to cellular and viral stresses, NPM1 is redistributed from the nucleoli to the nucleoplasm and cytoplasm (Hiscox, 2002; Rubbi and Milner, 2003). There is no evidence that phosphorylation of NPM1 is involved in its translocation during cellular and viral stresses (Kurki et al., 2004). Upon cellular stresses, NPM1 binds to HDM2 which inhibits its E3 ubiquitin ligase activity and protects p53 from HDM2-mediated degradation (Kurki et al., 2004). Also, NPM1 directly interacts with p53 and regulates its protein stability and transcriptional activity (Colombo et al., 2002). Thus, NPM1 functions as a critical regulator of ARF, HDM2 and p53 in the p53 pathway (Grisendi et al., 2006).
Notwithstanding these findings, the precise biological reasons why adenoviral pV redistributes NPM1 in vitro and why NPM1 is redistributed from the nucleoli to the nucleoplasm during infection are not understood. It is also unclear whether pV induces the NPM1 redistribution during infection. Here we demonstrate that pV triggers the relocalization of NPM1 from the nucleoli to the nucleoplasm in primary human endothelial cells by comparative analysis using a pV-deletion mutant, Ad5-dV/TSB, of Ad5. We also provide evidence that adenovirus replication in primary cells requires the relocalization of NPM1 from the nucleoli to the nucleoplasm. Moreover, we show that NPM1 knockdown inhibits Ad5 replication, demonstrating a direct role of NPM1 in adenoviral replication. While lack of pV causes a great restriction of adenoviral replication in primary cells, Ad5-dV/TSB replicates and induces tumoricidal effect in cancer cells where NPM1 is also abundant in the nucleoplasm. Additionally, transmission electron microscope based analysis demonstrates that lack of pV is linked to a defect in viral assembly in primary cells, but not in cancerous cells. Furthermore, NPM1 is co-purified with and can be directly detected in situ in empty adenovirus particles. Thus, to our knowledge, NPM1 is the first cellular protein to be identified as being involved in an event of adenoviral virion maturation. These findings imply that at least one significant biological function of pV is in the process of assembly of empty adenovirus particles during virion maturation, and NPM1 is involved in adenoviral biology. Also, our data provide an important new insight in adenoviral assembly and may aid vector design to achieve the selective replication of adenovirus between normal and cancer cells.
Materials and Methods
Cells
Human embryonic retinoblast cell line 911 was a kind gift of Dr. Alex J. van der Eb (Leiden University, Leiden, the Netherlands). Human umbilical vein endothelial cells (HUVEC) and pulmonary artery endothelial cells (HPAEC) were purchased from Lonza Walkersville, Inc. (Walkersville, MD). The other cell lines were obtained from American Type Culture Collection (ATCC; Rockville, MD). The 293 cells stably expressing human NPM1-EGFP (293-NPM1-EGFP) were generated with plasmid pEGFP-N1-NPM1 (Hindley, Davidson, and Matthews, 2007) and selected with 1 mg/ml geneticin (G418). All cell lines were cultured with media recommended by the depositor. Information of human cell lines used in this study was provided in Supplementary Table 1.
Adenoviruses
Ad5 (Ugai et al., 2007), Ad5-dV/TSB (Ugai et al., 2007), Ad5-wt-IX-mRFP1 (Le et al., 2006a), and Ad5-E3-V-EGFP (Le et al., 2006b) were previously reported. Adenoviral mutants, Ad5-dV/TSB, dE1B55K, and dE1B55K-dV/TSB, were generated from pTG3602-dV/TSB, pTG3602-dE1B55K, and pTG3602-dE1B55K-dV/TSB, respectively, as described in Supplementary Materials and Methods. However, dE1-dV/TSB which contains the deletion of both the E1 region and pV gene was not generated using pTG3602DS-dE1-dV/TSB. dE1B55K is an adenovirus mutant containing a deletion of the E1b-55k gene, and dE1B55K-dV/TSB is an adenovirus mutant carrying both the E1b-55k and the pV gene deletions. Ad5-dV/TSB-IX-mRFP1 which is a fluorescently labeled Ad5-dV/TSB was generated by the DNA-TPC method as described in Supplementary Information. The particles and infectious titers of adenoviruses used in this study were: Ad5 (4.94 × 1011 VP/ml and 1.58 × 1010 PFU/ml), Ad5-dV/TSB (4.35 × 1012 VP/ml and 2.33 × 1010 PFU/ml), Ad5-wt-IX-mRFP1 (2.09 × 1012 VP/ml and 2.17 × 109 PFU/ml), Ad5-E3-V-EGFP (5.71 × 1011 VP/ml and 1.03 × 1010 PFU/ml), dE1B55K (2.94 × 1012 VP/ml and 1.52 × 1010 PFU/ml), dE1B55K-dV/TSB (1.97 × 1012 VP/ml and 8.42 × 106 PFU/ml), and Ad5-dV/TSB-IX-mRFP1 (1.82 × 1012 VP/ml and 1.67 × 1010 PFU/ml).
Determination of protein concentration
Total protein concentration of cell lysates extracted from uninfected and adenoviral infected cells was determined using DC protein assay kit (Bio-Rad laboratories Inc.) according to the manufacturer’s instructions.
Antibodies
The primary antibodies were used against adenoviral pV [1:1000, rabbit polyclonal (Matthews and Russell, 1998)], Ad5 [1:1000, rabbit polyclonal anti-serum (Ugai et al., 2007) for Western blot analysis and 1:20 for immunoelectron microscopy], E1A (M58 at 1:200, mouse monoclonal, Calbiochem, San Diego, CA), E1B-55K (1:10, mouse monoclonal 2A6, from A. J. Levine, The Cancer Institute of New Jersey), E2A (1:10, mouse monoclonal B6, from A. J. Levine, The Cancer Institute of New Jersey), E3-ADP (1:500, rabbit polyclonal anti-ADP peptide antibodies p63–77, from W. S. M. Wold, St. Louis University), E4orf6, 6/7 (1:10, RSA-3, from T. Shenk, Princeton University). Also, we used p53 antibody (DO-1 at 1:200; Santa Cruz Technology Inc., Santa Cruz, CA) and HDM2 (SMP14 at 1:200; Santa Cruz Technology Inc., 2A10 at 1:100; Abcam Inc., Cambridge, MA), ARF (C-18 at 1:200; Santa Cruz Technology Inc., 4C6/4 at 1:1000; Abcam Inc.), β-actin (AC-15 at 1:1000; Abcam Inc.), and NPM1 (FC-61991 at 1:1000 for Western blot analysis and 1:20 for immunoelectron microscopy).
Western blot analysis, immunoprecipitation, and gel-staining analyses
Cells were cultured on 6-well plates and 15-cm dishes, respectively, for 2 days, and the number of cells was counted to determine a multiplicity of infection (MOI). Cells were infected with adenovirus at an MOI of 10 PFU/cell, or at MOIs specified in the Figure legend. The infected cells and the medium were harvested by using a cell lifter at various time points post-infection. The cell suspension was centrifuged at 1,000 × g for 5 min at 4 °C to remove medium. Infected cells were resuspended with 5 ml of phosphate-buffered saline (PBS) or 30 mM HEPES-buffered saline, and centrifuged at 1,000 × g for 5 min at 4 °C. The infected cancer and HPAEC cell pellets were resuspended with 100 µl and 400 µl of lysis buffer (30 mM HEPES-NaOH [pH 7.4], 1% Nonidet P-40, 0.5 mM EDTA, 150 mM NaCl) containing protease inhibitor (Sigma-Aldrich, St. Louis, MO), respectively. The cell lysate was centrifuged at 15,000 × g for 5 min at 4 °C to remove cell debris, and the supernatant was stored at −80 °C prior to Western blot analysis. Cell lysates extracted from adenovirus infected cells were separated by 10% (w/v) SDS-polyacrylamide gel electrophoresis (SDS-PAGE). Immunoprecipitation and/or Western blot analysis was carried out as described elsewhere (Fukuda et al., 2003; Ugai et al., 2005), and the signals were visualized by enhanced chemiluminescence (Amersham, Piscataway, NJ). Also, purified viral particles were analyzed by using GELCODE Blue reagent (Pierce Biotechnology Inc., Rockford IL). Pre-stained protein ladder of Kaleidoscope Standards (Bio-Rad Laboratories, Inc., Hercules, CA) was used.
Tracking of cellular and viral proteins
An equivalent of 1.0 × 104 cells was cultured on a collagen coated glass bottom dish (World Precision Instruments, Inc., Sarasota, FL) for 24 hours. For the tracking of fluorescently labeled adenovirus at the early stage of viral infection, HPAEC cells were infected by fluorescently labeled adenoviruses at an MOI of 10, 100 or 1,000 PFU/cell and the infected cells were incubated at 37 °C for 1 hour. Also, localization of cellular proteins in primary and cancer cell lines was verified by immunofluorescence staining. In order to observe localization of cellular and viral proteins at various time points in infected cells, cells were infected with adenovirus at an MOI of 10 PFU/cell, and localization of cellular and viral proteins was verified by immunofluorescence staining.
Immunofluorescence
Cells were cultured on a collagen coated glass bottom dish (World Precision Instruments, Inc., Sarasota, FL) for 24 hours. We first optimized immunofluorescence staining in cells and performed the staining procedure without a washing step with 1 ml of 30 mM HEPES-NaOH buffer (pH 7.4) before the fixation step. Because the release of nucleolar proteins from the nucleoli of primary cells was detected, we skipped the washing step for immunofluorescence staining. In brief, after growth medium was removed, cells were directly fixed with 1 ml of 30 mM HEPES-NaOH buffer (pH 7.4) containing 0.01% of glutaraldehyde and 2% paraformaldehyde for 10 min. After remove the fixative solution, the cells were two times washed with 1 ml of 30 mM HEPES-NaOH buffer (pH 7.4) and permeabilized with 1 ml of 30 mM HEPES-NaOH (pH 7.4) containing 1% Triton X-100 for 30 sec. The cells were two times washed with 1 ml of 30 mM HEPES-NaOH buffer (pH 7.4) and incubated with 1 ml of blocking solution (30 mM HEPES-NaOH buffer [pH 7.4], 2% FBS, and 0.2 M glycine) for 10 min. The cells were two times washed with 1 ml of 30 mM HEPES-NaOH buffer (pH 7.4) and incubated with a primary antibody for 1 hour at room temperature. The cells were two times washed with 1 ml of 30 mM HEPES-NaOH buffer (pH 7.4) and incubated with a second antibody conjugated with Alexa Fluor 488 or Alexa Fluor 594 (dilution range 1: 1,000–10,000) for overnight under dark condition. The cells were four times washed with 1 ml of 30 mM HEPES-NaOH buffer (pH 7.4) for 10 min each. DNA was stained with 1 ml of 30 mM HEPES-NaOH buffer (pH 7.4) containing 0.2 µg/ml Hoechst 33342 (Sigma-Aldrich), and the cells were four times washed with 1 ml of 30 mM HEPES-NaOH buffer (pH 7.4) for 10 min each. The fluorescent signals were detected by fluorescence microscopy.
siRNA
The siRNA duplexes for p53, and NPM1 mRNA sequences and non-silencing which are commercially available and had been validated were purchased from Qiagen Inc., Valencia, CA. The p53 siRNA duplex sequences are 5’-AGGUGGUUCUCUUCCCAAATT-3’ (sense strand) and 5’-UUUGGGAAGAGAACCACCUTT-3’ (antisense strand), and the NPM1 siRNA duplex sequences are 5’-GGAAAUUUGCGUGUGGAGUTT-3’ (sense strand), 5’-ACUCCACACGCAAAUUUCCTT-3’ (antisense strand). AllStars negative control siRNA was used for non-silencing of human gene expression. HPAEC cells were cultured at a density of 1.0 × 104 cells per dish on a collagen coated glass bottom dish or 6-well plate for 24 hours. Transfection was performed with Lipofection 2000 according to the manufacturer’s instructions, at a final concentration of 5 nM or 10 nM siRNA for 48 hours. The transfected cells were used for immunofluorescence or adenoviral infection.
PCR and quantitative PCR analyses
Adenoviral genome was extracted from purified infectious viral particles and capsid complex by method described previously (Saito et al., 1985), and an equivalent of 1.0 × 1010 viral particles was used for template of PCR amplification with FIBF31136 (5’-CCCAATGGGTTTCAAGAGAGT-3’) and FIBR32843 (5’-ATGACTTGAAATTTTCTGCAATTG-3’). PCR was performed by using KOD Hot Start Master Mix (EMD Millipore, Darmstadt, Germany) according to the manufacturer’s instructions. Cells were grown to 80% confluency in 6-well plate and the number of cells in a dish was counted before viral infection. We infected cells with adenovirus at an MOI of 10 PFU/cell and maintained infected cells in 3 ml of medium. The infected cells were harvested at various times post-infection by using a cell scraper. Total cell DNA containing the adenoviral genome was extracted by method described previously (Saito et al., 1985) and stored at −80 °C prior to quantitative PCR (qPCR) analysis. Oligonucleotides corresponding to the sense strand of adenoviral E4 region (5’-TGACACGCATACTCGGAGCTA-3’: 34,885–34,905 nucleotide position [nt]), the antisense strand of the E4 region (5’-TTTGAGCAGCACCTTGCATT-3’: 34,977–34,958 nt), and a TaqMan probe (5’-FAM-CGCCGCCCATGCAACAAGCTT-BHQ-1-3’: 34,930–34,951 nt) were synthesized and used as PCR primers and TaqMan probe for qPCR. The qPCR conditions were as follows: 35 cycles of denaturation (94 °C, 20 sec), annealing (55 °C, 20 sec), and extension (72 °C, 30 sec). The Ad5 genome was used to generate a standard curve for the adenoviral E4 DNA copy number. Three replicates were performed with 50 ng of total DNA extracted from infected cells and control sample without template. To determine lentiviral titer, (Delenda and Gaillard, 2005; Sastry et al., 2002) we used a primer set; 2875-2893F (5’-GCCGTCTTTTGGCAATGTG-3’) and 2921-2940R (5’-CCCCTAGGAATGCTCGTCAA-3’), and FastStart SYBR Green master (Roche Applied Science, Mannheim, Germany).
