Abstract
Curli are functional amyloids produced by enteric bacteria. The major curli fiber subunit, CsgA, self-assembles into an amyloid fiber in vitro. The minor curli subunit protein, CsgB, is required for CsgA polymerization on the cell surface. Both CsgA and CsgB are composed of five predicted β–strand-loop-β–strand-loop repeating units that feature conserved glutamine and asparagine residues. Because of this structural homology, we proposed that CsgB might form an amyloid template that initiates CsgA polymerization on the cell surface. To test this model, we purified wild-type CsgB, and found that it self-assembled into amyloid fibers in vitro. Preformed CsgB fibers seeded CsgA polymerization as did soluble CsgB added to the surface of cells secreting soluble CsgA. To define the molecular basis of CsgB nucleation, we generated a series of mutants that removed each of the five repeating units. Each of these CsgB deletion mutants was capable of self-assembly in vitro. In vivo, membrane-localized mutants lacking the 1st, 2nd or 3rd repeating units were able to convert CsgA into fibers. However, mutants missing either the 4th or 5th repeating units were unable to complement a csgB mutant. These mutant proteins were not localized to the outer membrane, but were instead secreted into the extracellular milieu. Synthetic CsgB peptides corresponding to repeating units 1, 2 and 4 self assembled into ordered amyloid polymers, while peptides corresponding to repeating units 3 and 5 did not, suggesting that there are redundant amyloidogenic domains in CsgB. Our results suggest a model where the rapid conversion of CsgB from unstructured protein to a β-sheet-rich amyloid template anchored to the cell surface is mediated by the C-terminal repeating units.
Introduction
Amyloid fibers are a common pathology associated with neurodegenerative diseases such as Alzheimer’s and Parkinson’s disease 1. These have classically been referred to as protein misfolding diseases because the amyloidogenesis is the result of an aberrant aggregation process in which a normally soluble protein self assembles into a highly ordered, structurally stable fiber. Biochemically, amyloid fibers are defined by unique tinctorial properties, resistance to proteases and detergents and a β-sheet rich secondary structure 2. Typically there are three phases to in vitro amyloid polymerization: a lag phase, a phase of rapid fiber growth and a stationary phase 3; 4; 5; 6; 7; 8. Rapid fiber growth is dependent on the formation of a nucleus 9. A common feature of nucleus-dependent polymerization reactions is that preformed fibers can act to seed the polymerization of soluble monomers. It is proposed that preformed fibers provide a template for monomer polymerization, and that this interaction mediates exit from the lag phase 9. Although amyloid nucleation and polymerization have been extensively studied in vitro, in vivo nucleation models are just now being developed. Functional amyloids provide a robust context to study in vivo amyloid nucleation and polymerization, especially in cases where the functional amyloid is produced by a genetically tractable model organism 10; 11; 12.
Curli fibers are extracellular amyloid fibers produced by E. coli and other enteric bacteria 10; 13. The discovery of a dedicated pathway for amyloid biogenesis in curli fiber assembly led to a paradigm shift in the amyloid field, which previously held that amyloids were exclusively the result of protein misfolding. Curli fibers are part of the extracellular matrix that is required for biofilm formation and for mediating host cell-bacteria interactions 14; 15; 16; 17; 18. Therefore, curli are virulence factors 19; 20; 21 and constitute potential novel targets for antibacterial agents 22; 23; 24. Curli are composed of a major and minor subunit, CsgA and CsgB, respectively 25; 26; 27. Purified CsgA polymerizes into an amyloid fiber in vitro, but CsgB is required for in vivo curli formation 8; 28. In the absence of CsgB, CsgA is secreted away from the cell as a soluble protein 26; 29. In a process referred to as interbacterial complementation, cell surface localized CsgB produced by a csgA mutant can convert secreted, soluble CsgA produced by a csgB mutant into an ordered amyloid fiber 10; 28. The CsgB expressed on the surface of csgA mutants can also nucleate exogenously added purified CsgA 30; 31. CsgB becomes incorporated into the fiber after initiating the polymerization of CsgA 25, yet the sequences or domains of CsgB that guide amyloid nucleation have yet to be elucidated.
In silico structural predictions suggest that both CsgA and CsgB contain a β-sheet rich domain that can be further divided into five imperfect β-strand-loop-β-strand repeats 26; 32; 33. The repeating unit domains of both CsgA and CsgB are 51% similar and they contain a number of conserved glutamines and asparagines 27. These observations led us to hypothesize that CsgB, like CsgA, may also adopt an amyloid-like fold and that this feature of CsgB may serve to template CsgA polymerization. We previously purified a truncated version of CsgB missing the fifth repeating unit (CsgBΔr5) 34. This truncated CsgB mutant self-assembled into β-sheet rich amyloid fibers that could template CsgA polymerization in vitro. However, unlike WT CsgB, CsgB missing the fifth repeating unit was secreted away from the cell surface and was unable to complement a csgB mutant in vivo 34.
We report here that wild-type (WT) CsgB polymerizes into an amyloid structure and does so faster than either CsgA or the previously characterized CsgB truncation mutant. Purified WT CsgB was able to template CsgA polymerization in vitro and in vivo when applied to csgB mutants. The contribution of each CsgB repeating unit to nucleator function in vivo and to CsgB polymerization in vitro was assessed. We found that the fourth repeating unit of CsgB is required for cell association suggesting that both the 4th and 5th repeating units function, directly or indirectly, to anchor CsgB to the cell. Our results support a model in which CsgB rapidly adopts a β-sheet-rich, amyloid fold at the cell surface that templates the conversion of slower-folding major subunit CsgA into cell-associated amyloid fibers. We propose that this process is mediated by the C-terminal repeating units of CsgB.
Results
Purified CsgB forms amyloid fibers that can seed CsgA polymerization
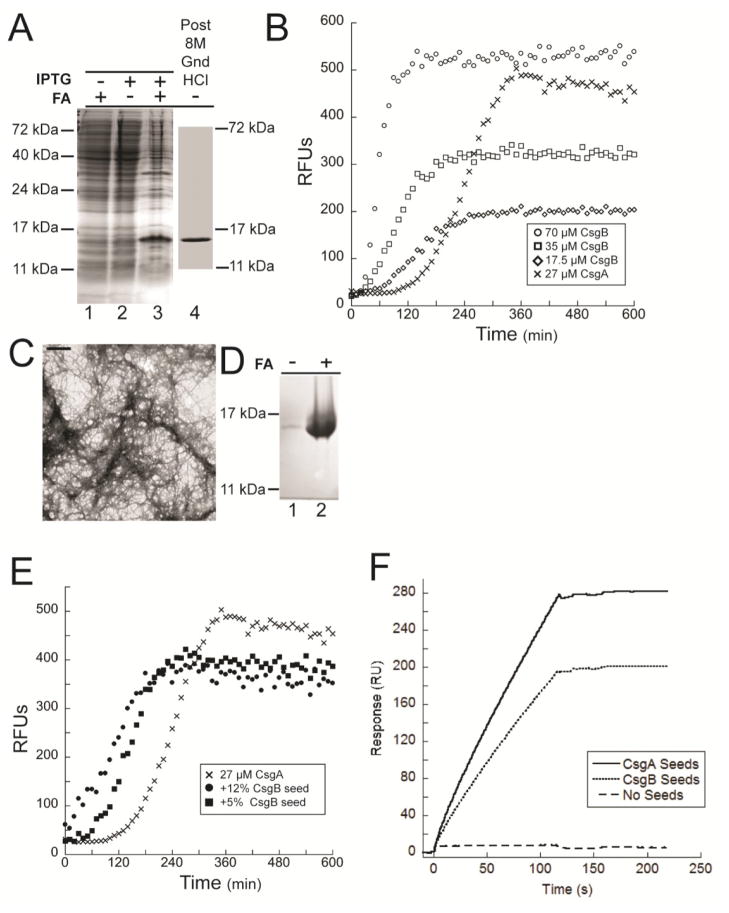
CsgB contains three domains. The first domain contains twenty-one amino acids and encodes a sec-signal sequence responsible for targeting CsgB to the periplasm. These amino acids are cleaved as the protein transverses the inner-membrane. The next twenty-two amino acids mark the N-terminus of the mature protein. This domain is predicted to be unstructured and cells expressing CsgB deleted for these amino acids produce WT fibers (data not shown). However, the synonymous region within CsgA is required for the secretion of the protein 35; 36. The last domain contains the amyloid-core of CsgB and can be further divided into five regions of imperfect homology referred to as repeating units. We previously purified a truncated CsgB molecule that lacked the C-terminal repeating unit by harvesting the supernatant of cells expressing the truncated protein. Attempts to purify WT CsgB from the extracellular milieu in a similar fashion were impeded by relatively low expression levels 34. Here, expression of mature WT CsgB (i.e. the WT sequence of CsgB lacking only the sec-signal sequence required for secretion across the inner membrane) was induced in the cytoplasm of cells. After induction, cells contained significant amounts of SDS insoluble CsgB protein (Fig. 1A lane 2 and 3). Soluble his-tagged CsgB was recovered by affinity purification after pellets of induced cells were resuspended in 8 M GndHCl (Fig. 1A lane 4) 33.
