Abstract
The C-terminal domain (CTD) of eukaryotic RNA polymerase II is an essential regulator for RNA polymerase II-mediated transcription. It is composed of multiple repeats of a consensus sequence Tyr1Ser2Pro3Thr4Ser5Pro6Ser7. Ser2 and Ser5 are the major phosphorylation sites in vivo while Pro3 and Pro6 can adopt either cis or trans conformations. CTD regulation of transcription is mediated both by phosphorylation of the serines and prolyl isomerization of the two prolines. Interestingly, the phosphorylation sites are typically close to prolines, thus the conformation of the adjacent proline could impact the specificity of the corresponding kinases and phosphatases.
Experimental evidence of cross-talk between these two regulatory mechanisms has been elusive. Pin1 is a highly conserved phosphorylation-specific peptidyl-prolyl isomerase (PPIase) that recognizes the phospho-Ser/Thr (pSer/Thr)-Pro motif with CTD as one of its primary substrates in vivo. In the present study, we provide structural snapshots and kinetic evidence that support the concept of cross-talk between prolyl isomerization and phosphorylation. We determined the structures of Pin1 bound with two substrate isosteres that mimic peptides containing pSer/Thr-Pro motifs in cis or trans conformations. The results unequivocally demonstrate the utility of both cis- and trans-locked alkene isosteres as close geometric mimics of peptides bound to a protein target. Building on this result, we identified a specific case in which Pin1 differentially affects the rate of dephosphorylation catalyzed by two phosphatases (Scp1 and Ssu72) that target the same serine residue in the CTD heptad repeat but that have different preferences for the isomerization state of the adjacent proline residue. These data exemplify for the first time how modulation of proline isomerization can kinetically impact signal transduction in transcription regulation.
The unique chemical structure of the proline residue makes it the only amino acid enabling the Xaa-Pro peptide bond (where Xaa is any amino acid residue) to adopt a cis conformation to any significant extent (10–30%).1 The transition between cis and trans conformations of the prolyl peptide bond occurs at a slow rate, and phosphorylation of the serine or threonine preceding the proline (pSer/Thr-Pro) can further slow the transition.2 Since protein kinases or phosphatases are specific for the proline isomeric state,3 it is possible that kinases and phosphatases could recognize the same Ser/Thr position but with different preferences for the isomerization state of the adjacent Pro. In other words, since proline isomerases can be dependent on the phosphorylated state, these enzymes may act as molecular 'switches' that govern the downstream recognition and kinetics of phosphatases.
To the extent this hypothesis is true, the conformation of the peptide bond should impact the substrate recognition of at least some modifying enzymes. Pin1 is a highly conserved peptidyl-prolyl isomerase (PPIase) that specifically recognizes the pSer/Thr-Pro motif and catalyzes faster transition between the two isomeric states, and thereby regulates protein functions.4 The isomerization of the pSer/Thr-Pro motif mediated by Pin1 is especially important for biological processes e.g. cancer and neurodegenerative diseases such as Alzheimer’s.4, 5 In humans, one of the most significant substrates of Pin1 is RNA polymerase II, the central molecule for eukaryotic transcription.6 The signature motif recognized by Pin1 is highly enriched in the C-terminal domain (CTD) of RNA polymerase II, which consists of 26–52 tandem heptapeptide repeats with the general consensus sequence from yeast to human, Tyr1Ser2Pro3Thr4Ser5Pro6Ser7.7 CTD phosphorylation is a major mechanism by which cells regulate gene expression, with serines at positions 2 and 5 as the primary phosphorylation sites.8 The conformational states of the prolines in the CTD also represent a critical regulatory checkpoint for transcription.9–11 By adjusting the cis-trans conformation of a proline adjacent to a phosphorylated serine, the interaction of the CTD and the binding partners it recruits can be modulated.12, 13 These CTD binding proteins are involved in a variety of processes during the transcription. However, nearly all the complex structures of CTD-binding proteins and the CTD peptides solved so far contain the pSer-Pro motif in trans conformation.14 Some examples of these CTD-binding proteins included Pcf11,15 a subunit of yeast cleavage and polyadenylation factor I, and mRNA capping enzyme Cgt1.16
Substantial evidence has been accumulated that Pin1 modulates the dephosphorylation of the CTD of RNA polymerase II.11, 13, 17, 18 Congruent with the hypothesis set out above, in the absence of Pin1-catalyzed cis/trans isomerization, a phosphatase might not be able to 'undo' the phosphorylation catalyzed by the kinase, even though they recognize the same Ser/Thr. In other words, Pin1 might significantly affect the steady-state phosphorylation level of a protein even though it has neither kinase nor phosphatase activity. Notably, the function of Pin1 itself is tightly regulated in normal tissues on both expression level and post-translational modification level,19 indicating Pin1 as a regulatory switch of the isomeric states of pSer/Thr-Pro in signal transduction.