Quantitative RT-PCR analysis
Total RNA was extracted from infected cells using Trizol reagent according to manufacturer’s instructions (Invitrogen, Carlsbad, CA) and incubated DNase (Promega Corporation, Madison, WI) to resolve host and viral DNA. DNase was inactivated and removed by phenol/chloroform extraction. Quantitative RT-PCR (qRT-PCR) was performed using the following primers designed by Primer 3 (http://frodo.wi.mit.edu/primer3/); NPM1F; 5’-TCTCTTCCCAAAGTGGAAGC-3’, NPM1R; 5’-CTCCACTGCCAGAGATCTTG-3’, IXF-RT-PCR; CGCGATGACAAGTTGACGG, IXR-RT-PCR; CCAACAGCTGCTGAGAAACG, E1B55KF-RT-PCR; CACGTAGCCAGCCACTCTC, E1B55KR-RT-PCR; CAAATGCAAGGAACAGCGGG, HX-RT-PCR; CATCATCGAAGGGGTAGCCAT, and TPL-3rd-RT-PCR; TAACCAGTCACAGTCGCAAG. Each reaction for qRT-PCR was performed with 100 ng of total RNA and control sample without template, and Brilliant SYBER Green QRT-PCR Master mix was utilized according to the manufacturer’s protocol (Strategene, La Jolla, CA). The following qRT-PCR condition applied: 30 min at 50 °C for reverse transcription, 1 min at 95 °C for denaturation reaction, and 45 cycles of 15 sec at 95 °C, and 30 sec at 72 °C for analyzing gene expression of NPM1 and hexon, and 45 cycles of 15 sec at 95 °C and 1 min at 60 °C for analyzing gene expression of E1b-55k and pIX. Analysis of qRT-PCR was performed by 2−ΔΔCT method after the threshold cycle (CT) of each sample was normalized to the intrinsic control (GAPDH). For the detection of GAPDH gene expression, the following primers were used: GAPDHF; 5’-GGTTTACATGTTCCAATATGATTCCA-3’, GAPDHR; 5’-ATGGGATTTCCATTGATGACAAG-3’, and a TaqMan probe; 5’-HEX-CGTTCTCAGCCTTGACGGTGCCAT-BHQ-1-3’.
Immunoelectron microscopy and transmission electron microscopy (TEM)
A total of 1.0 × 108 VP equivalents of purified viral particles were transferred into a 1.5-ml tube and vortexed for 20 seconds (sec) to break up aggregates. The first antibody was added the viral solution and stored at room temperature for 1 hour. Ten microliters of the solution were placed on parafilm, and a glow discharged grid (400-mesh, pure carbon support films; Electron Microscopy Sciences, Hatfield, PA) was placed on the viral solution for 1 hour. The carbon grids were washed for 30 sec with 100 µl of filtered 30 mM HEPES-NaOH buffer (pH 7.4) on parafilm ten times at room temperature, and blocked two times with 1% bovine serum albumin (BSA; Sigma-Aldrich) in 30 mM HEPES-NaOH buffer (pH 7.4) at room temperature for 20 min. Subsequently, the carbon grids were incubated with donkey anti-mouse IgG antibody conjugated with 10-nm gold (1:40) or donkey anti-rabbit IgG antibody conjugated with 25-nm gold (1:40) at room temperature for 1 hour. Next, the grids were washed for 30 sec on parafilm three times with 1% BSA in 30 mM HEPES-NaOH buffer (pH 7.4) at room temperature. The viral particles were fixed with 100 µl of 1% glutaraldehyde in 30 mM HEPES-NaOH buffer (pH 7.4) for 20 sec at room temperature, and the grids were washed for 30 sec three times with 100 µl of 30 mM HEPES-NaOH buffer (pH 7.4). The adenoviral particles on the grid were negatively stained with 1% (wt/vol) uranyl acetate at room temperature for 10 sec and, subsequently, the grids were air dried. Electron micrographs for the adenoviral particles were taken with FEI Tecnai F20 200 kV at the High-Resolution Imaging Facility in the University of Alabama at Birmingham and Molecular Microbiology Imaging Facility at Washington University in St. Louis. Also, we analyzed infected cells by TEM. In brief, cells cultured in 15-cm dish were infected with adenovirus at an MOI of 10 PFU/cell, and infected cells were incubated for 72 hours. Infected cells were harvested according to the manufacturer’s (Lonza Walkersville, Inc.) instructions. The harvested cells were fixed with a fixative solution (30 mM HEPES-NaOH buffer [pH 7.4] containing 1.25% of glutaraldehyde and 2% paraformaldehyde) for 1 hour at room temperature. The fixed cells were centrifuged at 1,000 × g for 5 min at room temperature and resuspended with the fixative solution. TEM was performed at School of Veterinary Medicine Microscopy Center in Louisiana State University.
Statistical analysis
Statistical analysis was performed with two-tailed unpaired Student’s t-tests between groups (* P<0.01, ** P<0.05, N.S. no significant difference).
Additional Materials and Methods
Additional materials and methods and any associated references were described as Supplemental Materials and Methods.
Results
Adenoviral pV is required for NPM1 redistribution from the nucleoli to the nucleoplasm
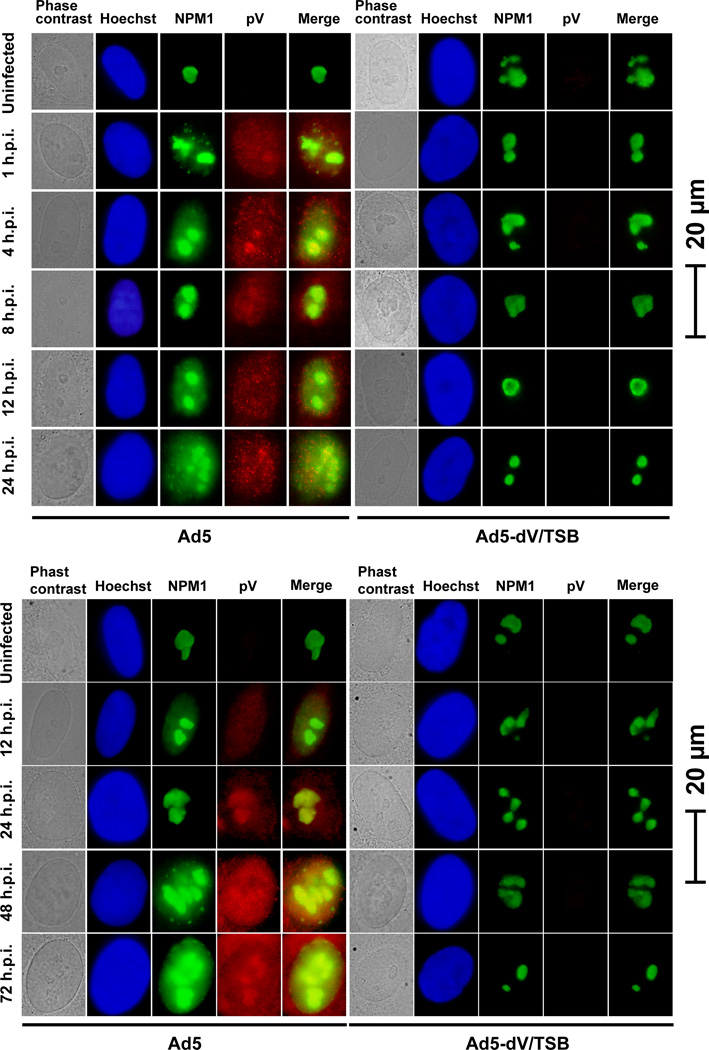
We genetically generated the adenoviral mutants except dE1-dV/TSB which contains both the E1 and pV deletions. A schematic representation of human adenovirus type 5 (Ad5) and the mutants used in this study was shown in Supplementary Figure S1. Although we previously reported that the Ad5-dV/TSB genome contains three suppressor mutations in the pre-Mu gene (Ugai et al., 2007), wild type pre-Mu and mature Mu are not associated with the NPM1 translocation (Lee et al., 2004). Also, production of infectious Ad5-dV which is the parental virus of Ad5-dV/TSB has been dramatically restricted in A549 cells (Ugai et al., 2007). Thus, we were not able to characterize Ad5-dV in primary cells. Moreover, we could not generate dE1-dV/TSB in host cell lines stably expressing E1 proteins, because Ad5-dV/TSB replication was severely restricted in 293 cells (Fig. S2). Therefore, we analyzed the pV function by comparing Ad5 with an array of adenovirus mutants including Ad5-dV/TSB. We first investigated whether adenoviral pV plays a role in translocation of NPM1 from the nucleoli to the nucleoplasm in human pulmonary endothelial cells (HPAEC). In our all experiments, we directly fixed cells after medium was removed as described in Materials and Methods section, because NPM1 was detected in the nucleoplasm as well as the nucleoli of HPAEC cells when we washed cells with 30 mM HEPES-NaOH buffer (pH7.4) (Fig. S3A). Under this condition, we also detected p53 at the nucleoli in HPAEC cells, but not in cancer cells (Fig. S3B). Thus, non-activated p53 which exists at very low levels in normal cells was localized in the nucleoli in normal cells, although wild type p53 is observed at the nucleus in malignant cells (Fig. S3B) (Koch et al., 2001). Localization of p53 may result from two essential regions for nucleolar localization contained in its C-terminal portion (Karni-Schmidt et al., 2007). Thus, we examined whether NPM1 is delocalized by adenoviral infection at an early time point by using Ad5-E3-V-EGFP (Ugai et al., 2010) containing fluorescently labeled pV (pV-EGFP). Fluorescent signal for pV-EGFP was detected in Ad5-E3-V-EGFP-infected cells, and pV-EGFP was colocalized with NPM1 in the nucleoli at 1 hour post-infection (h.p.i.) (Fig. S4A). Also, the partial NPM1 translocation was detected in an MOI-dependent manner of Ad5-E3-V-EGFP infection (Fig. S4A). It is unlikely that this translocation of NPM is a result of adenoviral early proteins expressed from the E1 region, since NPM1-EGFP expression was predominantly in the nucleoli of 293-NPM1-EGFP cells which are stably expressing both E1 and NPM1-EGFP (Fig. S4B). These results, in conjunction with our published findings (Hindley, Davidson, and Matthews, 2007; Matthews and Russell, 1998; Ugai et al., 2010), suggested that pV-EGFP released from the Ad5-E3-V-EGFP particles may move to the nucleoli, and Ad5-E3-V-EGFP infection induced the NPM1 redistribution at 1 h.p.i. (Fig. S4A). Indeed, Ad5 infection resulted in the partial disruption of NPM1 nucleolar localization during the course of infection, but Ad5-dV/TSB infection did not (Fig. 1). Collectively, these data indicate that wild type Ad5 infection, but not Ad5-dV/TSB infection, stimulates the NPM1 redistribution. Also, we conclude that pV is necessary for NPM1 translocation from the nucleoli during the course of infection as Ad5-dV/TSB is unable to induce such changes in contrast to wild type Ad5 (Fig. 1).
Fig. 1. Adenoviral protein V regulates subcellular localization of NPM1 during infection.
HPAEC cells were infected with adenovirus at an MOI of 10 PFU/cell for various times, followed by immunofluorescent staining. Subcellular localization of adenoviral pV and endogenous NPM1 was observed in infected cells at various times post-infection. Viral and endogenous cellular proteins in infected cells were visualized by staining with anti-pV and anti-NPM1 followed by secondary antibody conjugated with fluorescent probes. Uninfected and infected cells were stained for nuclear DNA using Hoechst 33342 and fluorescent signals were visualized by epifluorescence microscopy.