Figure 1. Biochemical and physiological properties of WT CsgB.
(A) Coomassie stained SDS-PAGE of OD600 normalized cell lysates pre-induction (IPTG −) or post-induction (IPTG +) lanes 1 through 3. Cells were resuspended in SDS loading buffer (FA −) or pretreated with formic acid (FA+). CsgB was solubilized post purification by incubation in 8 M guanidine HCl (Lane 4). (B) Representative ThT kinetic plot of 70 μM (○), 35 μM (□), or 17.5 μM (◇) purified CsgB and 27 μM purified CsgA (X). Relative fluorescent units (RFUs) emitted at 495 nm were recorded every 10 min. after excitation at 438 nm. (C) TEM of 65 μM CsgB incubated at room temperature for 24 hours. The scale bar represents 500 nm. (D) Coomassie stained SDS-PAGE of 65 μM CsgB polymerized into fibers, centrifuged and resuspended in SDS loading buffer with (+) or without (−) prior formic acid (FA) treatment. (E) ThT kinetic plot of 27 μM CsgA (X), 27 μM CsgA and +5% w/w WT CsgB seeds (●) and 27μM CsgA + 12% w/w WT CsgB seeds (■). (F) Surface plasmon resonance (SPR) sensorgrams of interactions between monomeric CsgA and CsgA seeds or CsgB seeds. 0.25μM fresh monomeric CsgA was flow over the CM5 chip immobilized with 3uM CsgA seeds, 3.5μM CsgB seeds or no seeds. The interaction was recorded in resonance units.
CsgB polymerization was monitored over time using the amyloid specific dye thioflavin-T (ThT). ThT fluorescence rapidly increased upon addition of freshly purified, soluble CsgB after only a short lag phase, and this increase in fluorescence was concentration dependent (Fig. 1B). Both CsgA and CsgB polymerization curves contained a lag phase, a fast phase, and a stationary phase. However, the CsgB lag phase was shorter than the CsgA lag phase at approximately equivalent protein concentrations (Fig. 1B). Transmission electron microscopy (TEM) analysis of fractions containing CsgB incubated overnight at room temperature revealed an ordered fibrous structure, similar to what has been observed for CsgA (Fig. 1C) 8. The fibrous CsgB aggregates were not soluble in SDS and pretreatment with formic acid (FA) was required to visualize monomeric protein on a SDS-PAGE gel (Fig. 1D lane 2).
To test whether CsgB fibers could template CsgA polymerization in vitro, we measured the polymerization of soluble CsgA in the presence of preformed CsgB fibers by ThT fluorescence. In the absence of CsgB, CsgA polymerization had a lag phase of approximately two hours. When 5% (w/w) preformed CsgB fibers were added to the reaction mix, a 1-hour lag phase was observed. When the amount of CsgB fibers was increased to 12% (w/w), CsgA polymerization proceeded without an apparent lag phase at this time scale (Fig. 1E). Interaction of soluble CsgA with CsgB fibers was also monitored by surface plasmon resonance (SPR). Preformed CsgB fibers were immobilized on a sensor chip. Soluble CsgA was able to interact with CsgB fibers shown by the increase of resonance units when CsgA was injected over the sensor chip. No obvious decay of resonance units was observed after injection, suggesting that the interaction between CsgA and CsgB fibers was non-reversible (Fig. 1F). These results suggest that WT CsgB rapidly polymerizes into an amyloid fiber that templates CsgA polymerization in vitro.
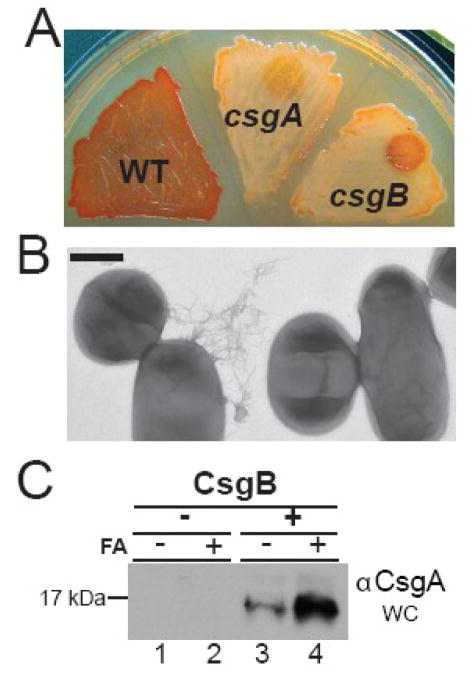
The ability of CsgB to template CsgA under physiologically relevant conditions was tested by adding purified, soluble CsgB to csgA and csgB mutant cells grown on YESCA plates. We have used this ‘overlay assay’ to measure the ability of cell surface localized CsgB to stimulate the polymerization of purified and exogenously added CsgA 30; 31. In this case, purified soluble CsgB was spotted on to bacterial lawns of csgA or csgB cells grown under curli-inducing conditions, followed by an overnight incubation. The cells were then stained with Congo red to visualize amyloid formation 30; 31. In comparison to the csgA mutant cells, the csgB mutants incubated with CsgB had a pronounced Congo red staining phenotype where the purified CsgB had been added (Fig. 2A). Thus, purified CsgB complemented the csgB mutant for Congo red binding when added to the cells exogenously. Using electron microscopy, we confirmed that fibers had formed when purified CsgB was incubated with the csgB mutants (Fig. 2B). To determine if the CsgA secreted from the CsgB-treated csgB mutants had formed detergent insoluble fibers, these cells were harvested and resuspended in the presence or absence of formic acid. The majority of CsgA in these samples required formic acid pretreatment to resolve the monomer, indicating that the protein was assembled into an SDS-insoluble and polymerized fiber form (Fig. 2C). Taken together, these results demonstrate that CsgB adopts an amyloid conformation more rapidly than the major subunit CsgA and this conformation is able to template CsgA polymerization near the cell surface.
Figure 2. Exogenously added CsgB functions as a nucleator.
(A) 10 ul of 37 μM purified CsgB was overlaid on csgA and csgB mutants that had been grown for 24 hours, and then stained with Congo red dye after an additional 24-hour incubation at 26°C. WT cells are shown as a positive staining control. (B) TEM of csgB cells that were incubated with 37 μM purified CsgB. The scale bar represents 500nm. (C) CsgA western blot analysis of a whole cell (WC) lysate of the csgB cells in (A) that were incubated in the absence of CsgB (Lanes 1 and 2) or presence of CsgB (lanes 3 and 4). Samples were resuspended in SDS loading buffer with (+) or without (−) formic acid (FA) pretreatment.