About a dozen CTD-specific kinases have been identified and characterized,20 but CTD phosphatases are understudied. Recently, our lab and others, have structurally characterized two CTD-specific phosphatases: small CTD phosphatase 1 (Scp1)21–23 and Ssu7224, and their interactions with phosphorylated CTD.25, 26 Scp1 was identified as a neuronal gene suppressor in non-neuronal cells as well as neuronal stem cells,21 where it epigenetically regulates the expression of a subset of genes. Scp1 is the first structurally characterized CTD-specific phosphatase with its substrate bound in humans.21–23 In our high-resolution structure of the Scp1-CTD complex (PDB code 2ght), it is obvious that pSer5 is the site that undergoes dephosphorylation, while the pSer2 side chain extends outwards from the protein surface, even though it is also phosphorylated. The Pro3 and Pro6 are both in the trans conformation, and the conversion from trans to cis would make the chain clash sterically with the protein unless dramatic conformational changes occurred.27 Ssu72, on the other hand, has been recognized as a housekeeping gene that is pivotal to the general transcription cycle. It exhibits substrate specificity toward the cis conformation of its bound CTD peptide.25, 26 It recognizes pSer5-Pro6 motif only if Pro6 adopts the cis conformation.26 Based on the structure, the high energy cis-proline substrate can be stabilized in part by the intramolecular hydrogen bond between the hydroxyl side chain of Thr4 and the carbonyl group of Pro6.25 This is surprising since all previously identified CTD binding proteins are trans specific. Since the cis conformation only accounts for ~20% of the peptide, the unambiguous observation in crystal structures of the cis conformation binding in Ssu72 indicates selectivity, rather than an averaging effect of crystallography. The structures strongly suggest that the cis or trans conformations of prolines upstream or downstream of a pSer site in the CTD can directly determine if the specific site can be subject to the dephosphorylation by the CTD phosphatases.
We therefore hypothesized that prolyl isomerization has a substantial impact on dephosphorylation rates by changing the suitability of CTD as the substrate for CTD phosphatases, and thus affects the phosphorylation patterns of the CTD.28 In particular, Scp1 and Ssu72 might respond differently to the Pin1-mediated prolyl isomerization. However, structural and kinetic evidence were lacking. In order to test this hypothesis, we determined the structures of two conformational-locked alkene isosteres bound to Pin1 that mimic the two endpoints of the isomerization reaction: pSer-cis-Pro and pSer-trans-Pro.29 These structures not only provide insight into how Pin1 recognizes its substrates, but also demonstrate unequivocally the utility of both cis- and trans-locked alkene isosteres as close geometric mimics of peptide bonds bound to a protein target. We further show that cis-specific phosphatase Ssu72 was highly activated by Pin1 activity, but that trans-specific phosphatase Scp1 was not affected substantially. These data illustrate how the ability of Pin1 to 'switch' the cis and trans conformation of its substrates may have significant implications for the regulation of RNA polymerase II-mediated transcription.
Results
The binding of cis and trans isosteric compounds to Pin1
In order to promote Pin1 crystals to endure prolonged chemical soaking, it was necessary to engineer Pin1 using an entropy reduction strategy.30 R14A mutation of Pin1 has been shown to dramatically stabilize the protein crystal, yet it has little impact on the PPIase activity or WW domain binding on the substrates of Pin1.31 The R14A mutant also shows identical binding modes as wild-type protein when bound to high-affinity inhibitor Ac-L-Phe-D-pThr-L-Pipecolic acid-L-Naphthylalanine-L-Gln-NH2,(D-PEPTIDE).30 Therefore, in our current investigation, crystals of this mutant were used to soak with cis and trans isosteric compounds. This strategy was extremely effective and the crystals diffracted to 2.1 and 2.3 Å on an in-house X-ray source for cis and trans complexes respectively.