Adenoviral pV dictates its viral replication in normal cells
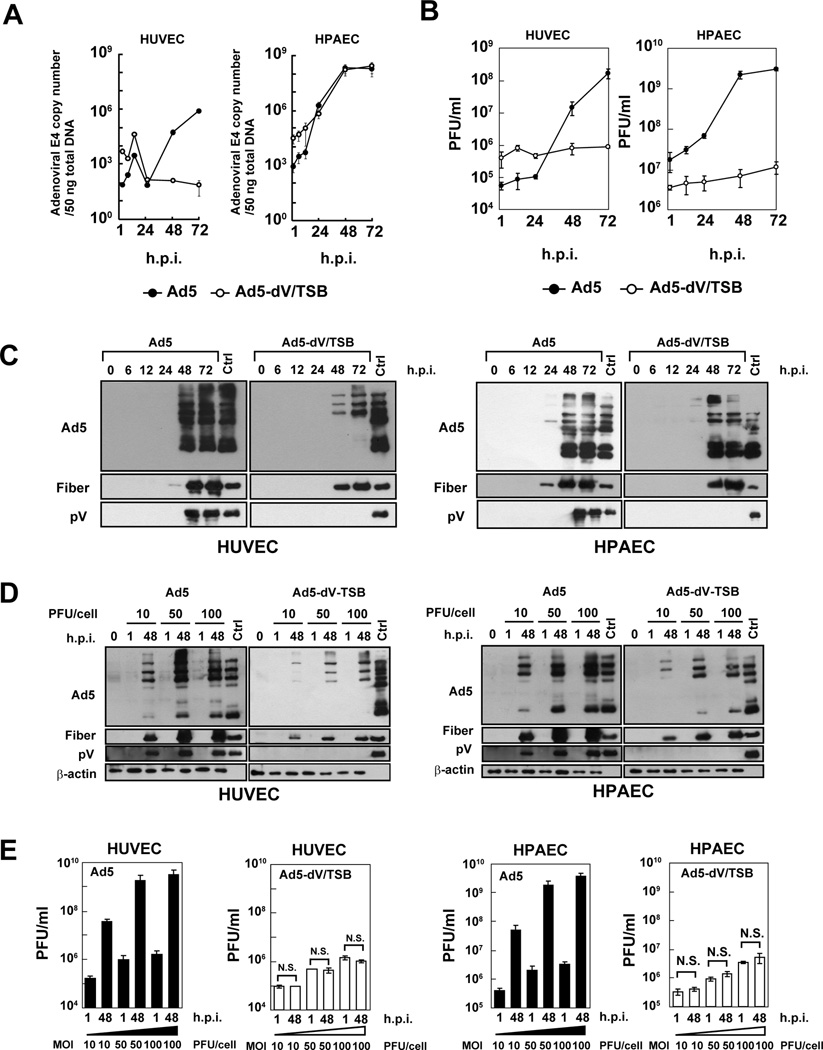
In order to examine whether pV is associated with adenoviral DNA replication in infected primary cell lines, HUVEC and HPAEC, we analyzed viral DNA replication by quantifying the copy number of the E4 region using the E4 primers. These cell lines express NPM1 at low levels as compared with lung carcinoma A549 cells (Fig. S5A). Ad5 DNA replication was readily detected in both primary and A549 cells (Figs. 2A and S5B). By contrast, Ad5-dV/TSB DNA replication was observed in HPAEC and A549 cells (Figs. 2A and S5B). Although we observed a severe defect in Ad5-dV/TSB DNA replication in HUVEC (Fig. 2A), we noted that Ad5 DNA replication was attenuated in HUVEC compared to HPAEC (Fig. 2A). Therefore, adenoviral DNA replication per se seemed to be restricted in HUVEC cells. Additionally, lack of pV did not seem to affect viral DNA replication in infected cells. We also performed one-step growth curve analysis of adenoviruses in order to examine the role of pV in viral replication in primary cells. Ad5 replication was observed in HUVEC and HPAEC cells, but Ad5-dV/TSB replication was severely restricted in those cell lines (Fig. 2B). Thus, pV was likely to be associated with viral replication. We next examined the kinetics of the late viral protein expression in infected primary cells. The expression of late viral proteins which were detected by anti-Ad5 antibody was impaired in Ad5-dV/TSB-infected HUVEC cells as compared to Ad5-infected HUVEC cells (Fig. 2C). In contrast to HUVEC, late viral proteins in both Ad5- and Ad5-dV/TSB-infected HPAEC cells were similarly expressed by 24 h.p.i. (Fig. 2C). On the other hand, the onset of the fiber expression was impaired in Ad5-dV/TSB-infected primary cells, since the expression of fiber protein was observed at an early time point in Ad5-infected primary cells (Fig. 2C). We also confirmed the lack of pV expression in Ad5-dV/TSB-infected primary cells (Fig. 2C). Thus, lack of pV affected the expression of fiber protein at an early time point in primary cells (Fig. 2C). Moreover, we investigated whether Ad5-dV/TSB infection at high MOIs reversed the viral replication restriction in primary cells. Although overexpression of late viral proteins was observed in both HUVEC and HPAEC cells infected with Ad5-dV/TSB at an MOI-dependent manner (Fig. 2D), Ad5-dV/TSB infection at higher MOIs did not compensate for the defect in the production of the Ad5-dV/TSB progeny (Fig. 2E). Further, we examined whether transient expression of pV affects adenoviral production in primary cells. Overexpression of pV fused with monomeric red fluorescent protein 1 (pV-mRFP1) was found to partially induce the NPM1 and p53 redistribution from nucleoli in primary cells compared to mock-infected cells (Fig. S6A). Both Ad5 and Ad5-dV/TSB production was enhanced in primary cells expressing pV (Fig. S6B and C). This data shows that transiently expressed pV compensates for the defect in Ad5-dV/TSB replication in normal cells (Fig. S6). Taken together, these data suggest that pV is essential for infectious adenovirus particle production in normal cells, but not adenovirus DNA replication (Figs. 2A, B, E, and S6).
Fig. 2. Protein V is required for adenoviral replication in normal cells.
(A–C) Primary cells were infected with adenovirus at an MOI of 10 PFU/cell for various times. (A) Total DNA was extracted from infected cells, and 50 ng of total DNA was analyzed by qPCR. Plots represent as the copy number of the E4 gene per 50 ng of total DNA. (B) One-step growth curve kinetics of adenoviruses was analyzed in primary cell lines. (C) Proteins were extracted from infected primary cells at various times post-infection and 40 µg of total proteins were subjected to Western blot with the indicated antibodies. (D) Primary cells were infected with adenovirus at an MOI of 10, 50 or 100 PFU/cell for 1 and 48 hours. Crude proteins were extracted from infected cells, and 10 µg of proteins was subjected to Western blot. Ctrl: 1.0 × 109 viral particles (VPs) of purified Ad5 were applied as a control for the pV expression. (E) Production level of infectious adenovirus in primary cell lines at 48 h.p.i. was measured by plaque assay. Results were reported as mean ± SD. N.S.; no significant difference
Protein V affects the E1B-55K and pIX expression
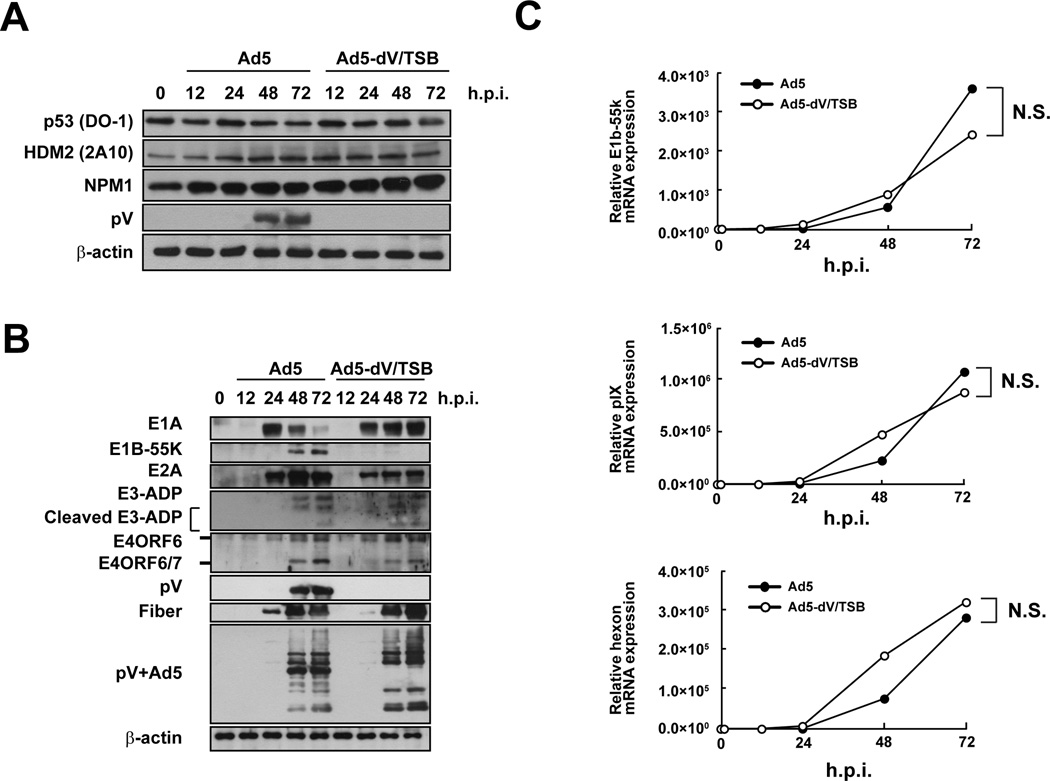
We next compared the kinetics of cellular and viral protein expression in infected primary cells. Ad5 and Ad5-dV-TSB infections did not affect the expression of NPM1, HDM2, and p53 during the eclipse stage of infection before viral replication, and ARF which is an alternative reading frame product of the p16INK4a locus was undetectable in uninfected or infected HPAEC cells compared to prostate adenocarcinoma DU145 cells which express low levels of ARF (Fig. S7) (Lee et al., 2005). The lack of ARF expression in HPAEC cells is consistent with previous data that ARF expression is not detectable in primary cells (Koch et al., 2001) and normal tissue (Sherr, 2006). Although ARF is induced by oncogenic stresses (Sherr, 2006) and stabilizes p53 (Zhang, Xiong, and Yarbrough, 1998), we did not observe the ARF expression in infected HPAEC cells (Fig. S7). Additionally, p53 expression was not dramatically changed in both Ad5- and Ad5-dV/TSB-infected cells (Fig. 3A). The onset of immediate early and early protein expression was comparable between Ad5- and Ad5-dV/TSB-infected HPAEC cells, except for the early gene product E1B-55K (Fig. 3B). Expression levels of the immediate early and early proteins at each time point were affected by the absence of pV (Fig. 3B). In particular, the lack of pV showed persisted E1A expression at 72 h.p.i., and this result was reproductive (Fig. 3B). Also, the lack of pV was markedly attenuated E1B55K expression in HPAEC cells (Fig. 3B). Therefore, we examined mRNA expression level of E1B-55K as well as pIX mRNA by qRT-PCR, since E1b-55k mRNA shares the polyadenylation signal with pIX mRNA (Le Moullec et al., 1983). However, lack of pV did not affect E1B-55K and pIX gene expression in infected primary cells (Fig. 3C). Furthermore, we tracked pIX expression by monitoring pIX-mRFP1 expression in cells infected with fluorescently labeled adenoviruses. The pIX-mRFP1 expression was attenuated in Ad5-wt-IX-mRFP1-infected primary and cancer cells as compared to Ad5-wt-IX-mRFP1-infected cells (Fig. S8). Therefore, this data also suggested that lack of pV repressed pIX expression in infected cells (Fig. S8). Although we determined the nucleotide sequence of the regions for the E1B-55K and the IX genes in the Ad5-dV/TSB genome, there are no mutations in the regions (data not shown). Taken together, there data suggested that pV was associated with the E1B-55K and pIX expression, but not gene expression of those mRNAs.
Fig. 3. Lack of pV affects the E1B-55K and pIX expression.
HPAEC cells were infected with adenovirus at an MOI of 10 PFU/cell for various times. (A and B) Proteins were extracted from infected primary cells at various times post-infection and 40 µg of total proteins were subjected to Western blot with the indicated antibodies. (C) Gene expression of E1b-55k, pIX, and hexon mRNAs. Gene expression levels of E1b-55k, pIX, and hexon mRNAs at various times post-infection were determined by qRT-PCR. The data are expressed as the mean values ± SD (n = 4). N.S.; no significant difference.