In vivo contribution of the repeating units to CsgB function
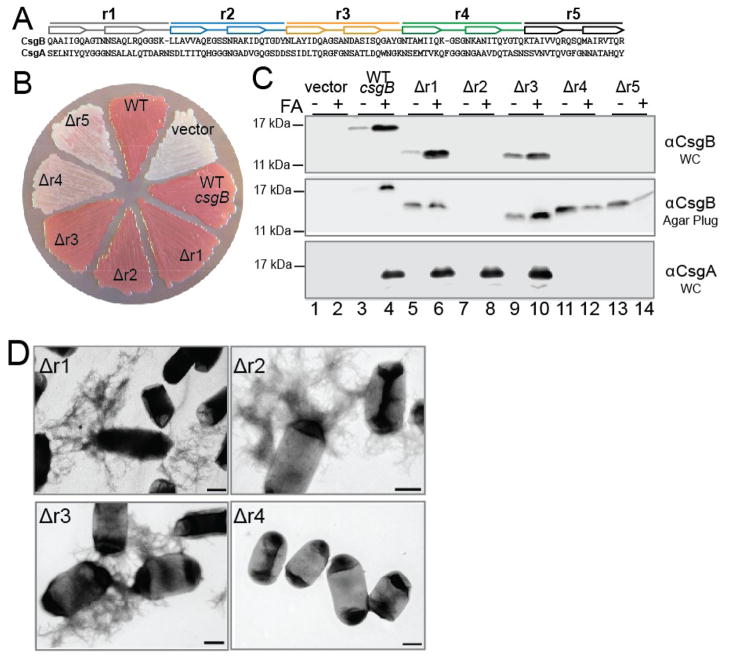
CsgB contains a glutamine-asparagine rich domain that can be divided into five repeat sequences called r1, r2, r3, r4 and r5 (Fig. 3A) 27; 28. Each repeating unit is predicted to contain two β-strands. The amino acid sequence of each repeating unit of CsgA and CsgB contain a high degree of similarity (Fig. 3A). In order to determine the contribution of each repeating unit to CsgB function in vivo, we constructed in-frame deletions of each repeating unit. When a csgB mutant strain expressing csgB constructs with deletions of r1, r2 or r3 (CsgBΔr1, CsgBΔr2, CsgBΔr3) were grown under curli-inducing conditions on Congo red-containing agar plates, these mutants bound similar levels of Congo red as both WT cells and a csgB mutant complemented with WT csgB (Fig. 3B). Both CsgBΔr1 and CsgBΔr3 could be detected by western blot and increased amounts of CsgBΔr1 and CsgBΔr3 monomers were seen when the whole-cell samples were pretreated with FA (Fig. 3C top panel, lanes 6 and 10). The CsgBΔr2 protein was not detected by western blot, because the CsgB antibody used here was raised using a peptide fragment found within r2 36. However, like the WT CsgB, CsgBΔr1 and CsgBΔr3 constructs, CsgA monomers could only be detected in whole-cell samples when the CsgBΔr2 samples were pretreated with FA (Fig. 3.2C bottom panel, lanes 4, 6, 8, and 10). These results suggest the CsgBΔr1, CsgBΔr2, and CsgBΔr3 constructs were able to convert soluble CsgA into a cell-associated SDS insoluble fiber. To confirm the presence of fibers, we prepared csgB mutants harboring each repeating unit deletion mutant for TEM. Consistent with the Congo red and western blot data, fibers were seen when csgB mutants harboring the CsgBΔr1, CsgBΔr2 and CsgBΔr3 constructs were viewed by TEM, and these fibers were morphologically similar to WT curli fibers (Fig. 3D). Therefore, the r1, r2, and r3 deletion mutants were able to effectively complement the csgB mutant to produce cell-associated amyloid fibers.
Figure 3. Contribution of each repeating unit to CsgB function in vivo.
(A) Amino acid sequence alignment of the CsgA and CsgB repeating units. Each repeating unit contains two predicted β-sheets (arrows). (B) Congo red binding phenotype of MC4100 (WT), or csgB harboring a vector control plasmid (vector), a plasmid vector containing WT csgB, csgBΔr1 (Δr1), csgBΔr2 (Δr2), csgBΔr3 (Δr3), csgBΔr4 (Δr4) or csgBΔr5 (Δr5). (C) Western blot analysis of a csgB mutant strain harboring vector control (vector lanes 1 and 2), a plasmid vector containing WT csgB (lanes 3 and 4), csgBΔr1 (Δr1 lanes 5 and 6), csgBΔr2 (Δr2 lanes 7 and 8), csgBΔr3 (Δr3 lanes 9 and 10), csgBΔr4 (Δr4 lanes 11 and 12) or csgBΔr5 (Δr5 lanes 13 and 14). Samples were resuspended in SDS loading buffer with (+) or without (−) formic acid (FA) pretreatment. The top two panels are blots probed with anti-CsgB antibody. The bottom panel is a blot probed with anti-CsgA antibody. Whole cells (WC) samples are represented in the first and third panel, while samples containing whole cells and the underlying agar are represented in the second panel. (D) TEM of csgB mutants harboring CsgBΔr1 (Δr1), CsgBΔr2 (Δr2), CsgBΔr3 (Δr3), or CsgBΔr4 (Δr4) grown under curli-inducing conditions. The scale bars represent 500 nm.
Unlike the r1, r2 and r3 deletions, both the CsgBΔr4 and CsgBΔr5 constructs were unable to complement a csgB mutant for Congo red binding (Fig. 3B). To test for the stability of CsgBΔr4 and CsgBΔr5, cell lysates were probed with anti-CsgB antibody and no cell-associated CsgBΔr4 or CsgBΔr5 protein could be detected (Fig. 3C top panel, lanes 11–14). However, when whole cells and the underlying agar were collected, bands corresponding to the CsgBΔr4 and CsgBΔr5 constructs were observed (Fig. 3C middle panel, lanes 11–14). This indicated that both CsgBΔr4 and CsgBΔr5 were not cell associated, but were instead secreted into the underlying agar. The soluble nature of these mutant proteins also renders them more sensitive to formic acid, which is manifested by an apparent decrease in protein stability (Fig. 3C middle panel, lanes 12 and 14). These results are also consistent with our previous report characterizing CsgBΔr5 (previously referred to as CsgBtrunc) 34. Soluble CsgA was detected when the underlying agar was collected along with the csgB cells harboring CsgBΔr4 or CsgBΔr5 (data not shown). Thus the CsgBΔr4 and CsgBΔr5 constructs do not alter CsgA stability. No fibers could be detected by TEM in samples of csgB cells expressing either csgBΔr4 or csgBΔr5 (Fig. 3D) 34. These results suggest that the C-terminal portion of CsgB required for nucleator function in vivo includes both CsgB r4 and r5. The observation that both csgBΔr4 and csgBΔr5 are secreted away from the cell suggests that the last two CsgB repeating units are required to facilitate anchoring of the nucleator to the outer membrane.
The repeating unit deletion mutants polymerize in vitro
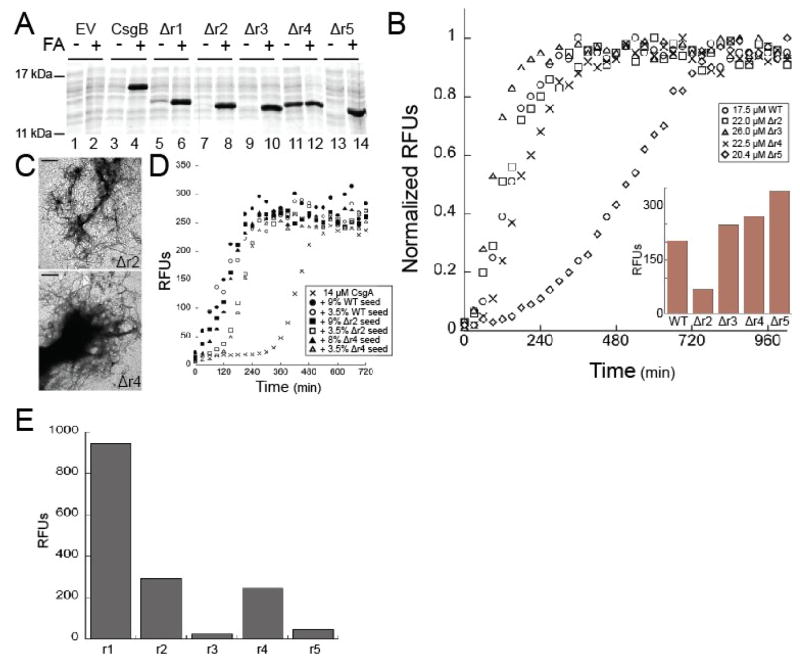
A possible explanation for the in vivo defect of the CsgBΔr4 construct was that the 4th repeating unit is required for interaction with CsgA. To determine if CsgBΔr4 could act as a template for CsgA polymerization and to characterize the in vitro characteristics of the other repeating unit mutants, we constructed expression plasmids for cytoplasmic purification of each of the CsgB repeating unit deletion mutants. Each repeating unit deletion, with the exception of CsgBΔr4, formed SDS-insoluble cytoplasmic aggregates after induction (Fig. 4A). Soluble CsgBΔr2, CsgBΔr3, CsgBΔr4 and CsgBΔr5 were purified after treatment with 8 M GndHCl (data not shown). We were unable to purify CsgBΔr1 (see materials and methods).
Figure 4. In vitro characterization of the repeating unit deletions.