The overall structure of Pin1 is highly consistent with previously reported structures. Briefly, human Pin1 has two distinctive domains, a WW domain that recognizes the signature motif pSer/Thr-Pro, and a PPIase domain that catalyzes the reaction of the prolyl-peptide. The linker between the two domains is highly flexible and disordered in all of the structures of Pin1 published thus far (Supplementary Figure 1).30, 32, 33 The flexible nature of the linker is inherited throughout the Pin1 family. The mobility of the linker and the inter-domain movement is proposed to be an integral regulatory mechanism for the communication between WW and PPIase domains and essential for the biological function of the protein.34, 35 A PEG400 molecule, used as additive in the crystallization buffer, was found as usual in the groove between the two domains. This PEG400 molecule stabilizes the mobility between the two domains and enables the crystallization of Pin1 molecules. The only exception is the structure of Pin1 with the phosphoryl-peptide derived from the CTD of RNA polymerase II, in which case the peptide replaced the PEG molecule (PDB code: 1f8a).33
The complex structures show that both cis and trans alkene compounds (Figure 1) bind to the PPIase domain of the Pin1. The structures indicate that Pin1 recognizes both cis and trans substrates in very similar conformations, even though the mode of the proline 5-membered ring analogue bound to the proline binding pocket is slightly different. The PPIase domain has two distinctive binding areas for each residue of the signature motif, pSer/Thr-Pro. Three essential residues of the PPIase domain, Lys63, Arg68 and Arg69, form a positive triad pocket that specifically binds to the phosphate group (Supplementary Figure 1). The elimination of any of these residues greatly diminishes the activity of the enzyme but does not totally abolish the isomerization activity.31 However, eliminating two out of the three residues reduces the activity of the enzyme to an undetectable level.31 These residues, embracing the phosphate group of the substrate with electrostatic interactions, form a roomy and elastic pocket that can accommodate “rolling” of the phosphate. When not occupied, the positive triad loop preserves an open conformation that can close up upon inhibitor binding (Supplementary Figure 2).33 A hydrophobic pocket that binds to the proline residue of the substrate is also accountable for the unique selectivity of Pin1 isomerase. This greasy pocket highly prefers hydrophobic residues like proline. When a proline-containing compound is not provided in solution, density from additives sometimes can be found in the crystal structure, most likely due to non-specific binding. This implies a strong preference for hydrophobic interactions in this binding area and provides clues for inhibitor design.30
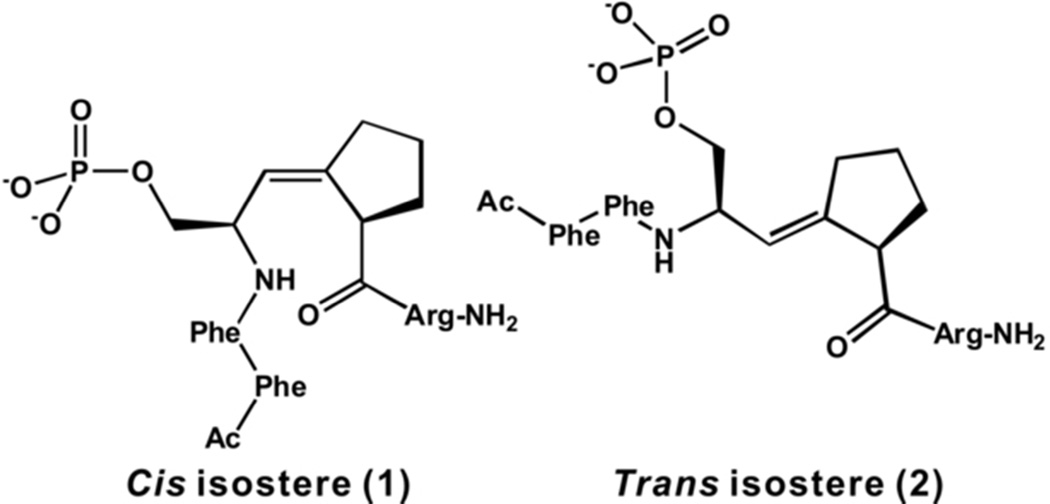
Figure 1.
Chemical structures of the cis and trans peptidomimetic inhibitors of Pin1: cis isostere (1) and trans isostere (2).
The Pin1 R14A-cis-isostere complex structure
The cis isosteric inhibitor binds to the PPIase domain, as predicted based on the kinetic results that the compound is a competitive PPIase inhibitor.29 Three residues of the inhibitor are ordered and modeled in the density at the active site (Figure 2a). Even though it has been proposed that the presence of hydrophobic residues N-terminal to the signature motif can enhance binding,6 the two phenylalanine residues in the cis peptidomimetics are not visible in our structure. This is similar to another complex structure of Pin1 (PDB code: 2itk) with high affinity peptide inhibitor (Ki of 19 nM) where N-terminal hydrophobic residue (Phe) immediately preceding the pSer/Thr-Pro motif (PDB code: 2itk) was also disordered.30 The consistent lack of order in these amino acids suggests that the residue(s) N-terminal to the pSer/Thr-Pro motif might contribute very little structurally to the tight binding between Pin1 and those inhibitors. We took advantage of this observation in our ensuing design of a reduced amide inhibitor of Pin1, Ac–pSer–Ψ[CH2N]-Pro–tryptamine.36 In this case, no hydrophobic residues were placed in front of the pSer, the resultant compound exhibited much improved solubility that allowed structural determination.