Protein V determines the replication property of adenovirus
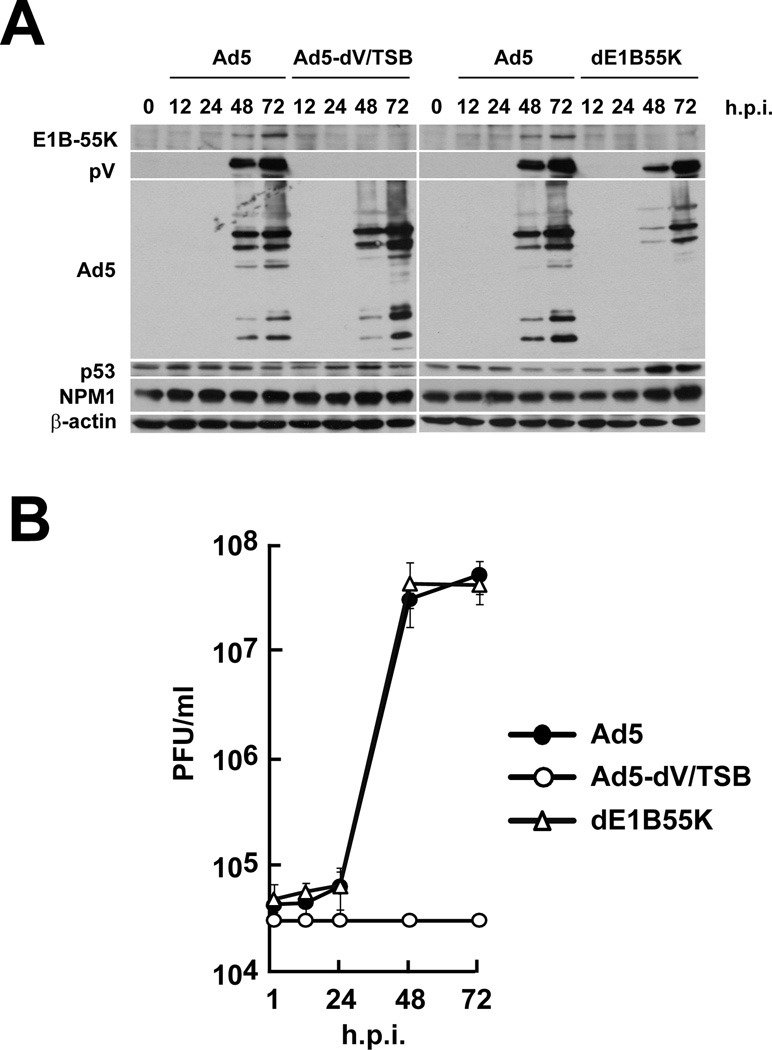
Lack of E1B-55K mediates a defect in late viral RNA export and late viral protein expression in primary cells as shown by a study on the E1B-55K deletion mutant virus, ONYX-015 (dl1520) (O'Shea et al., 2004). However, dl1520 is an E1b-55K gene deletion mutant derived from an E3-deleted derivative virus called dl309 which is a hybrid of Ad2 and Ad5 sequences in the E1 region (Barker and Berk, 1987). Thus, dl1520 has a different genetic background compared to the Ad5 genome used in this study. Therefore, we genetically generated dE1B55K in order to determine whether restriction of Ad5-dV/TSB replication depends on reduced expression of E1B-55K. Deletion of the E1B-55K gene showed a dramatic attenuation of late viral protein expression in dE1B55K-infected HPAEC as compared to Ad5- and Ad5-dV/TSB-infected HPAEC cells (Fig. 4A). Despite lack of the E1B-55K expression and a dramatic attenuation of the late viral protein expression in dE1B55K-infected HAPEC cells (Fig. 4A), dE1B55K replication closely paralleled Ad5 replication in HPAEC cells and had a distinct different profile compared to Ad5-dV/TSB replication (Fig. 4B). Thus, concomitant reduction of E1B55K expression observed in Ad5-dV/TSB-infected cells is not directly responsible for the restriction of Ad5-dV/TSB replication in primary cells. Although we were able to generate dE1B55K-dV/TSB (i.e. deleted for E1B-55K and pV shown in Fig. S1), dE1B55K-dV/TSB virus production was dramatically restricted in A549 cells (see Materials and Methods section). Thus, we were unable to analyze the growth kinetics and protein expression of dE1B55K-dV/TSB. Also, we did not detect attenuated expression of late viral proteins in Ad5-dV/TSB-infected HPAEC cells as similar to that in dE1B55K-infected HPAEC cells (Fig. 4A). Therefore, the lack of pV does not seem to completely shut off E1B-55K expression in Ad5-dV/TSB-infected HPAEC cells. Based on the kinetics of dE1B55K replication in primary cells (Fig. 4B), we conclude that the attenuation of E1B-55K is not a major determinant of the replication property of Ad5-dV/TSB. Collectively, these data illustrate that pV dictates adenoviral replication in normal cells. Also, these data indicate that the lack of pV, rather than the E1B-55K status determines phenotype of Ad5-dV/TSB replication (Fig. 4).
Fig. 4. Protein V determines property of adenoviral replication in primary cells.
HPAEC cells were infected with adenovirus at an MOI of 10 PFU/cell for various times. (A) An equivalent of 40 µg of total proteins was analyzed by Western blot with the indicated antibodies. (B) One-step growth curve kinetics of adenoviruses was analyzed in HPAEC cells. Infectious titer was determined by plaque assay.
NPM1 promotes adenoviral replication
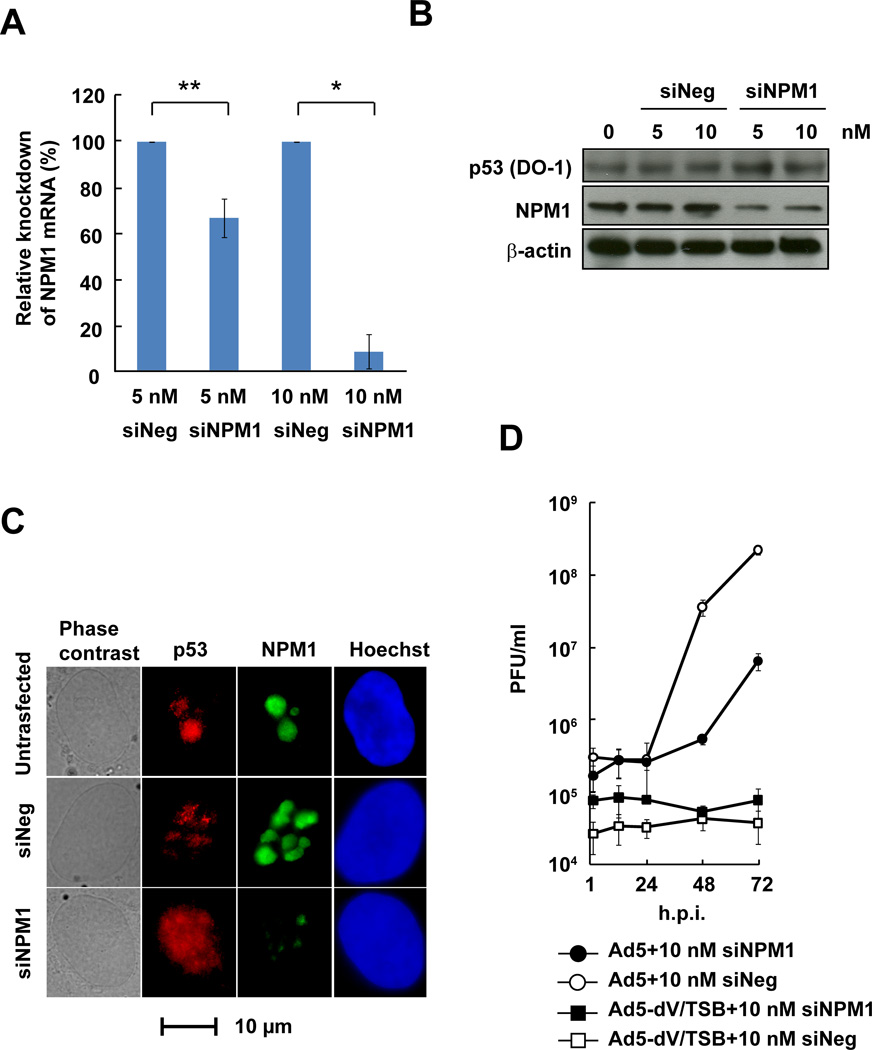
Analysis of Ad5-dV/TSB-infected cells indicates that pV induces NPM1 translocation from the nucleoli to the nucleoplasm (Fig. 1), and transient expression of pV-mRFP1 also triggers NPM1 translocation to the nucleoplasm compared to mock-infected cells (Fig. S6). Therefore, we tested whether NPM1 knockdown inhibits adenovirus replication in primary cells. We observed a reduction of NPM1 mRNA level after NPM1 siRNA treatment (Fig. 5A). Also, Western blot analysis revealed that NPM1 expression was reduced in NPM siRNA-transfected cells compared to control siRNA-transfected cells, but NPM1 knockdown did not alter p53 expression level (Fig. 5B). Immunofluorescence analysis after NPM1 knockdown showed attenuated NPM1 staining in the nucleoli, while p53 appeared to be partially translocated from the nucleoli to the nucleoplasm as compared to the controls (Fig. 5C). As we expected, NPM1 knockdown dramatically inhibited Ad5 replication compared to control siRNA (Fig. 5D). However, low levels of Ad5 replication were observed by 48 h.p.i. in siNPM1-treated HPAEC cells, possibly because adenovirus-associated (VA) RNAs inhibit RNAi machinery (Andersson et al., 2005; de Vries and Berkhout, 2008; Lu and Cullen, 2004; Xu et al., 2007). Also, NPM1 knockdown did not recover Ad5-dV/TSB replication in primary cells (Fig. 5D). Therefore, these results indicate that NPM1 is required for adenoviral replication in primary cells.
Fig. 5. Knockdown of NPM1 inhibits adenoviral replication.
(A and B) HPAEC cells were transfected with siRNA for NPM1 mRNA (siNPM1) or a control non-targeting RNA (siNeg) for 48 hours, followed by qRT-PCR to determine knockdown level of NPM1 mRNA (A); Data was shown as mean ± S.E.M., * P<0.01, ** P<0.05; two-tailed unpaired Student’s t-tests between groups, and Western blot analysis (B). (C) HPAEC cells were transfected with 10 nM siRNA for 48 hours, followed by immunofluorescent staining. (D) One-step growth curve analysis of adenoviruses in siNPM1-treated primary cells. Primary cells transfected with siRNA for 48 hours were infected with adenovirus at an MOI of 10 PFU/cell for various times. Infectious titer was determined by plaque assay.
Ad5-dV/TSB replicates in cancerous cells expressing nucleoplasmic NPM1
Ad5-dV/TSB replicates well in lung carcinoma A549 cells which are deleted the gene for ARF and therefore have a defective p53 pathway (Ugai et al., 2007). In order to understand why pV is not required for replication in malignant cells, we examined the expression and localization of NPM1 in cancer cell lines. NPM1 was delocalized in all cancer cell lines assayed, and overexpressed in all cancer cell lines except BXPC-3 when compared to HPAEC (Fig. S9A and B), confirming previous reports that NPM1 is altered in cancer cells (Grisendi et al., 2006). We further examined whether Ad5-dV/TSB replicates in a range of cancer cell lines and observed Ad5-dV/TSB replication in those cancer cell lines but not in HPAEC cells (Fig. S9C). Also, Ad5-dV/TSB infection was as effective as Ad5 in inducing cell killing in cancer cells (Fig. S9D). Moreover, we confirmed that Ad5-dV/TSB production was observed in a broad range of cancer cell lines, those cancer lines containing nucleoplasmic NPM1 (Fig. S9E–G). Collectively, these data showed that in contrast to primary cells, Ad5-dV/TSB replicates well in a range of cancer cells with readily detectable nucleoplasmic NPM1.