(A) Coomassie stained SDS-PAGE of OD600 normalized cell lysates of cells harboring the empty vector (EV) pET11d (EV lanes 1 and 2), WT CsgB (lanes 3 and 4), CsgBΔr1 (Δr1 lanes 5 and 6), CsgBΔr2 (Δr2 lanes 7 and 8), CsgBΔr3 (Δr3 lanes 9 and 10), CsgBΔr4 (Δr4 lanes 11 and 12), or CsgBΔr5 (Δr5 lanes 13 and 14) after 1 hour induction with 0.5 mM IPTG. Samples were resuspended in SDS loading buffer with (+) or without (−) formic acid (FA) pretreatment. (B) Representative ThT kinetic plot of purified WT CsgB (○), CsgBΔr2 (Δr2□), CsgBΔr3 (Δr3△), CsgBΔr4 (Δr4 X), and CsgBΔr5 (Δr5 ◇). The inset is a representative bar graph of the RFUs for each protein after 12 hours of incubation at room temperature with ThT. (C) TEM of purified CsgBΔr2 (Δr2 top panel) or purified CsgBΔr4 (Δr4 bottom panel) after 24 hours of incubation at room temperature. The scale bars represent 500nm. (D) Representative ThT kinetic plot monitoring the polymerization of 14 μM CsgA (X), 14 μM seeded with 9% w/w WT CsgB (●), seeded with 3.5% w/w WT CsgB (○), seeded with 9% w/w CsgBΔr2 (Δr2 ■), seeded with 3.5% w/w CsgBΔr2 (Δr2 □), seeded with 8% w/w CsgBΔr4 (Δr4 ▲), or seeded with 3.5% w/w CsgBΔr4 (Δr4 △). (E) Relative fluorescent units (RFUs) produced by synthetic peptides composed of the amino acids in r1 (145 μM), r2 (102 μM), r3 (138 μM), r4 (176 μM), and r5 (167 μM ) after incubation at room temperature for 24 hours in the presence of ThT.
We monitored the in vitro polymerization of CsgBΔr2, CsgBΔr3, CsgBΔr4 and CsgBΔr5 using the ThT time course assay. The polymerization curve of CsgBΔr5 displayed a two-hour lag phase, consistent with previously published results (Fig. 4B) 34. The lag phase for CsgBΔr5 was noticeably longer than that of WT, CsgBΔr2, CsgBΔr3, and CsgBΔr4, which all exhibited no apparent lag phase on this time scale. This suggests that repeating unit r5 is required for rapid, WT-like polymerization of CsgB in vitro.
The normalized CsgBΔr2 ThT polymerization profile demonstrated that CsgBΔr2 polymerized as quickly as WT CsgB (Fig. 4B). However, the relative fluorescent units (RFUs) recorded for CsgBΔr2 after twelve hours of polymerization were consistently lower than WT CsgB at the same time point (Fig. 4B inset), suggesting that CsgBΔr2 might adopt an altered conformation. No gross morphological differences between CsgBΔr2 fibers and WT CsgB fibers were observed when the fibers were viewed utilizing TEM (Fig. 1C and Fig. 4C top panel). We next tested the ability of the CsgBΔr2 fibers to seed CsgA polymerization. Using a 14 μM concentration of CsgA, we observed a lag phase of five hours for CsgA by itself, which is consistent with previous reports that the CsgA lag phase is concentration-dependent 8. At this concentration, only 3.5% (w/w) seed of WT CsgB was needed to eliminate the lag phase (Fig. 4D). A short lag phase of roughly 30 minutes was seen when 3.5% (w/w) CsgBΔr2 fiber was added to the reaction mix, but no apparent lag phase was observed when the amount of seed was increased to 9% (w/w) CsgBΔr2 pre-formed fiber (Fig. 4D). These observations suggest CsgBΔr2 may be less efficient than WT CsgB at converting CsgA into SDS-insoluble fibers, however they are consistent with the in vivo results demonstrating SDS-insoluble CsgA fibers are formed in the presence of CsgBΔr2 (Fig. 3).
Despite the inability to produce curli in vivo, the CsgBΔr4 had a ThT fluorescence profile similar to WT CsgB in vitro (Fig. 4B). Fractions containing purified CsgBΔr4 were incubated overnight and viewed by TEM to assess aggregate morphology. By this analysis, CsgBΔr4 fibers appeared similar to WT CsgB fibers (Fig. 4C bottom panel). To determine if the in vivo defect of CsgBΔr4 could be attributed to an inability of CsgBΔr4 to seed CsgA polymerization in vitro, pre-formed CsgBΔr4 fibers were added to a CsgA polymerization reaction. A short lag phase of 30 minutes was observed when 3.5% (w/w) CsgBΔr4 fibers were added to CsgA and no apparent lag phase could be detected when the amount of preformed fiber was increased to 8% (w/w) (Fig. 4D). These results demonstrate that CsgBΔr4 formed amyloid fibers in vitro and that these CsgBΔr4 fibers template in vitro CsgA polymerization. Thus the inability of CsgBΔr4 to complement a csgB mutant cannot be fully explained by a defect of CsgBΔr4 to template CsgA polymerization in vitro. These results suggest that like CsgA, CsgB may have more than one domain that acts to template CsgA polymerization in vivo. Alternatively, CsgB r4 may play a role in anchoring CsgB to the cell surface as CsgBΔr4 was detected in the underlying agar in vivo (Fig. 2).
In order to gain a better understanding of the repeating unit(s) that have the potential to act as a template for CsgA polymerization we determined the ability of the individual CsgB repeating units to self-assemble using synthetic peptides composed of the amino acids found within each repeating unit. A similar approach was used to determine the repeating units of CsgA that facilitated CsgA polymerization. Synthetic peptides composed of the repeating units CsgA r1 and r5 were able to polymerize in vitro. CsgA r5 peptide polymerized with at a remarkable rate. Deleting CsgA r1 or CsgA r5 abrogated curli biogenesis in vivo highlighting the importance of these repeating units to CsgA polymerization 30. The template-mediated polymerization hypothesis predicts that domains of CsgB that self-assemble will contribute to nucleator function. Therefore, we hypothesized that repeating units CsgB r5 and CsgB r4 would self assemble given their importance to in vivo curli biogenesis.
Synthetic peptides composed of amino acids found in each CsgB repeating unit were incubated with ThT to assess the self-assembly capability of each CsgB repeating unit. Peptides composed of the corresponding amino acid sequence of CsgB r1, CsgB r2 and CsgB r4 increased ThT fluorescence after a 24-hour incubation at room temperature, while peptides composed of the corresponding amino acid sequence of CsgB r3 and CsgB r5 did not result in ThT fluorescence over a 24 hour time period (Fig. 4E). Interestingly, deleting a repeating unit incapable of self-assembly into amyloid, CsgB r5, resulted in significant delay in CsgB polymerization in vitro (Fig. 4B). These results suggest that the initiation of CsgB polymerization via CsgB r5 does not require an amyloid-like self-assembly of this repeating unit. Taken together with the in vivo data demonstrating that CsgBΔr5 is defective for nucleation, these findings imply that CsgB r5 might aid in the nucleating properties of CsgB by promoting CsgB polymerization in addition to its role of ensuring that CsgB is properly localized.
Discussion
Shewmaker et al. recently purified full-length CsgA and CsgB (i.e. versions of the proteins that would be expressed in the cytoplasm before processing by the Sec machinery) and demonstrated that both proteins self-assembled into fibers that contain the biochemical features of amyloid fibers 33. Solid state NMR revealed that both CsgA and CsgB fibers are composed of parallel β–sheets that are not in-register. Together with the solid state NMR data, electron microscopy analysis suggests CsgA fibers and CsgB fibers consist of β–helix-like structure. This structure is different from the in-register parallel β–sheet structure shared among the yeast prions and human disease-associated amyloids. In their studies with CsgA and CsgB, Shewmaker et al. suggest that a functional amyloid may adopt a similar final amyloid structure 33. Despite the conserved β-helix structure and high degree of similarity at the amino acid level our results demonstrate that CsgB has several unique features that distinguish it from CsgA. We demonstrate that these unique features facilitate the initial step of curli fiber biogenesis, a process that occurs at the cell surface and shortens the lag phase of CsgA polymerization, thus increasing the efficiency of curli assembly at the correct cellular location.
WT CsgB polymerizes rapidly in vitro
At equal concentrations the lag phase of WT CsgB is shorter (20–30 minutes) than the two-hour lag phases that are observed when polymerization of both CsgA and CsgBΔr5 is monitored (Fig. 1B and Fig. 3B). Our model predicts that CsgB is secreted to the cell surface where it rapidly adopts an amyloid fold that templates CsgA polymerization. CsgB amyloid fibers seeded CsgA polymerization in vitro (Fig. 1E) and freshly purified, soluble CsgB applied to csgB mutant cells was able to nucleate the CsgA secreted from these csgB mutant cells (Fig. 2). These data support the hypothesis that CsgB rapidly adopts an amyloid fold that templates CsgA polymerization at the cell surface.