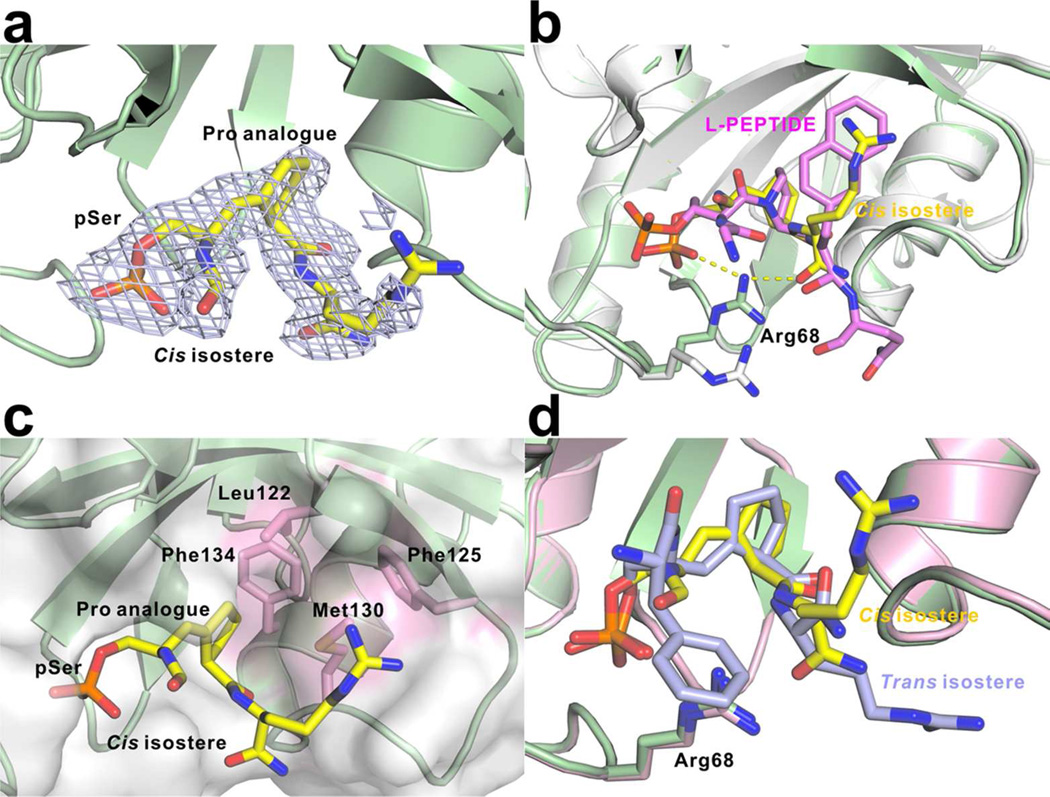
Figure 2.
Complex structures of Pin1 bound with cis or trans isosteres. (a) Electron density map (2Fo-Fc) of cis isostere contoured at 1σ. (b) Superimposition of Pin1 bound with cis isostere (yellow) and L-PEPTIDE (magenta, PDB code: 2q5a). Pin1 bound with cis isostere is shown in pale green, and the Pin bound with L-PEPTIDE is shown in white. Arg68 in both structures is shown as sticks. The yellow dashed lines indicate the hydrogen bonds. (c) Hydrophobic pocket (pink surface) which recognizes Pro analogue of the peptidomimetic inhibitor. The key hydrophobic residues are shown in pink as sticks. (d) Superimposition of Pin1 bound with cis (yellow) and trans (light blue) isosteres.