NPM1 interacts with empty adenovirus particles
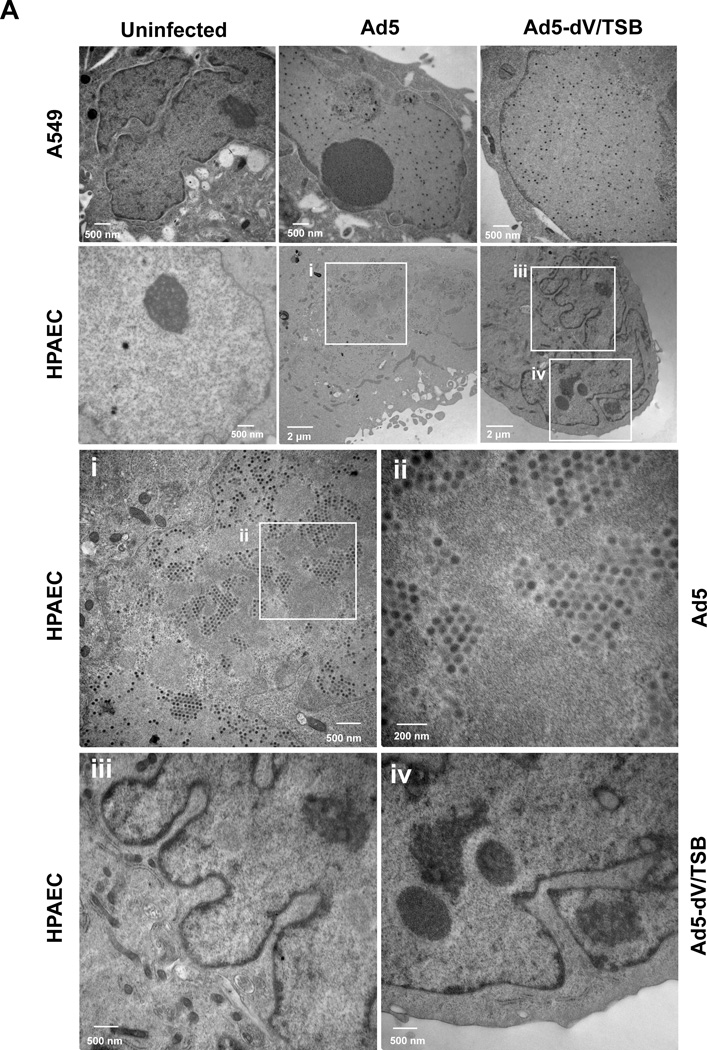
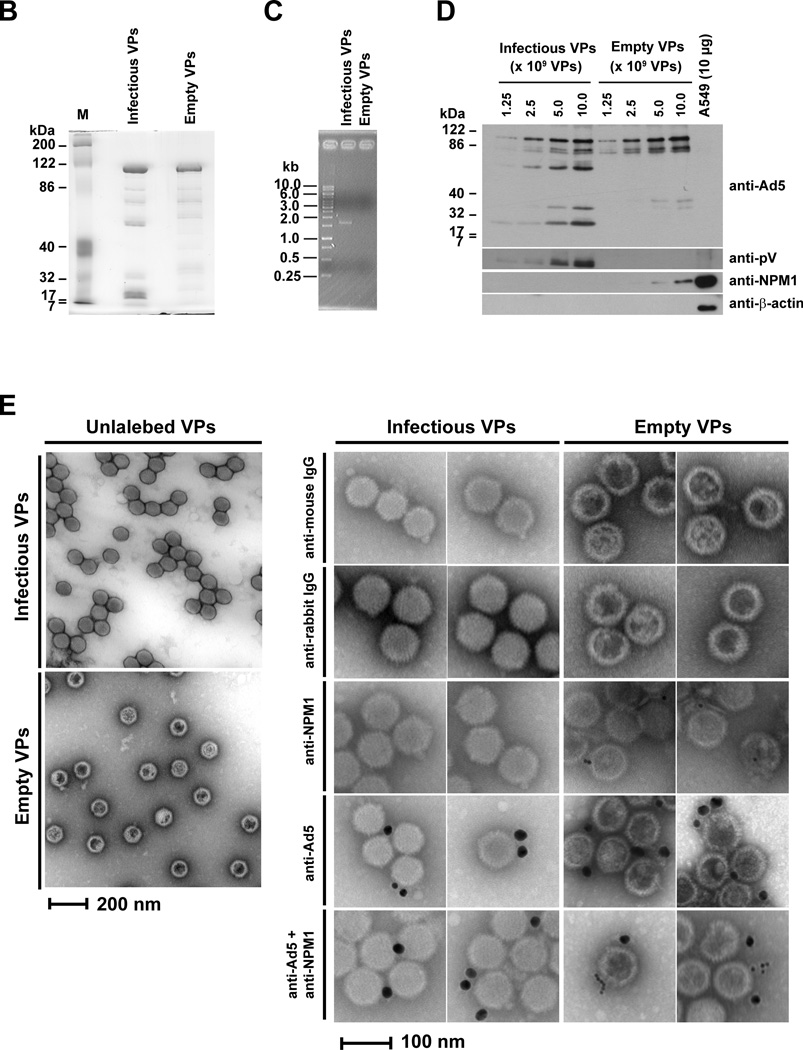
Adenoviral protein V is required for NPM1 redistribution from the nucleoli to the nucleoplasm (Fig. 1), and NPM1 promotes adenoviral replication (Fig. 5). Therefore, we investigated whether NPM1 is recruited to adenoviral replication center. We first examined the location of nucleolar NPM1 during infection by analyzing Ad5-E3-V-EGFP. We tracked fluorescent signals for pV-EGFP and NPM1 under short time of exposure in order to detect newly synthesized pV-EGFP in infected primary cells. Therefore, we did not detect virion-associated pV-EGFP in infected cells at 12 h.p.i., but did detect partial translocation of NPM1 at that time (Fig. S10A, upper panels for 12 h.p.i.). Newly synthesized pV-EGFP was detected at the nucleoli in the majority of infected cells by 12 h.p.i. (Fig. S10A and B), and at this time NPM1 was exclusively associated with pV-EGFP in the nucleoli (Fig. S10A, lower panels for 12 h.p.i.). At 24 h.p.i., pV-EGFP was translocated from the nucleoli to the nucleoplasm and involved in virus-induced specific nuclear structures; in many cases NPM1 was still restricted to the nucleolus (Fig. S10A, upper panels for 24 h.p.i.). However, in other cells at this time point, we observed flecked distribution of pV-EGFP in the nucleoplasm, and in those cases NPM1 localized in the nucleoplasmic pV-EGFP-rich centers (Fig. S10A, lower panels for 24 h.p.i.). The redistribution of pV-EGFP and NPM1 continued during viral replication (Fig. S10A, panel for 36 h.p.i.). Subsequently, NPM1 was also detected in the cytoplasm at 48 h.p.i. and excluded from the nucleus at 72 h.p.i., while pV-EGFP was localized in both the nucleus and the cytoplasm (Fig. S10A). These results agree with a previous report that pV indirectly induces the NPM1 translocation from the nucleoli to the nucleoplasm (Matthews, 2001). Although pV-EGFP was colocalized with NPM1 in the nucleoli and the nucleoplasm (Fig. S10A), we did not detect this association between pV and NPM1 in infected HPAEC and 293 cells by immunoprecipitation analysis (data not shown). We also examined the localization of NPM1 and pIX-mRFP1 in Ad5-wt-IX-mRFP1-infected HPAEC cells, and found that NPM1 was partially colocalized with pIX-mRFP1 in the nucleoplasm of infected HPAEC cells (Fig. S10C). These data indicate that a proportion of NPM1 is present in the same location, as both capsid (pIX) and core (pV) structural proteins accumulate in the nucleoplasm (Figs. S10A and S10C). Collectively, these data imply that nucleoplasmic NPM1 may play a role in adenoviral assembly. Adenoviral mature virions are produced through three intermediates during maturation: empty virus particles, incomplete virus particles, and young virions (Edvardsson et al., 1976; Ishibashi and Maizel, 1974). Protein V is included in young and mature virions, but not empty virus particles and incomplete virus particles (Ishibashi and Maizel, 1974). Our findings prompted us to investigate by transmission electron microscope (TEM) whether assembled Ad5-dV/TSB particles are detected in infected primary cells. While wild type Ad5 particles were readily detected in infected primary cells, the Ad5-dV/TSB particles were not (Fig. 6A). In contrast to primary cells, both wild type Ad5 and Ad5-dV/TSB particles were readily observed in infected A549 cells (Fig. 6A). Thus, TEM analysis showed that Ad5-dV/TSB was defective for assembly in primary cells (Fig. 6A). Because we could not detect Ad5-dV/TSB particles in infected HPAEC cells (Fig. 6A), we next examined whether NPM1 plays a role during viral assembly. Therefore, we purified empty and infectious virus particles from the nuclei of infected A549 cells (Figs. 6B and C) and analyzed whether NPM1 is contained in the fraction of those virus particles. Western blot analysis showed NPM1 was co-fractionated with empty virus particles (Fig. 6D), and immunoelectron microscopic analysis demonstrated that NPM1 is detected around empty virus particles, but not infectious virus particles (Fig. 6E). Taken together, our data suggest that pV is essential for viral assembly in normal cells, but not cancer cells, and that NPM1 interacts with empty virus particles and may be involved in a process of virion maturation.
Fig. 6. NPM1 is involved in empty adenovirus particles.
(A) HPAEC and A549 cells were infected with adenovirus at an MOI of 10 PFU/cell, followed by cell harvesting at 72 h.p.i. Uninfected and infected cells were fixed in 30 mM HEPES-NaOH buffer (pH 7.4) containing 1.25% glutaraldehyde and 2% paraformaldehyde, embedded, and sectioned for transmission electron microscopy. (i – iv) White framed areas display enlarged areas. (B) Analysis of protein composition of viral particles by GELCODE blue staining. Infectious and empty virus particles (VPs) of Ad5 were CsCl-purified from the nuclei fraction of infected A549 cells. A total of 1.0 × 1010 VPs of purified infectious and empty VPs was separated by electrophoresis in 10% SDS-PAGE, and the gel was stained with GELCODE Blue Stain Reagent. (C) Validation of purified infectious and empty VPs by PCR analysis. The fiber gene was amplified using a template which is equivalent to 1.0 × 1010 VPs of purified VPs. (D) Detection of NPM1 in the fraction of the Ad5 empty VPs. Infectious and empty VPs were denatured and separated by electrophoresis in 10% SDS-PAGE, followed by Western blot analysis using indicated antibodies. An equivalent of 10 µg total proteins which were extracted from A549 cells was applied for a control to detect cellular proteins. (E) Immunoelectron microscopic analysis of NPM1 accessibility on empty VPs. To test the accessibility of NPM1 on empty VPs, infectious and empty VPs were bound on copper grids with a carbon-coated Formvar film. The NPM1 epitope was detected with indicated antibodies, followed by donkey anti-mouse immunoglobulin conjugated with 10-nm gold and donkey anti-rabbit immunoglobulin conjugated with 25-nm gold. Electron micrographs for the adenoviral particles were taken with JEOL 1200 EX II Transmission Electron Microscope.
Discussion
In response to cellular stress, p53 is rapidly stabilized and activated by posttranslational modification at serines 15 and 20 mediated through ATM and CHK2, respectively, and CHK2 is itself activated by ATM (Toledo and Wahl, 2006). However, adenoviral proteins such as the E1B-55K/E4ORF6 complex not only degrade p53 (Querido et al., 1997) but also block signaling through degradation of the MRN (Mre11/RAD50/NBS1) complex (Carson et al., 2003) which activates ATM (Lee and Paull, 2005). Also, E1A and E4ORF3 inactivates p53-mediated transactivation (Somasundaram and El-Deiry, 1997) and p53 independently of E1B-55K by preventing p53-DNA binding (Soria et al., 2010), respectively. Thus, degradation of p53 (Querido et al., 1997) and elimination of p53 activity (Soria et al., 2010) are considered essential to maximize adenoviral replication (Levine, 2009). Although adenovirus utilizes the p53 pathway for infection (Sherr and McCormick, 2002), p53 is not stabilized, and its target genes are not activated throughout wild type adenoviral infection (Soria et al., 2010). Therefore, adenoviral mutants which are incapable of antagonizing p53 and/or its activity were considered to be useful for cancer gene therapy (Bischoff et al., 1996; Soria et al., 2010). However, there is no direct proof that p53 and/or its activity inhibit wild type adenoviral replication (Koch et al., 2001). Importantly, adenoviral replication observed by virion maturation is clearly detected in p53-negative cells (Harada and Berk, 1999), and adenoviral assembly does not require p53 and its activity (Koch et al., 2001; Levine, 2009). Thus, adenoviral assembly occurs independently of p53 and its activity (Harada and Berk, 1999; Koch et al., 2001; Levine, 2009).
Adenovirus interacts with and disrupts host nucleoli during viral lytic growth (Lam et al., 2009; Lawrence, McStay, and Matthews, 2006) and disrupts NPM1 nucleolar localization (Matthews, 2001; Walton et al., 1989). Thus, adenovirus infection reorganizes nucleolar NPM1 for productive infection. Our comparative experiments with Ad5-dV/TSB revealed that pV is essential for disrupting the delocalization of nucleolar NPM1 during infection (Fig. 1), and this redistribution of NPM1 was correlated with adenoviral replication (Figs. 1, 2, 4, and S10). Also, our result revealed that the pV translocation from the nucleoli to the nucleoplasm coincided with the delocalization of nucleolar NPM1 during infection (Figs. 1 and S10). Therefore, NPM1 is considered to be associated with adenoviral replication.
NPM1 colocalizes with pre-terminal protein (pTP) and DNA-binding protein (DBP) involved in adenoviral DNA replication during infection (Hindley, Davidson, and Matthews, 2007), and a cellular fraction containing NPM1 which was purified from HeLa cells stably expressing HPV E6 and E7 proteins stimulates adenoviral DNA replication in vitro (Okuwaki et al., 2001). Although NPM1 appeared to associate with adenoviral DNA replication, recombinant protein NPM1 did not enhance adenoviral DNA replication in vitro (Okuwaki et al., 2001). On the other hand, HPV E6 and E7 proteins enhance adenoviral DNA replication in vitro and in vivo (Steinwaerder, Carlson, and Lieber, 2001). Therefore, the contribution of NPM1 to enhancement of adenoviral DNA replication observed in vitro still remains to be elucidated. Importantly, NPM1 seemed to be dispensable for adenoviral DNA replication, because Ad5-dV/TSB DNA replication was observed at an absence of nucleoplasmic NPM1 in infected HPAEC cells (Figs. 1 and 3A). In our result, NPM1 knockdown showed that it promoted adenoviral replication (Fig. 4), suggesting an involvement of NPM1 in a process of adenoviral replication. Although NPM1 phosphorylation, involved in its relocalization in response to cellular and viral stresses, is not observed (Kurki et al., 2004), significantly, we have identified a viral factor, pV, to trigger the NPM1 delocalization in infected normal cells by comparing with Ad5-dV/TSB.
Adenoviral E1A gene products activate the E1b promoter (Imperiale, Feldman, and Nevins, 1983) as well as other early promoters (Nevins, 1987). Since E1A was abundantly expressed in Ad5-dV/TSB-infected primary cells by 24 h.p.i. (Fig. 3B), it seems to activate the E1b promoter. NPM1 is selectively and specifically deposited on E1b mRNA during its 3’-end processing in vitro (Palaniswamy et al., 2006), and a reduction of NPM1 expression by knockdown causes accumulation of E1b mRNA in the nucleus in vitro (Sagawa et al., 2011). Accordingly, an absence of NPM1 observed in the nucleoplasm of Ad5-dV/TSB-infected primary cells (Fig. 1) seems to result in accumulation of E1b-55k and pIX mRNAs in the nucleoplasm. Thus, we might detect attenuated expression of E1B-55K and pIX-mRFP1 in Ad5-dV/TSB-infected cells (Figs. 3B and 4A, and S8).
We have previously reported that pIX was similarly incorporated in purified Ad5-dV/TSB as well as purified Ad5 (Ugai et al., 2007). The pIX gene is deleted from the Ad5 genome and the lack of pIX does not affect its viral replication (Sargent, Meulenbroek, and Parks, 2004). On the other hand, our experiments with several adenoviral mutants revealed that attenuation of the E1B-55K expression mediated by lack of pV is not responsible for the Ad5-dV/TSB replication in primary cells (Fig. 3). Attenuation of the E1B-55K expression may induce p53 stabilization in Ad5-dV/TSB-infected primary cells as observed in dl1520-infected primary cells (O'Shea et al., 2004), and p53 stabilization may suppress Ad5-dV/TSB replication. However, active and non-degradable p53 does not inhibit adenoviral replication (Koch et al., 2001) and overexpression of p53 mediated by a p53-recombinant adenovirus does not impair its production (Sauthoff et al., 2002). Therefore, p53 stabilization does not seem to restrict Ad5-dV/TSB replication in primary cells. Collectively, lack of pV, but not p53 stabilization, E1B-55K and pIX attenuation, resulted in restriction of Ad5-dV/TSB replication in primary cells.