Proteins associated with amyloid diseases aggregate relatively slowly in vitro, and the longer lag phases associated with the polymerization of these proteins have led to the hypothesis that cytotoxic amyloid precursors accumulate in vivo 37; 38; 39; 40; 41; 42; 43; 44; 45. These precursors have been shown to compromise membrane integrity, leading to cell death 46; 47; 48; 49. Functional amyloid biogenesis pathways proceed without compromising cell physiology. One strategy cells employ to reduce the accumulation of cytotoxic precursors is to promote the rapid conversion of innocuous monomers to stable amyloid fibrils. An example of this is the mammalian functional amyloid protein Pmel that polymerizes without a lag phase in vitro 11. Our previous studies have demonstrated that CsgA polymerization in vitro has a consistent two-hour lag phase that is concentration dependent 8. A potentially toxic intermediate is formed during the lag phase, but the addition of preformed CsgA fibers to freshly purified CsgA can eliminate the lag phase and the formation of this folding intermediate 8. Therefore, we speculate that CsgB-mediated nucleation of CsgA promotes the conversion of CsgA from a soluble monomer to an ordered amyloid aggregate without allowing CsgA to sample the toxic oligomeric state that many amyloids are known to populate 8; 48; 50; 51. Consistent with this, a mutant variant of CsgA that polymerizes independently of CsgB in vivo has potent cytotoxic activity 52.
The C-terminal repeating units of CsgB are essential for nucleator function
CsgA r1 and CsgA r5 deletion mutants are defective for polymerization in vivo and peptides composed of CsgA r1 and r5 self assemble in vitro 31. CsgA r1 and r5 peptides also seed in vitro CsgA polymerization 31. These findings supported our model that proposes that CsgA polymerization is mediated by the interaction of the terminal amyloidogenic repeating units CsgA r1 and CsgA r5.
In-frame deletions of CsgB repeating unit r4 and r5 are the most defective CsgB mutants (Fig. 3) suggesting the C-terminal portion of CsgB directs curli nucleation. We previously reported that the CsgBΔr5 mutant was defective for nucleator activity in vivo due to mislocalization of the protein 34. Synthetic peptides composed of CsgB r5 did not self assemble in vitro (Fig. 4E). The CsgBΔr5 mutant also had the longest polymerization lag phase when compared to WT CsgB and the other CsgB repeating unit deletion mutants. These results suggest CsgB r5, which does not self-assemble, directs CsgB polymerization in vitro.
CsgA polymerization is inhibited when repeating units that can self-assemble are deleted 31. However, deleting self-assembling CsgB repeating units, CsgB r2 and CsgB r4, did not affect CsgB polymerization in vitro (Fig. 4). The CsgBΔr2 and CsgBΔr4 mutants also seeded CsgA polymerization in vitro (Fig. 4). Deleting CsgB r4 abolished nucleator activity in vivo despite having no effect on in vitro CsgB polymerization (Fig. 3). The CsgBΔr4 mutant, like CsgBΔr5, was secreted away from the cell (Fig. 3). Synthetic peptides composed of CsgB r 4 self-assembled in vitro (Fig. 4E). Taken together, these results underscore another difference between CsgA and CsgB. CsgA polymerization is directed by amyloidogenic repeating units located on either side of the predicted amyloid core domain (CsgA r1 and r5). CsgB nucleator function is directed by two repeating units located at the C-terminal portion of the amyloid core. Only one of these repeating units self-assembles in vitro (CsgB r4). Interestingly, the repeating unit that directs CsgB polymerization in vitro, CsgB r5, does not self-assemble into amyloid (Fig. 4). This property of CsgB r5 is not surprising given the importance of membrane localization to the function of CsgB, and the sensitivity membranes have to the toxic effect of amyloids in general. An amyloid-independent mechanism to facilitating the initiation of an amyloid fold would ensures membrane integrity is not compromised during polymerization. Future experiments will define the mechanism by which CsgB r5 facilitates the folding of CsgB.
CsgB self-assembles in vivo
In a csgA mutant CsgB remains SDS-soluble at the cell surface where it can initiate the polymerization of CsgA secreted from cells grown in close proximity26; 53. Bian and Normark demonstrated that CsgB fibers can be formed at the cell surface when CsgB is overexpressed 25. These results suggest that CsgB expression is under transcriptional control that keeps the protein from self-assembling in vivo. We have shown that purified CsgB rapidly self-assembles in vitro (Fig. 1). When purified CsgB was added to csgA mutants, there was a modest increase of Congo red binding in the area where CsgB had been applied (Fig. 2A). However, within this area fibers were not observed by EM and CsgB remained SDS-soluble (data not shown). These results suggest that interactions with the membrane or a membrane-localized protein may keep CsgB from polymerizing in vivo. The chaperone-like protein CsgF ensures the proper localization of curli biogenesis 53. CsgF is the most likely candidate for interacting with CsgB, but a direct CsgB-CsgF interaction has not been demonstrated experimentally. Determining the molecular details of the CsgB-membrane interaction will increase our understanding of how E. coli keeps CsgB from polymerizing on the cell surface.
Curli are among a class of microbial functional amyloids that promote the formation of multicellular communities13; 54; 55; 56; 57. During amyloid formation, soluble peptides can assemble into structurally conserved oligomers before adopting the final amyloid fiber conformation 48. Lipid membranes are particularly sensitive to the oligomeric intermediates formed during amyloid polymerization 58. We speculate that dedicated amyloid nucleator proteins like CsgB stimulate the transition of CsgA from the soluble monomeric state to the final amyloid fiber, thus minimizing the chance for membrane-toxic oligomers to form that might compromise the integrity of the outer membrane. To date, a dedicated amyloidogenic nucleator of the extracellular Gram positive and yeast functional amyloids has not been identified 56; 59. Future experiments will define the nucleation events that initiate the polymerization of this class of functional amyloids, but it is tempting to speculate that a dedicated amyloidogenic nucleator is essential for Gram negative bacteria to assemble a nontoxic, cell-associated amyloid fiber. The molecular basis of the CsgB-membrane interaction will lead to novel insights into how Gram negative bacteria avoid the accumulation of cytotoxic intermediates produced during amyloid fiber biogenesis.
The molecular events that initiate amyloid biogenesis in vivo are not entirely understood. Curli biogenesis represents an elegant functional amyloid biosynthetic pathway that faithfully assembles an amyloid on the cell surface without causing appreciable cytotoxicity. Our results support the template-mediated polymerization model of curli biogenesis, where CsgB rapidly adopts an amyloid-like fold that initiates the folding of soluble CsgA as it comes in contact with the cell surface. This process is directed by the C-terminal repeating units of CsgB. Further study of the nucleation event in curli biogenesis, from both in vivo and in vitro perspectives, may provide alternative approaches for inhibiting disease associated amyloid biogenesis.
Materials and methods
Bacterial Growth
For protein expression and general strain propagation bacteria were cultured in LB broth. To induce curli production, bacteria were grown for 48 hours at 26° C on yeast extract casamino acids (YESCA) plates 10. Curli production was monitored by using Congo red-containing YESCA plates 10. When necessary growth media was supplemented with antibiotics at the following concentrations: kanamycin 50 μg/ml or ampillicin, 100 μg/ml.
Bacterial Strains and Plasmids
MC4100 was the WT strain used 60. The other strains and plasmids used in this study can be found in table S1. Primers used to construct the plasmids in this study can be found in table S2. The repeating unit deletions and point mutants were constructed using synthesis by overlapping ends PCR. PCR was performed using standard techniques and the primers listed in table S3. To test the ability of the csgB mutants to complement the csgB mutant strain the mutated sequences were all subcloned into pLR2, a plasmid that contains the native csgBA promoter, using NcoI and BamHI restriction sites found at the 5′ and 3′ end respectively 36. To express and purify cytoplasmic (i.e., lacking the N-terminal sec-signal sequence) WT CsgB and the repeating unit deletions, PCR-amplified sequences including 6 histidine residues at the C-terminus were subcloned into pET11d (Novagen, Darmstadt, Germany) using NcoI and BamHI restriction sites. Our attempts to purify proteins using a denaturing protocol described below were hindered by a contaminating protein of approximately 30 kDa. We identified this protein as SlyD, a protein enriched with histidine residues at the C-terminus, by mass spectrophotometry analysis (Michigan Proteome Consortium). In order to eliminate this contamination we P1 transduced the slyD::aph allele from the Keio collection strain, JW3311, into our expression strain NEB 3016 61; 62. We named the resulting strain NDH 471
CsgA and CsgB Protein purification
NDH 471 (NEB 3016 slyD::aph) cells harboring a pET11d vector containing polyhistidine-tagged cytoplasmic versions of CsgA, CsgB, or the CsgB repeating unit deletions mutants were grown at 37° C to 0.9 OD600. The cells were induced with 0.5 mM isopropyl-β-D-thiogalactoside (IPTG) and induction proceeded at 37° C for an hour. Cells were collected by centrifugation and the pellets were stored at −80° C. The cells were chemically lysed using 8 M GndHCl buffered with 50 mM potassium phosphate buffer (KPi) pH7.2. A total of 75 ml of lysis solution was used per pellet generated from a 500 ml culture. The lysate was incubated at room temperature with magnetic stirring for 24 hours. The insoluble portion of the lysate was removed by centrifuging at 10,500 × g for 15 minutes and the resulting supernatant was sonicated 5 times, each time for 15 seconds. Sonication was applied at 5 amplitudes. The samples were incubated for 2 minutes on ice in between bursts. The supernatant was then centrifuged again at 10,500 × g for 15 min. HIS-Select ™ HF NiNTA (Sigma Aldrich, Atlanta, GA) was added to the supernatant and this mixture was incubated for 1 hour at room temperature with rocking. The polyhistidine-tagged proteins were affinity purified by collecting the nickel beads as the mixture passed through a Kontes column. The beads were washed with 50 mM KPi pH7.2 to eliminate the GndHCl and then washed with 50 mM KPi containing 12.5 mM imidazole pH7.2 to elute contaminating proteins that bound to the nickel beads. His-tagged proteins were eluted from the column using 125 mM imidazole in 50 mM KPi pH 7.2. Fractions were collected and analyzed for the presence of protein by SDS-PAGE. Protein concentration was determined by the BCA assay (Thermo Scientific, Rockford, IL). We were not able to purify CsgBΔr1. A significant amount of CsgBΔr1 protein was found to be SDS insoluble even after the cells were resuspended in 8 M GndHCl (data not shown).
Thioflavin-T (ThT) Assay
Proteins were mixed with 20 μM ThT in 96-well plates in duplicate and incubated at room temperature. Every 10 minutes samples were excited at 438 nm and fluorescence emitted 495 nm with a 475 nm cutoff was measured using a Spectramax M2 plate reader (Molecular Devices, Sunnyvale, CA). Samples were shaken for 5 seconds before each read. Due to the differences in preparation–to-preparation protein yield, a representative ThT kinetic graph taken from a series of at least three ThT assays is shown. The net RFUs generated by a protein at a specific concentration after a 24 hour incubation from a given preparation were consistent from protein preparation to protein preparation. ThT fluorescence was normalized by averaging the duplicate samples and using (Fi – Fo)/(Fmax- Fo) were Fo was the ThT background intensity (fluorescence arbitrary unit), Fi was the ThT intensity of samples, and Fmax was the maximum ThT intensity of the reaction. Samples used to seed CsgA were sonicated using a sonic dismembrator (Fisher Model 100; Fisher, Pittsburg, PA) for three 15-second bursts on ice.
BIAcore Binding Assay
A BIAcore3000 (GE Appliance) was used to analyze the seeding reaction. Mature CsgB or CsgA fibers were sonicated (Sonicator XL2020, Misonix) on ice with 4 times of 15 second burst at power two and 50 second pause and diluted with 10 mM sodium acetate buffer, pH5 by 10 fold. Sonicated fibers were then immobilized on a CM5 chip (GE Appliance) pre-activated with 1:1 (v/v) mixture of 0.4 M of 1-ethyl-3-(3-dimethylpropyl)-carboiimide (EDC) and 0.1 M N-hydroxysuccinimide (NHS) to reach the immobilization of 3000 resonance units. A blank flow cell was prepared in absence of seeds. This flow cell was used as the negative control. Excessive reactive groups were deactivated with 1M ethanolamine-HCl, pH 8.5. To test interactions between monomeric CsgA and the CsgA or CsgB seeds, 40 μl 0.25 μM monomeric CsgA was injected over the sensor chip at a flow rate of 50 μl/minute and the response was recorded in resonance units.
Overlay assay
Cells were grown under curli inducing conditions as previously described for 24 hours on YESCA plates. Purified CsgB (10 ul) was pipetted onto the bacterial lawn and incubated at 26° C for an additional 24 hours. Congo red dye (10 mg/ml) was then applied to the bacterial lawn-CsgB mixture for 10 minutes. The dye was decanted and the bacterial lawns were carefully washed twice with 50 mM KPi buffer pH 7.2.
Transmission Electron Microscopy (TEM)
Samples (10 μL) were placed on glow discharged, Formvar-coated copper grids (Ernest F. Fullman, Inc., Latham, NY) for 2 minutes, washed twice briefly (10 seconds) with deionized water, and negatively stained with 2% uranyl acetate for 90 seconds. Samples were viewed using a Phillips CM-100 transmission electron microscope at 60kv.
SDS/PAGE and Western Blotting
Bacteria whole cell lysates and agar plugs were prepared and probed for both CsgA and CsgB by using previously described methods 34; 36.
Peptide preparation
Peptides were chemically synthesized (Proteintech Group, Chicago, IL). Purity was greater than 90% as measured by high pressure liquid chromatography, and size was confirmed by mass spectroscopy (Proteintech Group, Chicago, IL). Lyophilized peptide (1 mg) was dissolved in 8 M GndHCl (1ml) buffered by 50 mM KPi pH 7.2. The suspension was incubated at room temperature for 1 hour with rocking. In order to remove the 8 M GndHCl and collect the soluble peptide, the sample was passed through a Sephadex G10 (GE Healthcare, Brentwood, TN) column that was balanced in 50 mM KPi pH 7.2. The BCA assay was used to assess peptide concentration and polymerization was monitored by ThT.
Supplementary Material
Highlights.
The polymerization of wild type CsgB has not been biochemically characterized.
Wild type CsgB polymerizes with rapid kinetics.
The fourth and fifth repeating units of CsgB are required for in vivo function.
Synthetic peptide composed of the fifth repeating unit domain do not self assemble.
The rapid self assembly of CsgB is dependent upon a non-aggregative domain.
Acknowledgments
We would like to thank members of the Chapman and Scott Hultgren laboratories for helpful discussions and review of this manuscript. We also thank the Almqvist lab and Emma Andersson for technical help with the BIAcore. This work was supported by NIH AI 073847 and by the Umeå University Linnaeus Foundation.