NPM1 is apparently localized to the nucleoplasm during infection (Hindley, Davidson, and Matthews, 2007; Walton et al., 1989) and is likely to be relocalized from the nucleoli to the nuclear matrix to form pseudonucleoli (Walton et al., 1989) nucleated around adenoviral genomes (Bodnar et al., 1989; Schaack et al., 1990), mRNA (Gallinaro et al., 1983), and proteins (Hindley, Davidson, and Matthews, 2007). We also observed that NPM1 was reorganized from the nucleoli to the nucleoplasm and colocalized with capsid and core proteins, pIX and pV, during productive infection (Figs. 1, S10A, and S10C). Therefore, our results suggested that NPM1 was involved in an event of adenoviral assembly. The average molecular mass of empty adenovirus particles is approximately 123 MDa (Tibbetts and Giam, 1979). On the other hand, NPM1 is present as monomeric and hexameric forms in cells, and the molecular weights of those forms are approximately 37 kDa and 230 kDa, respectively (Yung and Chan, 1987). The sedimentation rates (550–650S) of adenoviral intermediates are different from that (10S) of NPM1 purified from cells (Ostapchuk and Hearing, 2005; Yung and Chan, 1987). Thus, our data demonstrated that NPM1 is co-purified with empty adenovirus particles from infected cells and specifically interacts with empty adenovirus particles (Fig. 6D and E). To our knowledge, NPM1 is the first cellular protein shown to be integrated into empty adenovirus particles. We detected the Ad5-dV/TSB viral particles in infected cancer cells, but did not detect them in infected HPAEC cells (Fig. 6A). Also, NPM1 knockdown inhibited production of infectious Ad5 progeny (Fig. 5D). Therefore, our data suggest that NPM1 is required for promoting assembly of capsid proteins. Additionally, our results suggested that NPM1 was released from infectious viral particles after virion maturation, because it was not incorporated in purified infectious viral particles (Fig. 6D and E). Therefore, NPM1 seems to play a role in capsid formation during virion maturation.
Our study is the first to show that adenoviral minor core protein V mediates the NPM1 translocation from the nucleoli to the nucleoplasm during productive infection in normal cells and that this event is required for productive infection in normal cells. Moreover, we propose that as infection progresses a pV-mediated NPM1 redistribution is associated with capsid formation during virion maturation. Therefore, the pV-mediated build-up of nucleoplasmic NPM1 is essential for adenoviral replication in normal cells. Also, lack of pV seems to function as a novel determinant for the selective replication between cancer and normal cells. Our data shows that pV-deleted adenoviruses may offer a novel approach to the development of oncolytic adenoviruses for cancer therapy that is distinct from current approaches through E1A and E1B-55K.
Supplementary Material
(A) Schematic representation of the core protein gene V region in the adenovirus genomes. The genome of Ad5-dV/TSB was created by deleting nucleotides at positions 16,545–17,651 of Ad5 and incorporating suppressor mutations at 17,714 (G to A; silent), 17,716 (G to A; G13E) and 17,728 (G to T; R17I) in the precursor region of polypeptide X (Mu). Position 1 of the nucleotide sequence refers to the left end of the Ad5 genome (Genbank ID: AY339865), for the precursor X. mu; map units, VII; polypeptide VII, V; polypeptide V, X; polypeptide X (Mu), VI; polypeptide VI, HX; hexon, L2 and L3; major late transcript units. (B) Schematic representation of the physical map of Ad5 and the adenoviral mutants used in this study. TP; terminal protein, E1; early gene 1, IX; polypeptide IX, mRFP1; monomeric red fluorescent protein 1, EGFP; enhanced green fluorescent protein, E3; early region 3, E1b19k; the gene for early gene 1b (E1b) 19 kilodalton protein, dE1; the E1 deletion, dV; the pV deletion, and dE3; the E3 deletion.
293 cells were infected with Ad5 or Ad5-dV/TSB at an MOI of 10 PFU/cell for various times. One-step growth curve analysis of adenoviruses was performed, and infectious titer was determined by plaque assay.
(A) After medium was removed from HPAEC cells, cells were washed with 30 mM HEPES-NaOH buffer (pH 7.4) and fixed with fixation solution. HPAEC cells were stained with anti-NPM1 or anti-p53 followed by a secondary antibody conjugated with fluorescent probe. (B) After medium was removed from HPAEC and MCF-7 cells, cells were directly fixed with fixation solution without washing. Subcellular localization of NPM1 and p53 was determined by immunofluorescent staining. Hoechst 33342 staining was performed for 10 min.
(A) Detection of the fluorescently labeled adenoviral particles in infected HPAEC cells. HPAEC cells were infected with Ad5-E3-V-EGFP at MOIs 10, 100 or 1,000 PFU/cell for 1 hour. The trafficking of virion-associated core pV-EGFP was monitored in infected cells at 1 h.p.i. Uninfected and infected HPAEC cells were stained with anti-NPM1 followed by a secondary antibody conjugated with fluorescent probe. (B) Localization of NPM1-EGFP in 293 cells stably expressing NPM1-EGFP. Hoechst 33342 staining was performed for 10 min, and the signals for NPM1-EGFP and DNA staining were detected by fluorescence microscopy.
(A) Crude proteins were extracted from HUVEC, HPAEC, and lung carcinoma A549 cells, and 10 µg of total proteins were separated on 10% (w/v) SDS-PAGE, and NPM1 expression level was validated by Western blot analysis. (B) DNA replication of Ad5-dV/TSB in cancer cells. A549 cells were infected with Ad5 or Ad5-dV/TSB at an MOI of 10 PFU/cell, and harvested at various times post-infection. Total DNA was extracted from infected cells, and viral DNA replication was analyzed by using 50 ng of total DNA and E4 primers. Plots represent as the copy number of the E4 gene per 50 ng of total DNA.
(A) HPAEC was infected with pV-mRFP1-lentivirus at an MOI of 50 proviral DNA/cell for 60 hours. Subcellular localization of NPM1 and p53 was detected in HPAEC infected with mock (LV-empty) or pV-recombinant lentivirus (LV-V). (B) Western blot analysis of pV expression in pV-lentivirus-infected HPAEC cells. (C) HPAEC cells were infected with the pV-recombinant lentivirus at an MOI of 2, 10 or 50 proviral DNA/cell for 60 hours. The pV-transduced cells were infected with adenovirus at an MOI of 10 PFU/cell, and harvested at 1 and 48 h.p.i. Production level of infectious adenovirus was measured by plaque assay. Results were reported as mean ± SD. * P<0.01, ** P<0.05; two-tailed unpaired Student’s t-tests between groups. N.S.; no significance.
HPAEC cells were infected with adenovirus at an MOI of 10 PFU/cell for various times, followed by Western blot analysis. Proteins extracted from infected cells at various time points were subjected to Western blot, and 40 µg of total proteins was analyzed with the indicated antibodies. Lysates extracted from DU145 cells which lowly express p14ARF were used as a positive control for detecting tumor suppressor p14ARF.
Primary (A) and A549 (B) cells were infected with Ad5-wt-IX-mRFP1 or Ad5-dV/TSB-IX-mRFP1 at an MOI of 10 PFU/cell for various times post-infection. Signal for pIX-mRFP1 was detected by fluorescence microscopy.
(A) Proteins extracted from cancer or primary cell lines were validated by Western blot analysis with the indicated antibodies. (B) Subcellular localization of endogenous NPM1 in cancer and primary cells was determined by immunofluorescent staining. (C) One-step growth curve analysis of Ad5-dV/TSB in infected cancer cells. Cancer cells were infected with Ad5 or Ad5-dV/TSB at an MOI of 10 PFU/cell for various times, and infectious adenoviruses were titrated by plaque assay. (D) Cancer cells were infected with Ad5 or Ad5-dV/TSB at an MOI of 10 PFU/cell for 4 days, and tumoricidal effect induced by adenovirus was observed by microscopy. Bar = 200 µm. (E) Comparison of endogenous NPM1 expression profiles in cancer cell lines. An equivalent of 10 µg of total protein extracted from cancer and HPAEC cells was separated on 10% (w/v) SDS-PAGE, and NPM1 expression level was validated by Western blot analysis. (F) Abnormality of NPM1 subcellular localization in a broad range of cancer cell lines. Subcellular localization of endogenous NPM1 in cancer and primary cells was visualized by staining with anti-NPM1 followed by secondary antibody conjugated with a fluorescent probe. Cells were stained for nuclear DNA using Hoechst 33342 followed by visualization of the fluorescent probe for NPM1 detection by epifluorescence microscopy. (G) Cancer cells were infected with adenovirus at an MOI 10 PFU/cell for 1 and 48 hours, and infected cells were harvested. Production level of infectious adenovirus was measured by plaque assay. Results were reported as mean ± SD. Black bar; Ad5, white bar; Ad5-dV/TSB.
(A) HPAEC cells were infected with Ad5-E3-V-EGFP at an MOI of 10 PFU/cell for various times post-infection. NPM1 was visualized by staining with anti-NPM1 followed by secondary antibody conjugated with a fluorescent probe. Uninfected and infected cells were stained for nuclear DNA using Hoechst 33342, and localization of pV-EGFP and NPM1 was detected by fluorescence microscopy. (B) Viral growth kinetics of Ad5-E3-V-EGFP in primary cells. HPAEC cells were infected with Ad5 or Ad5-E3-V-EGFP at an MOI of 10 PFU/cell for various times post-infection. Infectious titer of adenovirus was determined by plaque assay. (C) HPAEC cells were infected with Ad5-wt-IX-mRFP1 at an MOI of 10 PFU/cell for various times post-infection. NPM1 was visualized by staining with anti-NPM1 followed by secondary antibody conjugated with a fluorescent probe. Uninfected and infected cells were stained for nuclear DNA using Hoechst 33342, and fluorescent signals for pIX-mRFP1 and NPM1 were detected by fluorescence microscopy. pIX-mRFP1 was initially expressed in the cytoplasm and accumulated in the nucleus for virus assembly at the late stage of infection, and the cytoplasm of infected cells was undetectable because infected cells shrank during the late stage of infection as previous reported by Ugai et al. (Ugai et al., 2010). Arrows show colocalization of pIX-mRFP1 and NPM1.
Acknowledgements
We thank Dr. Roger Y. Tsien (University of California at San Diego) for providing the mRFP1 construct, and Drs A. J. Levine (The Cancer Institute of New Jersey), W. S. M. Wold (St. Louis University), T. Shenk (Princeton University) for providing antibodies against adenoviral proteins. We are also grateful to Drs. Melissa F. Chimento and Olga Borkhsenious for technical support with transmission electron microscopy at the High Resolution Imaging Facility in University of Alabama at Birmingham and School of Veterinary Medicine Microscopy Center in Louisiana State University, respectively. This work was supported by grants from Susan G. Komen for the Cure PDF0707736 (Dr. Hideyo Ugai), Susan G. Komen for the Cure KG100194 (Drs. David T. Curiel and Hideyo Ugai), Grant number 083604 from the Wellcome Trust (Dr. David A. Matthews), grant T32-NS048039 from the National Institutes of Health (Dr. George C. Dobbins), and grant R01CA121187 from the National Institutes of Health (Dr. David T. Curiel).