Abbreviations
- FA
formic acid
- WT
wild type
- Gnd HCl
Guandine Hydrochloride
- TEM
Transmission electron microscopy
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Cohen FE, Kelly JW. Therapeutic approaches to protein-misfolding diseases. Nature. 2003;426:905–9. doi: 10.1038/nature02265. [DOI] [PubMed] [Google Scholar]
- 2.Chiti F, Dobson CM. Protein misfolding, functional amyloid, and human disease. Annu Rev Biochem. 2006;75:333–66. doi: 10.1146/annurev.biochem.75.101304.123901. [DOI] [PubMed] [Google Scholar]
- 3.Jarrett JT, Lansbury PT., Jr Seeding “one-dimensional crystallization” of amyloid: a pathogenic mechanism in Alzheimer’s disease and scrapie? Cell. 1993;73:1055–8. doi: 10.1016/0092-8674(93)90635-4. [DOI] [PubMed] [Google Scholar]
- 4.Lomakin A, Chung DS, Benedek GB, Kirschner DA, Teplow DB. On the nucleation and growth of amyloid beta-protein fibrils: detection of nuclei and quantitation of rate constants. Proc Natl Acad Sci U S A. 1996;93:1125–9. doi: 10.1073/pnas.93.3.1125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pedersen JS, Christensen G, Otzen DE. Modulation of S6 fibrillation by unfolding rates and gatekeeper residues. J Mol Biol. 2004;341:575–88. doi: 10.1016/j.jmb.2004.06.020. [DOI] [PubMed] [Google Scholar]
- 6.Serio TR, Cashikar AG, Kowal AS, Sawicki GJ, Moslehi JJ, Serpell L, Arnsdorf MF, Lindquist SL. Nucleated conformational conversion and the replication of conformational information by a prion determinant. Science. 2000;289:1317–21. doi: 10.1126/science.289.5483.1317. [DOI] [PubMed] [Google Scholar]
- 7.Uversky VN, Li J, Souillac P, Millett IS, Doniach S, Jakes R, Goedert M, Fink AL. Biophysical properties of the synucleins and their propensities to fibrillate: inhibition of alpha-synuclein assembly by beta- and gamma-synucleins. J Biol Chem. 2002;277:11970–8. doi: 10.1074/jbc.M109541200. [DOI] [PubMed] [Google Scholar]
- 8.Wang X, Smith DR, Jones JW, Chapman MR. In Vitro Polymerization of a Functional Escherichia coli Amyloid Protein. J Biol Chem. 2007;282:3713–9. doi: 10.1074/jbc.M609228200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rochet JC, Lansbury PT., Jr Amyloid fibrillogenesis: themes and variations. Curr Opin Struct Biol. 2000;10:60–8. doi: 10.1016/s0959-440x(99)00049-4. [DOI] [PubMed] [Google Scholar]
- 10.Chapman MR, Robinson LS, Pinkner JS, Roth R, Heuser J, Hammar M, Normark S, Hultgren SJ. Role of Escherichia coli curli operons in directing amyloid fiber formation. Science. 2002;295:851–5. doi: 10.1126/science.1067484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fowler DM, Koulov AV, Alory-Jost C, Marks MS, Balch WE, Kelly JW. Functional amyloid formation within mammalian tissue. PLoS Biol. 2006;4:e6. doi: 10.1371/journal.pbio.0040006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Serio TR, Lindquist SL. The yeast prion [PSI+]: molecular insights and functional consequences. Adv Protein Chem. 2001;59:391–412. doi: 10.1016/s0065-3233(01)59012-9. [DOI] [PubMed] [Google Scholar]
- 13.Wang X, Rochon M, Lamprokostopoulou A, Lunsdorf H, Nimtz M, Romling U. Impact of biofilm matrix components on interaction of commensal Escherichia coli with the gastrointestinal cell line HT-29. Cell Mol Life Sci. 2006;63:2352–63. doi: 10.1007/s00018-006-6222-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Barnhart MM, Chapman MR. Curli biogenesis and function. Annu Rev Microbiol. 2006;60:131–47. doi: 10.1146/annurev.micro.60.080805.142106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gerstel U, Romling U. The csgD promoter, a control unit for biofilm formation in Salmonella typhimurium. Res Microbiol. 2003;154:659–67. doi: 10.1016/j.resmic.2003.08.005. [DOI] [PubMed] [Google Scholar]
- 16.Olsen A, Jonsson A, Normark S. Fibronectin binding mediated by a novel class of surface organelles on Escherichia coli. Nature. 1989;338:652–5. doi: 10.1038/338652a0. [DOI] [PubMed] [Google Scholar]
- 17.Uhlich GA, Cooke PH, Solomon EB. Analyses of the red-dry-rough phenotype of an Escherichia coli O157:H7 strain and its role in biofilm formation and resistance to antibacterial agents. Appl Environ Microbiol. 2006;72:2564–72. doi: 10.1128/AEM.72.4.2564-2572.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Vidal O, Longin R, Prigent-Combaret C, Dorel C, Hooreman M, Lejeune P. Isolation of an Escherichia coli K-12 mutant strain able to form biofilms on inert surfaces: involvement of a new ompR allele that increases curli expression. J Bacteriol. 1998;180:2442–9. doi: 10.1128/jb.180.9.2442-2449.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cegelski L, Marshall GR, Eldridge GR, Hultgren SJ. The biology and future prospects of antivirulence therapies. Nat Rev Microbiol. 2008;6:17–27. doi: 10.1038/nrmicro1818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Clatworthy AE, Pierson E, Hung DT. Targeting virulence: a new paradigm for antimicrobial therapy. Nat Chem Biol. 2007;3:541–8. doi: 10.1038/nchembio.2007.24. [DOI] [PubMed] [Google Scholar]
- 21.Lee YM, Almqvist F, Hultgren SJ. Targeting virulence for antimicrobial chemotherapy. Curr Opin Pharmacol. 2003;3:513–9. doi: 10.1016/j.coph.2003.04.001. [DOI] [PubMed] [Google Scholar]
- 22.Cegelski L, Pinkner JS, Hammer ND, Cusumano CK, Hung CS, Chorell E, Aberg V, Walker JN, Seed PC, Almqvist F, Chapman MR, Hultgren SJ. Small-molecule inhibitors target Escherichia coli amyloid biogenesis and biofilm formation. Nat Chem Biol. 2009;5:913–9. doi: 10.1038/nchembio.242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chorell E, Bengtsson C, Banchelin TS, Das P, Uvell H, Sinha AK, Pinkner JS, Hultgren SJ, Almqvist F. Synthesis and application of a bromomethyl substituted scaffold to be used for efficient optimization of anti-virulence activity. Eur J Med Chem. 2011;46:1103–1116. doi: 10.1016/j.ejmech.2011.01.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Horvath I, Weise CF, Andersson EK, Chorell E, Sellstedt M, Bengtsson C, Olofsson A, Hultgren SJ, Chapman M, Wolf-Watz M, Almqvist F, Wittung-Stafshede P. Mechanisms of Protein Oligomerization: Inhibitor of Functional Amyloids Templates alpha-Synuclein Fibrillation. J Am Chem Soc. 2012 doi: 10.1021/ja209829m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bian Z, Normark S. Nucleator function of CsgB for the assembly of adhesive surface organelles in Escherichia coli. Embo J. 1997;16:5827–36. doi: 10.1093/emboj/16.19.5827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hammar M, Bian Z, Normark S. Nucleator-dependent intercellular assembly of adhesive curli organelles in Escherichia coli. Proc Natl Acad Sci U S A. 1996;93:6562–6. doi: 10.1073/pnas.93.13.6562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.White AP, Collinson SK, Banser PA, Gibson DL, Paetzel M, Strynadka NC, Kay WW. Structure and characterization of AgfB from Salmonella enteritidis thin aggregative fimbriae. J Mol Biol. 2001;311:735–49. doi: 10.1006/jmbi.2001.4876. [DOI] [PubMed] [Google Scholar]
- 28.Hammar M, Arnqvist A, Bian Z, Olsen A, Normark S. Expression of two csg operons is required for production of fibronectin- and congo red-binding curli polymers in Escherichia coli K-12. Mol Microbiol. 1995;18:661–70. doi: 10.1111/j.1365-2958.1995.mmi_18040661.x.. [DOI] [PubMed] [Google Scholar]
- 29.Loferer H, Hammar M, Normark S. Availability of the fibre subunit CsgA and the nucleator protein CsgB during assembly of fibronectin-binding curli is limited by the intracellular concentration of the novel lipoprotein CsgG. Mol Microbiol. 1997;26:11–23. doi: 10.1046/j.1365-2958.1997.5231883.x. [DOI] [PubMed] [Google Scholar]
- 30.Wang X, Chapman MR. Sequence determinants of bacterial amyloid formation. J Mol Biol. 2008;380:570–80. doi: 10.1016/j.jmb.2008.05.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wang X, Hammer ND, Chapman MR. The molecular basis of functional bacterial amyloid polymerization and nucleation. J Biol Chem. 2008;283:21530–9. doi: 10.1074/jbc.M800466200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Collinson SK, Parker JM, Hodges RS, Kay WW. Structural predictions of AgfA, the insoluble fimbrial subunit of Salmonella thin aggregative fimbriae. J Mol Biol. 1999;290:741–56. doi: 10.1006/jmbi.1999.2882. [DOI] [PubMed] [Google Scholar]
- 33.Shewmaker F, McGlinchey RP, Thurber KR, McPhie P, Dyda F, Tycko R, Wickner RB. The functional curli amyloid is not based on in-register parallel beta-sheet structure. J Biol Chem. 2009;284:25065–76. doi: 10.1074/jbc.M109.007054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hammer ND, Schmidt JC, Chapman MR. The curli nucleator protein, CsgB, contains an amyloidogenic domain that directs CsgA polymerization. Proc Natl Acad Sci U S A. 2007;104:12494–9. doi: 10.1073/pnas.0703310104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nenninger AA, Robinson LS, Hammer ND, Epstein EA, Badtke MP, Hultgren SJ, Chapman MR. CsgE is a curli secretion specificity factor that prevents amyloid fibre aggregation. Mol Microbiol. 2011;81:486–99. doi: 10.1111/j.1365-2958.2011.07706.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Robinson LS, Ashman EM, Hultgren SJ, Chapman MR. Secretion of curli fibre subunits is mediated by the outer membrane-localized CsgG protein. Mol Microbiol. 2006;59:870–81. doi: 10.1111/j.1365-2958.2005.04997.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bucciantini M, Giannoni E, Chiti F, Baroni F, Formigli L, Zurdo J, Taddei N, Ramponi G, Dobson CM, Stefani M. Inherent toxicity of aggregates implies a common mechanism for protein misfolding diseases. Nature. 2002;416:507–11. doi: 10.1038/416507a. [DOI] [PubMed] [Google Scholar]
- 38.Larson J, Lynch G, Games D, Seubert P. Alterations in synaptic transmission and long-term potentiation in hippocampal slices from young and aged PDAPP mice. Brain Res. 1999;840:23–35. doi: 10.1016/s0006-8993(99)01698-4. [DOI] [PubMed] [Google Scholar]
- 39.Lue LF, Kuo YM, Roher AE, Brachova L, Shen Y, Sue L, Beach T, Kurth JH, Rydel RE, Rogers J. Soluble amyloid beta peptide concentration as a predictor of synaptic change in Alzheimer’s disease. Am J Pathol. 1999;155:853–62. doi: 10.1016/s0002-9440(10)65184-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Malisauskas M, Ostman J, Darinskas A, Zamotin V, Liutkevicius E, Lundgren E, Morozova-Roche LA. Does the cytotoxic effect of transient amyloid oligomers from common equine lysozyme in vitro imply innate amyloid toxicity? J Biol Chem. 2005;280:6269–75. doi: 10.1074/jbc.M407273200. [DOI] [PubMed] [Google Scholar]
- 41.McLean CA, Cherny RA, Fraser FW, Fuller SJ, Smith MJ, Beyreuther K, Bush AI, Masters CL. Soluble pool of Abeta amyloid as a determinant of severity of neurodegeneration in Alzheimer’s disease. Ann Neurol. 1999;46:860–6. doi: 10.1002/1531-8249(199912)46:6<860::aid-ana8>3.0.co;2-m. [DOI] [PubMed] [Google Scholar]
- 42.Moechars D, Dewachter I, Lorent K, Reverse D, Baekelandt V, Naidu A, Tesseur I, Spittaels K, Haute CV, Checler F, Godaux E, Cordell B, Van Leuven F. Early phenotypic changes in transgenic mice that overexpress different mutants of amyloid precursor protein in brain. J Biol Chem. 1999;274:6483–92. doi: 10.1074/jbc.274.10.6483. [DOI] [PubMed] [Google Scholar]
- 43.Sirangelo I, Malmo C, Iannuzzi C, Mezzogiorno A, Bianco MR, Papa M, Irace G. Fibrillogenesis and cytotoxic activity of the amyloid-forming apomyoglobin mutant W7FW14F. J Biol Chem. 2004;279:13183–9. doi: 10.1074/jbc.M308207200. [DOI] [PubMed] [Google Scholar]
- 44.Sousa MM, Cardoso I, Fernandes R, Guimaraes A, Saraiva MJ. Deposition of transthyretin in early stages of familial amyloidotic polyneuropathy: evidence for toxicity of nonfibrillar aggregates. Am J Pathol. 2001;159:1993–2000. doi: 10.1016/s0002-9440(10)63050-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wang J, Dickson DW, Trojanowski JQ, Lee VM. The levels of soluble versus insoluble brain Abeta distinguish Alzheimer’s disease from normal and pathologic aging. Exp Neurol. 1999;158:328–37. doi: 10.1006/exnr.1999.7085. [DOI] [PubMed] [Google Scholar]
- 46.Bokvist M, Lindstrom F, Watts A, Grobner G. Two types of Alzheimer’s beta-amyloid (1–40) peptide membrane interactions: aggregation preventing transmembrane anchoring versus accelerated surface fibril formation. J Mol Biol. 2004;335:1039–49. doi: 10.1016/j.jmb.2003.11.046. [DOI] [PubMed] [Google Scholar]
- 47.Cecchi C, Baglioni S, Fiorillo C, Pensalfini A, Liguri G, Nosi D, Rigacci S, Bucciantini M, Stefani M. Insights into the molecular basis of the differing susceptibility of varying cell types to the toxicity of amyloid aggregates. J Cell Sci. 2005;118:3459–70. doi: 10.1242/jcs.02473. [DOI] [PubMed] [Google Scholar]
- 48.Kayed R, Head E, Thompson JL, McIntire TM, Milton SC, Cotman CW, Glabe CG. Common structure of soluble amyloid oligomers implies common mechanism of pathogenesis. Science. 2003;300:486–9. doi: 10.1126/science.1079469. [DOI] [PubMed] [Google Scholar]
- 49.Stefani M, Dobson CM. Protein aggregation and aggregate toxicity: new insights into protein folding, misfolding diseases and biological evolution. J Mol Med. 2003;81:678–99. doi: 10.1007/s00109-003-0464-5. [DOI] [PubMed] [Google Scholar]
- 50.Bieschke J, Herbst M, Wiglenda T, Friedrich RP, Boeddrich A, Schiele F, Kleckers D, Lopez Del Amo JM, Gruning BA, Wang Q, Schmidt MR, Lurz R, Anwyl R, Schnoegl S, Fandrich M, Frank RF, Reif B, Gunther S, Walsh DM, Wanker EE. Small-molecule conversion of toxic oligomers to nontoxic beta-sheet-rich amyloid fibrils. Nat Chem Biol. 2011;8:93–101. doi: 10.1038/nchembio.719. [DOI] [PubMed] [Google Scholar]
- 51.Shorter J, Lindquist S. Hsp104 catalyzes formation and elimination of self-replicating Sup35 prion conformers. Science. 2004;304:1793–7. doi: 10.1126/science.1098007. [DOI] [PubMed] [Google Scholar]
- 52.Wang X, Zhou Y, Ren JJ, Hammer ND, Chapman MR. Gatekeeper residues in the major curlin subunit modulate bacterial amyloid fiber biogenesis. Proc Natl Acad Sci U S A. 107:163–8. doi: 10.1073/pnas.0908714107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Nenninger AA, Robinson LS, Hultgren SJ. Localized and efficient curli nucleation requires the chaperone-like amyloid assembly protein CsgF. Proc Natl Acad Sci U S A. 2009;106:900–5. doi: 10.1073/pnas.0812143106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ramsook CB, Tan C, Garcia MC, Fung R, Soybelman G, Henry R, Litewka A, O’Meally S, Otoo HN, Khalaf RA, Dranginis AM, Gaur NK, Klotz SA, Rauceo JM, Jue CK, Lipke PN. Yeast cell adhesion molecules have functional amyloid-forming sequences. Eukaryot Cell. 2010;9:393–404. doi: 10.1128/EC.00068-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Otoo HN, Lee KG, Qiu W, Lipke PN. Candida albicans Als adhesins have conserved amyloid-forming sequences. Eukaryot Cell. 2008;7:776–82. doi: 10.1128/EC.00309-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Romero D, Vlamakis H, Losick R, Kolter R. An accessory protein required for anchoring and assembly of amyloid fibres in B. subtilis biofilms. Mol Microbiol. 2011;80:1155–68. doi: 10.1111/j.1365-2958.2011.07653.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Romero D, Aguilar C, Losick R, Kolter R. Amyloid fibers provide structural integrity to Bacillus subtilis biofilms. Proc Natl Acad Sci U S A. 2010;107:2230–4. doi: 10.1073/pnas.0910560107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kayed R, Sokolov Y, Edmonds B, McIntire TM, Milton SC, Hall JE, Glabe CG. Permeabilization of lipid bilayers is a common conformation-dependent activity of soluble amyloid oligomers in protein misfolding diseases. J Biol Chem. 2004;279:46363–6. doi: 10.1074/jbc.C400260200. [DOI] [PubMed] [Google Scholar]
- 59.de Jong W, Wosten HA, Dijkhuizen L, Claessen D. Attachment of Streptomyces coelicolor is mediated by amyloidal fimbriae that are anchored to the cell surface via cellulose. Mol Microbiol. 2009;73:1128–40. doi: 10.1111/j.1365-2958.2009.06838.x. [DOI] [PubMed] [Google Scholar]
- 60.Casadaban MJ. Transposition and fusion of the lac genes to selected promoters in Escherichia coli using bacteriophage lambda and Mu. J Mol Biol. 1976;104:541–55. doi: 10.1016/0022-2836(76)90119-4. [DOI] [PubMed] [Google Scholar]
- 61.Baba T, Ara T, Hasegawa M, Takai Y, Okumura Y, Baba M, Datsenko KA, Tomita M, Wanner BL, Mori H. Construction of Escherichia coli K-12 in-frame, single-gene knockout mutants: the Keio collection. Mol Syst Biol. 2006;2:2006 0008. doi: 10.1038/msb4100050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Goldberg RB, Bender RA, Streicher SL. Direct selection for P1-sensitive mutants of enteric bacteria. J Bacteriol. 1974;118:810–4. doi: 10.1128/jb.118.3.810-814.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.