References
- Anderson CW, Young ME, Flint SJ. Characterization of the adenovirus 2 virion protein, mu. Virology. 1989;172(2):506–512. doi: 10.1016/0042-6822(89)90193-1. [DOI] [PubMed] [Google Scholar]
- Andersson MG, Haasnoot PC, Xu N, Berenjian S, Berkhout B, Akusjarvi G. Suppression of RNA interference by adenovirus virus-associated RNA. J Virol. 2005;79(15):9556–9565. doi: 10.1128/JVI.79.15.9556-9565.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barker DD, Berk AJ. Adenovirus proteins from both E1B reading frames are required for transformation of rodent cells by viral infection and DNA transfection. Virology. 1987;156(1):107–121. doi: 10.1016/0042-6822(87)90441-7. [DOI] [PubMed] [Google Scholar]
- Bischoff JR, Kirn DH, Williams A, Heise C, Horn S, Muna M, Ng L, Nye JA, Sampson-Johannes A, Fattaey A, McCormick F. An adenovirus mutant that replicates selectively in p53-deficient human tumor cells. Science. 1996;274(5286):373–376. doi: 10.1126/science.274.5286.373. [DOI] [PubMed] [Google Scholar]
- Bodnar JW, Hanson PI, Polvino-Bodnar M, Zempsky W, Ward DC. The terminal regions of adenovirus and minute virus of mice DNAs are preferentially associated with the nuclear matrix in infected cells. J Virol. 1989;63(10):4344–4353. doi: 10.1128/jvi.63.10.4344-4353.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carson CT, Schwartz RA, Stracker TH, Lilley CE, Lee DV, Weitzman MD. The Mre11 complex is required for ATM activation and the G2/M checkpoint. Embo J. 2003;22(24):6610–6620. doi: 10.1093/emboj/cdg630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chatterjee PK, Vayda ME, Flint SJ. Interactions among the three adenovirus core proteins. J Virol. 1985;55(2):379–386. doi: 10.1128/jvi.55.2.379-386.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chatterjee PK, Vayda ME, Flint SJ. Identification of proteins and protein domains that contact DNA within adenovirus nucleoprotein cores by ultraviolet light crosslinking of oligonucleotides 32P-labelled in vivo. J Mol Biol. 1986;188(1):23–37. doi: 10.1016/0022-2836(86)90477-8. [DOI] [PubMed] [Google Scholar]
- Colombo E, Marine JC, Danovi D, Falini B, Pelicci PG. Nucleophosmin regulates the stability and transcriptional activity of p53. Nat Cell Biol. 2002;4(7):529–533. doi: 10.1038/ncb814. [DOI] [PubMed] [Google Scholar]
- Dalenc F, Drouet J, Ader I, Delmas C, Rochaix P, Favre G, Cohen-Jonathan E, Toulas C. Increased expression of a COOH-truncated nucleophosmin resulting from alternative splicing is associated with cellular resistance to ionizing radiation in HeLa cells. Int J Cancer. 2002;100(6):662–668. doi: 10.1002/ijc.10558. [DOI] [PubMed] [Google Scholar]
- Davison AJ, Benko M, Harrach B. Genetic content and evolution of adenoviruses. J Gen Virol. 2003;84(Pt 11):2895–2908. doi: 10.1099/vir.0.19497-0. [DOI] [PubMed] [Google Scholar]
- de Vries W, Berkhout B. RNAi suppressors encoded by pathogenic human viruses. Int J Biochem Cell Biol. 2008;40(10):2007–2012. doi: 10.1016/j.biocel.2008.04.015. [DOI] [PubMed] [Google Scholar]
- Delenda C, Gaillard C. Real-time quantitative PCR for the design of lentiviral vector analytical assays. Gene Ther. 2005;12(Suppl 1):S36–S50. doi: 10.1038/sj.gt.3302614. [DOI] [PubMed] [Google Scholar]
- Edvardsson B, Everitt E, Jornvall H, Prage L, Philipson L. Intermediates in adenovirus assembly. J Virol. 1976;19(2):533–547. doi: 10.1128/jvi.19.2.533-547.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukuda K, Abei M, Ugai H, Seo E, Wakayama M, Murata T, Todoroki T, Tanaka N, Hamada H, Yokoyama KK. E1A, E1B double-restricted adenovirus for oncolytic gene therapy of gallbladder cancer. Cancer Res. 2003;63(15):4434–4440. [PubMed] [Google Scholar]
- Gallinaro H, Puvion E, Kister L, Jacob M. Nuclear matrix and hnRNP share a common structural constituent associated with premessenger RNA. EMBO J. 1983;2(6):953–960. doi: 10.1002/j.1460-2075.1983.tb01527.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grisendi S, Mecucci C, Falini B, Pandolfi PP. Nucleophosmin and cancer. Nat Rev Cancer. 2006;6(7):493–505. doi: 10.1038/nrc1885. [DOI] [PubMed] [Google Scholar]
- Harada JN, Berk AJ. p53-Independent and -dependent requirements for E1B-55K in adenovirus type 5 replication. J Virol. 1999;73(7):5333–5344. doi: 10.1128/jvi.73.7.5333-5344.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herrera JE, Savkur R, Olson MO. The ribonuclease activity of nucleolar protein B23. Nucleic Acids Res. 1995;23(19):3974–3979. doi: 10.1093/nar/23.19.3974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hindley CE, Davidson AD, Matthews DA. Relationship between adenovirus DNA replication proteins and nucleolar proteins B23.1 and B23.2. J Gen Virol. 2007;88(Pt 12):3244–3248. doi: 10.1099/vir.0.83196-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hingorani K, Szebeni A, Olson MO. Mapping the functional domains of nucleolar protein B23. J Biol Chem. 2000;275(32):24451–24457. doi: 10.1074/jbc.M003278200. [DOI] [PubMed] [Google Scholar]
- Hiscox JA. The nucleolus--a gateway to viral infection? Arch Virol. 2002;147(6):1077–1089. doi: 10.1007/s00705-001-0792-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imperiale MJ, Feldman LT, Nevins JR. Activation of gene expression by adenovirus and herpesvirus regulatory genes acting in trans and by a cis-acting adenovirus enhancer element. Cell. 1983;35(1):127–136. doi: 10.1016/0092-8674(83)90215-5. [DOI] [PubMed] [Google Scholar]
- Ishibashi M, Maizel JV., Jr The polypeptides of adenovirusVYoung virions, structural intermediate between top components and aged virions. Virology. 1974;57(2):409–424. doi: 10.1016/0042-6822(74)90181-0. [DOI] [PubMed] [Google Scholar]
- Itahana K, Bhat KP, Jin A, Itahana Y, Hawke D, Kobayashi R, Zhang Y. Tumor suppressor ARF degrades B23, a nucleolar protein involved in ribosome biogenesis and cell proliferation. Mol Cell. 2003;12(5):1151–1164. doi: 10.1016/s1097-2765(03)00431-3. [DOI] [PubMed] [Google Scholar]
- Karni-Schmidt O, Friedler A, Zupnick A, McKinney K, Mattia M, Beckerman R, Bouvet P, Sheetz M, Fersht A, Prives C. Energy-dependent nucleolar localization of p53 in vitro requires two discrete regions within the p53 carboxyl terminus. Oncogene. 2007;26(26):3878–3891. doi: 10.1038/sj.onc.1210162. [DOI] [PubMed] [Google Scholar]
- Koch P, Gatfield J, Lober C, Hobom U, Lenz-Stoppler C, Roth J, Dobbelstein M. Efficient replication of adenovirus despite the overexpression of active and nondegradable p53. Cancer Res. 2001;61(15):5941–5947. [PubMed] [Google Scholar]
- Kurki S, Peltonen K, Latonen L, Kiviharju TM, Ojala PM, Meek D, Laiho M. Nucleolar protein NPM interacts with HDM2 and protects tumor suppressor protein p53 from HDM2-mediated degradation. Cancer Cell. 2004;5(5):465–475. doi: 10.1016/s1535-6108(04)00110-2. [DOI] [PubMed] [Google Scholar]
- Lam YW, Evans VC, Heesom KJ, Lamond AI, Matthews DA. Proteomic analysis of the nucleolus in adenovirus-infected cells. Mol Cell Proteomics. 2009 doi: 10.1074/mcp.M900338-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawrence FJ, McStay B, Matthews DA. Nucleolar protein upstream binding factor is sequestered into adenovirus DNA replication centres during infection without affecting RNA polymerase I location or ablating rRNA synthesis. J Cell Sci. 2006;119(Pt 12):2621–2631. doi: 10.1242/jcs.02982. [DOI] [PubMed] [Google Scholar]
- Le LP, Le HN, Dmitriev IP, Davydova JG, Gavrikova T, Yamamoto S, Curiel DT, Yamamoto M. Dynamic monitoring of oncolytic adenovirus in vivo by genetic capsid labeling. J Natl Cancer Inst. 2006a;98(3):203–214. doi: 10.1093/jnci/djj022. [DOI] [PubMed] [Google Scholar]
- Le LP, Le HN, Nelson AR, Matthews DA, Yamamoto M, Curiel DT. Core labeling of adenovirus with EGFP. Virology. 2006b;351(2):291–302. doi: 10.1016/j.virol.2006.03.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Moullec JM, Akusjarvi G, Stalhandske P, Pettersson U, Chambraud B, Gilardi P, Nasri M, Perricaudet M. Polyadenylic acid addition sites in the adenovirus type 2 major late transcription unit. J Virol. 1983;48(1):127–134. doi: 10.1128/jvi.48.1.127-134.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee C, Smith BA, Bandyopadhyay K, Gjerset RA. DNA damage disrupts the p14ARF-B23(nucleophosmin) interaction and triggers a transient subnuclear redistribution of p14ARF. Cancer Res. 2005;65(21):9834–9842. doi: 10.1158/0008-5472.CAN-05-1759. [DOI] [PubMed] [Google Scholar]
- Lee JH, Paull TT. ATM activation by DNA double-strand breaks through the Mre11-Rad50-Nbs1 complex. Science. 2005;308(5721):551–554. doi: 10.1126/science.1108297. [DOI] [PubMed] [Google Scholar]
- Lee TW, Lawrence FJ, Dauksaite V, Akusjarvi G, Blair GE, Matthews DA. Precursor of human adenovirus core polypeptide Mu targets the nucleolus and modulates the expression of E2 proteins. J Gen Virol. 2004;85(Pt 1):185–196. doi: 10.1099/vir.0.19352-0. [DOI] [PubMed] [Google Scholar]
- Lehmberg E, Traina JA, Chakel JA, Chang RJ, Parkman M, McCaman MT, Murakami PK, Lahidji V, Nelson JW, Hancock WS, Nestaas E, Pungor E., Jr Reversed-phase high-performance liquid chromatographic assay for the adenovirus type 5 proteome. J Chromatogr B Biomed Sci Appl. 1999;732(2):411–423. doi: 10.1016/s0378-4347(99)00316-3. [DOI] [PubMed] [Google Scholar]
- Levine AJ. The common mechanisms of transformation by the small DNA tumor viruses: The inactivation of tumor suppressor gene products: p53. Virology. 2009;384(2):285–293. doi: 10.1016/j.virol.2008.09.034. [DOI] [PubMed] [Google Scholar]
- Lu S, Cullen BR. Adenovirus VA1 noncoding RNA can inhibit small interfering RNA and MicroRNA biogenesis. J Virol. 2004;78(23):12868–12876. doi: 10.1128/JVI.78.23.12868-12876.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mattern KA, Humbel BM, Muijsers AO, de Jong L, van Driel R. hnRNP proteins and B23 are the major proteins of the internal nuclear matrix of HeLa S3 cells. J Cell Biochem. 1996;62(2):275–289. doi: 10.1002/(sici)1097-4644(199608)62:2<275::aid-jcb15>3.0.co;2-k. [DOI] [PubMed] [Google Scholar]
- Matthews DA. Adenovirus protein V induces redistribution of nucleolin and B23 from nucleolus to cytoplasm. J Virol. 2001;75(2):1031–1038. doi: 10.1128/JVI.75.2.1031-1038.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matthews DA, Russell WC. Adenovirus core protein V is delivered by the invading virus to the nucleus of the infected cell and later in infection is associated with nucleoli. J Gen Virol. 1998;79(Pt 7):1671–1675. doi: 10.1099/0022-1317-79-7-1671. [DOI] [PubMed] [Google Scholar]
- Nevins JR. Regulation of early adenovirus gene expression. Microbiol Rev. 1987;51(4):419–430. doi: 10.1128/mr.51.4.419-430.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nozawa Y, Van Belzen N, Van der Made AC, Dinjens WN, Bosman FT. Expression of nucleophosmin/B23 in normal and neoplastic colorectal mucosa. J Pathol. 1996;178(1):48–52. doi: 10.1002/(SICI)1096-9896(199601)178:1<48::AID-PATH432>3.0.CO;2-Y. [DOI] [PubMed] [Google Scholar]
- O'Shea CC, Johnson L, Bagus B, Choi S, Nicholas C, Shen A, Boyle L, Pandey K, Soria C, Kunich J, Shen Y, Habets G, Ginzinger D, McCormick F. Late viral RNA export, rather than p53 inactivation, determines ONYX-015 tumor selectivity. Cancer Cell. 2004;6(6):611–623. doi: 10.1016/j.ccr.2004.11.012. [DOI] [PubMed] [Google Scholar]
- Okuwaki M, Iwamatsu A, Tsujimoto M, Nagata K. Identification of nucleophosmin/B23, an acidic nucleolar protein, as a stimulatory factor for in vitro replication of adenovirus DNA complexed with viral basic core proteins. J Mol Biol. 2001;311(1):41–55. doi: 10.1006/jmbi.2001.4812. [DOI] [PubMed] [Google Scholar]
- Ostapchuk P, Hearing P. Control of adenovirus packaging. J Cell Biochem. 2005;96(1):25–35. doi: 10.1002/jcb.20523. [DOI] [PubMed] [Google Scholar]
- Palaniswamy V, Moraes KC, Wilusz CJ, Wilusz J. Nucleophosmin is selectively deposited on mRNA during polyadenylation. Nat Struct Mol Biol. 2006;13(5):429–435. doi: 10.1038/nsmb1080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Querido E, Marcellus RC, Lai A, Charbonneau R, Teodoro JG, Ketner G, Branton PE. Regulation of p53 levels by the E1B 55-kilodalton protein and E4orf6 in adenovirus-infected cells. J Virol. 1997;71(5):3788–3798. doi: 10.1128/jvi.71.5.3788-3798.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubbi CP, Milner J. Disruption of the nucleolus mediates stabilization of p53 in response to DNA damage and other stresses. Embo J. 2003;22(22):6068–6077. doi: 10.1093/emboj/cdg579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sagawa F, Ibrahim H, Morrison AL, Wilusz CJ, Wilusz J. Nucleophosmin deposition during mRNA 3' end processing influences poly(A) tail length. Embo J. 2011 doi: 10.1038/emboj.2011.272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saito I, Oya Y, Yamamoto K, Yuasa T, Shimojo H. Construction of nondefective adenovirus type 5 bearing a 2.8-kilobase hepatitis B virus DNA near the right end of its genome. J Virol. 1985;54(3):711–719. doi: 10.1128/jvi.54.3.711-719.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sargent KL, Meulenbroek RA, Parks RJ. Activation of adenoviral gene expression by protein IX is not required for efficient virus replication. J Virol. 2004;78(10):5032–5037. doi: 10.1128/JVI.78.10.5032-5037.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sastry L, Johnson T, Hobson MJ, Smucker B, Cornetta K. Titering lentiviral vectors: comparison of DNA RNA and marker expression methods. Gene Ther. 2002;9(17):1155–1162. doi: 10.1038/sj.gt.3301731. [DOI] [PubMed] [Google Scholar]
- Sauthoff H, Pipiya T, Heitner S, Chen S, Norman RG, Rom WN, Hay JG. Late expression of p53 from a replicating adenovirus improves tumor cell killing and is more tumor cell specific than expression of the adenoviral death protein. Hum Gene Ther. 2002;13(15):1859–1871. doi: 10.1089/104303402760372954. [DOI] [PubMed] [Google Scholar]
- Sautkina EN, Potapenko NA, Bulycheva TI, Vladimirova NM. [Isolation of the protein B23/nucleophosmin from HeLa cell nuclei] Prikl Biokhim Mikrobiol. 2008;44(3):287–295. [PubMed] [Google Scholar]
- Savkur RS, Olson MO. Preferential cleavage in pre-ribosomal RNA byprotein B23 endoribonuclease. Nucleic Acids Res. 1998;26(19):4508–4515. doi: 10.1093/nar/26.19.4508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaack J, Ho WY, Freimuth P, Shenk T. Adenovirus terminal protein mediates both nuclear matrix association and efficient transcription of adenovirus DNA. Genes Dev. 1990;4(7):1197–1208. doi: 10.1101/gad.4.7.1197. [DOI] [PubMed] [Google Scholar]
- Sherr CJ. Divorcing ARF and p53: an unsettled case. Nat Rev Cancer. 2006;6(9):663–673. doi: 10.1038/nrc1954. [DOI] [PubMed] [Google Scholar]
- Sherr CJ, McCormick F. The RB and p53 pathways in cancer. Cancer Cell. 2002;2(2):103–112. doi: 10.1016/s1535-6108(02)00102-2. [DOI] [PubMed] [Google Scholar]
- Somasundaram K, El-Deiry WS. Inhibition of p53-mediated transactivation and cell cycle arrest by E1A through its p300/CBP-interacting region. Oncogene. 1997;14(9):1047–1057. doi: 10.1038/sj.onc.1201002. [DOI] [PubMed] [Google Scholar]
- Soria C, Estermann FE, Espantman KC, O'Shea CC. Heterochromatin silencing of p53 target genes by a small viral protein. Nature. 2010;466(7310):1076–1081. doi: 10.1038/nature09307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steinwaerder DS, Carlson CA, Lieber A. Human papilloma virus E6 and E7 proteins support DNA replication of adenoviruses deleted for the E1A and E1B genes. Mol Ther. 2001;4(3):211–216. doi: 10.1006/mthe.2001.0447. [DOI] [PubMed] [Google Scholar]
- Subong EN, Shue MJ, Epstein JI, Briggman JV, Chan PK, Partin AW. Monoclonal antibody to prostate cancer nuclear matrix protein (PRO: 4-216) recognizes nucleophosmin/B23. Prostate. 1999;39(4):298–304. doi: 10.1002/(sici)1097-0045(19990601)39:4<298::aid-pros11>3.0.co;2-m. [DOI] [PubMed] [Google Scholar]
- Tibbetts C, Giam CZ. In vitro association of empty adenovirus capsids with double-stranded DNA. J Virol. 1979;32(3):995–1005. doi: 10.1128/jvi.32.3.995-1005.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toledo F, Wahl GM. Regulating the p53 pathway: in vitro hypotheses, in vivo veritas. Nat Rev Cancer. 2006;6(12):909–923. doi: 10.1038/nrc2012. [DOI] [PubMed] [Google Scholar]
- Ugai H, Borovjagin AV, Le LP, Wang M, Curiel DT. Thermostability/infectivity defect caused by deletion of the core protein V gene in human adenovirus type 5 is rescued by thermo-selectable mutations in the core protein X precursor. J Mol Biol. 2007;366(4):1142–1160. doi: 10.1016/j.jmb.2006.11.090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ugai H, Wang M, Le LP, Matthews DA, Yamamoto M, Curiel DT. In vitro dynamic visualization analysis of fluorescently labeled minor capsid protein IX and core protein V by simultaneous detection. J Mol Biol. 2010;395(1):55–78. doi: 10.1016/j.jmb.2009.10.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ugai H, Yamasaki T, Hirose M, Inabe K, Kujime Y, Terashima M, Liu B, Tang H, Zhao M, Murata T, Kimura M, Pan J, Obata Y, Hamada H, Yokoyama KK. Purification of infectious adenovirus in two hours by ultracentrifugation and tangential flow filtration. Biochem Biophys Res Commun. 2005;331(4):1053–1060. doi: 10.1016/j.bbrc.2005.03.227. [DOI] [PubMed] [Google Scholar]
- van Oostrum J, Burnett RM. Molecular composition of the adenovirus type 2 virion. J Virol. 1985;56(2):439–448. doi: 10.1128/jvi.56.2.439-448.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vayda ME, Rogers AE, Flint SJ. The structure of nucleoprotein cores released from adenovirions. Nucleic Acids Res. 1983;11(2):441–460. doi: 10.1093/nar/11.2.441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walton TH, Moen PT, Jr, Fox E, Bodnar JW. Interactions of minute virus of mice and adenovirus with host nucleoli. J Virol. 1989;63(9):3651–3660. doi: 10.1128/jvi.63.9.3651-3660.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang D, Umekawa H, Olson MO. Expression and subcellular locations of two forms of nucleolar protein B23 in rat tissues and cells. Cell Mol Biol Res. 1993;39(1):33–42. [PubMed] [Google Scholar]
- Xu N, Segerman B, Zhou X, Akusjarvi G. Adenovirus virus-associated RNAII-derived small RNAs are efficiently incorporated into the rna-induced silencing complex and associate with polyribosomes. J Virol. 2007;81(19):10540–10549. doi: 10.1128/JVI.00885-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yung BY, Chan PK. Identification and characterization of a hexameric form of nucleolar phosphoprotein B23. Biochim Biophys Acta. 1987;925(1):74–82. doi: 10.1016/0304-4165(87)90149-8. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Xiong Y, Yarbrough WG. ARF promotes MDM2 degradation and stabilizes p53: ARF-INK4a locus deletion impairs both the Rb and p53 tumor suppression pathways. Cell. 1998;92(6):725–734. doi: 10.1016/s0092-8674(00)81401-4. [DOI] [PubMed] [Google Scholar]
- Zink D, Fischer AH, Nickerson JA. Nuclear structure in cancer cells. Nat Rev Cancer. 2004;4(9):677–687. doi: 10.1038/nrc1430. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(A) Schematic representation of the core protein gene V region in the adenovirus genomes. The genome of Ad5-dV/TSB was created by deleting nucleotides at positions 16,545–17,651 of Ad5 and incorporating suppressor mutations at 17,714 (G to A; silent), 17,716 (G to A; G13E) and 17,728 (G to T; R17I) in the precursor region of polypeptide X (Mu). Position 1 of the nucleotide sequence refers to the left end of the Ad5 genome (Genbank ID: AY339865), for the precursor X. mu; map units, VII; polypeptide VII, V; polypeptide V, X; polypeptide X (Mu), VI; polypeptide VI, HX; hexon, L2 and L3; major late transcript units. (B) Schematic representation of the physical map of Ad5 and the adenoviral mutants used in this study. TP; terminal protein, E1; early gene 1, IX; polypeptide IX, mRFP1; monomeric red fluorescent protein 1, EGFP; enhanced green fluorescent protein, E3; early region 3, E1b19k; the gene for early gene 1b (E1b) 19 kilodalton protein, dE1; the E1 deletion, dV; the pV deletion, and dE3; the E3 deletion.
293 cells were infected with Ad5 or Ad5-dV/TSB at an MOI of 10 PFU/cell for various times. One-step growth curve analysis of adenoviruses was performed, and infectious titer was determined by plaque assay.
(A) After medium was removed from HPAEC cells, cells were washed with 30 mM HEPES-NaOH buffer (pH 7.4) and fixed with fixation solution. HPAEC cells were stained with anti-NPM1 or anti-p53 followed by a secondary antibody conjugated with fluorescent probe. (B) After medium was removed from HPAEC and MCF-7 cells, cells were directly fixed with fixation solution without washing. Subcellular localization of NPM1 and p53 was determined by immunofluorescent staining. Hoechst 33342 staining was performed for 10 min.
(A) Detection of the fluorescently labeled adenoviral particles in infected HPAEC cells. HPAEC cells were infected with Ad5-E3-V-EGFP at MOIs 10, 100 or 1,000 PFU/cell for 1 hour. The trafficking of virion-associated core pV-EGFP was monitored in infected cells at 1 h.p.i. Uninfected and infected HPAEC cells were stained with anti-NPM1 followed by a secondary antibody conjugated with fluorescent probe. (B) Localization of NPM1-EGFP in 293 cells stably expressing NPM1-EGFP. Hoechst 33342 staining was performed for 10 min, and the signals for NPM1-EGFP and DNA staining were detected by fluorescence microscopy.
(A) Crude proteins were extracted from HUVEC, HPAEC, and lung carcinoma A549 cells, and 10 µg of total proteins were separated on 10% (w/v) SDS-PAGE, and NPM1 expression level was validated by Western blot analysis. (B) DNA replication of Ad5-dV/TSB in cancer cells. A549 cells were infected with Ad5 or Ad5-dV/TSB at an MOI of 10 PFU/cell, and harvested at various times post-infection. Total DNA was extracted from infected cells, and viral DNA replication was analyzed by using 50 ng of total DNA and E4 primers. Plots represent as the copy number of the E4 gene per 50 ng of total DNA.
(A) HPAEC was infected with pV-mRFP1-lentivirus at an MOI of 50 proviral DNA/cell for 60 hours. Subcellular localization of NPM1 and p53 was detected in HPAEC infected with mock (LV-empty) or pV-recombinant lentivirus (LV-V). (B) Western blot analysis of pV expression in pV-lentivirus-infected HPAEC cells. (C) HPAEC cells were infected with the pV-recombinant lentivirus at an MOI of 2, 10 or 50 proviral DNA/cell for 60 hours. The pV-transduced cells were infected with adenovirus at an MOI of 10 PFU/cell, and harvested at 1 and 48 h.p.i. Production level of infectious adenovirus was measured by plaque assay. Results were reported as mean ± SD. * P<0.01, ** P<0.05; two-tailed unpaired Student’s t-tests between groups. N.S.; no significance.
HPAEC cells were infected with adenovirus at an MOI of 10 PFU/cell for various times, followed by Western blot analysis. Proteins extracted from infected cells at various time points were subjected to Western blot, and 40 µg of total proteins was analyzed with the indicated antibodies. Lysates extracted from DU145 cells which lowly express p14ARF were used as a positive control for detecting tumor suppressor p14ARF.
Primary (A) and A549 (B) cells were infected with Ad5-wt-IX-mRFP1 or Ad5-dV/TSB-IX-mRFP1 at an MOI of 10 PFU/cell for various times post-infection. Signal for pIX-mRFP1 was detected by fluorescence microscopy.
(A) Proteins extracted from cancer or primary cell lines were validated by Western blot analysis with the indicated antibodies. (B) Subcellular localization of endogenous NPM1 in cancer and primary cells was determined by immunofluorescent staining. (C) One-step growth curve analysis of Ad5-dV/TSB in infected cancer cells. Cancer cells were infected with Ad5 or Ad5-dV/TSB at an MOI of 10 PFU/cell for various times, and infectious adenoviruses were titrated by plaque assay. (D) Cancer cells were infected with Ad5 or Ad5-dV/TSB at an MOI of 10 PFU/cell for 4 days, and tumoricidal effect induced by adenovirus was observed by microscopy. Bar = 200 µm. (E) Comparison of endogenous NPM1 expression profiles in cancer cell lines. An equivalent of 10 µg of total protein extracted from cancer and HPAEC cells was separated on 10% (w/v) SDS-PAGE, and NPM1 expression level was validated by Western blot analysis. (F) Abnormality of NPM1 subcellular localization in a broad range of cancer cell lines. Subcellular localization of endogenous NPM1 in cancer and primary cells was visualized by staining with anti-NPM1 followed by secondary antibody conjugated with a fluorescent probe. Cells were stained for nuclear DNA using Hoechst 33342 followed by visualization of the fluorescent probe for NPM1 detection by epifluorescence microscopy. (G) Cancer cells were infected with adenovirus at an MOI 10 PFU/cell for 1 and 48 hours, and infected cells were harvested. Production level of infectious adenovirus was measured by plaque assay. Results were reported as mean ± SD. Black bar; Ad5, white bar; Ad5-dV/TSB.
(A) HPAEC cells were infected with Ad5-E3-V-EGFP at an MOI of 10 PFU/cell for various times post-infection. NPM1 was visualized by staining with anti-NPM1 followed by secondary antibody conjugated with a fluorescent probe. Uninfected and infected cells were stained for nuclear DNA using Hoechst 33342, and localization of pV-EGFP and NPM1 was detected by fluorescence microscopy. (B) Viral growth kinetics of Ad5-E3-V-EGFP in primary cells. HPAEC cells were infected with Ad5 or Ad5-E3-V-EGFP at an MOI of 10 PFU/cell for various times post-infection. Infectious titer of adenovirus was determined by plaque assay. (C) HPAEC cells were infected with Ad5-wt-IX-mRFP1 at an MOI of 10 PFU/cell for various times post-infection. NPM1 was visualized by staining with anti-NPM1 followed by secondary antibody conjugated with a fluorescent probe. Uninfected and infected cells were stained for nuclear DNA using Hoechst 33342, and fluorescent signals for pIX-mRFP1 and NPM1 were detected by fluorescence microscopy. pIX-mRFP1 was initially expressed in the cytoplasm and accumulated in the nucleus for virus assembly at the late stage of infection, and the cytoplasm of infected cells was undetectable because infected cells shrank during the late stage of infection as previous reported by Ugai et al. (Ugai et al., 2010). Arrows show colocalization of pIX-mRFP1 and NPM1.