One interesting aspect of our structure is the conformation of Arg68 of Pin1, whose side chain was disordered in the previous PPIase complex structure.30 The loop containing the positively charged triad (Arg68, Arg69 and Lys63) forms favorable electrostatic interactions with phosphate group, stabilizing the interaction between protein and peptide inhibitor. The loop can adopt two dramatically different conformations, a closed conformation when a negatively charged group, such as inorganic sulfate or a phosphate group from substrate or substrate analogues, occupies the active site, and an open conformation when the site is unoccupied. Compared with the closed conformation, the tip of the loop swings 23.5 Å away when the pocket is empty (Supplementary Figure 2). Furthermore, even in the closed conformation, the side chain of Arg68 is highly flexible in different structures, and it is usually totally disordered. However, in this pair of structures of Pin1 with substrate analogues, Arg68 covers the entrance of the active site cavity and provides a lid with hydrogen bonds to the amides of both inhibitors’ C-terminal arginine residues and phosphates (Figure 2b). The position of the Arg68 side chain is also very close to the alkene bond that mimics the peptidyl-prolyl bond in the substrate. The highly mobile Arg68 side chain enables a very elastic binding pocket for phosphate, and permits the rotation of the phosphate group upon isomerization.
The proline-binding pocket, on the other hand, is less flexible and favors certain positioning of proline over others. This pocket is composed of Phe125, Phe134, Met130 and Leu122 (Figure 2c), which provides hydrophobic interactions with the proline-mimic moiety of the inhibitors. Hydrophobic interactions usually grant high-affinity binding for inhibitors. For example, rapamycin presents a 0.2 nM Kd toward another PPIase, FKBP12 of the FK506 binding protein (FKBP) family, yet all the strong interactions are driven by hydrophobic interactions with only one potential hydrogen bond between the ligand and the enzyme (PDB code: 2dg3). Exploitation of the proline-binding pocket in Pin1 will help us to design inhibitors for human Pin1 with stronger affinity and selectivity. Indeed, the high affinity of D- and L-PEPTIDE is at least partially attributed to the exchange of proline residue by a 6-membered ring analogue of proline, pipecolic acid.30 The natural substrate of Pin1 at this site, proline, actually has relatively weak binding to the PPIase domain, which is mimicked by the position of the proline analogue in our structure (Figure 2c). Instead, the WW domain shows a strong affinity for pSer/Thr-Pro sequences, and may function as a recruiter for natural substrates. Pin1 PPIase only binds substrate with Kd in millimolar range, therefore it is believed that the PPIase domain can only take the substrate after the WW domain targets the protein to the substrate.31, 32 This dual mode of action has been the center of investigation for human Pin1 function,35, 37 but how the two domains communicate and coordinate catalysis remains to be elucidated.
The Pin1 R14A-trans-isostere complex structure
The structure of Pin1 bound to the inhibitor mimicking trans-proline exhibits a very similar conformation as its cis counterpart (Figure 2d). Consistently, Arg68 is ordered in this structure and covers the active site entrance. However, one interesting distinction from the cis complexes is the much weaker density at the alkene bond that mimics the prolyl peptide bond even though the densities of the compound at the phosphate binding site and proline binding pocket are rather strong. Considering that both complexes were obtained with similar amounts of soaking time at similar resolutions, this suggests that the isosteric bond is more ordered in the cis compound compared to the trans. The Ki of the trans compound is 23-fold higher than that of the cis compound, possibly due to the different binding modes of the proline residues as restricted by the cis or trans conformation. Alternatively, it is possible that the binding of the trans conformation of carbocyclic proline analogue exerts strain upon binding to Pin1, as evidenced by the protein dynamics measured by NMR.35 In contrast, the cis conformation of the alkene bond introduces less distortion, resulting in more favorable binding.
Impacts on CTD dephosphorylation mediated by Scp1 and Ssu72
Even though both cis- and trans-proline are suitable substrates for Pin1, as mimicked in our structures, the impact of isomerase activity is not the same on different enzymes recognizing different prolines. Since cis-proline is only a minor component in naturally occurring proteins, enzymes recognizing cis-proline as substrate will have their substrate pool greatly affected by Pin1 activity. Phosphatases targeting the same substrate sequence motif, but requiring different proline isomers, represent the best system to test this. Scp1 and Ssu72 are eukaryotic phosphatases recognizing the Ser5 position of the CTD. However, their complex structures suggest that Scp1 and Ssu72 prefer different proline conformations. We have examined how Pin1 activity affects the phosphatase activity of these enzymes.
In order to investigate whether Pin1 can regulate the activity of CTD-specific phosphatases, we tested the dephosphorylation of a CTD-derived peptide using the malachite green assay. This peptide includes four repeats, (YSPTpSPS)4, with all four Ser5 phosphorylated. Previously, Ssu72 has been shown to be more active upon the addition of yeast homologue of Pin1, Ess1.25 We asked whether similar effects would be observed when human Pin1 was used. The isomerization effects of Pin1 on another CTD phosphatase, transcription factor IIF-interacting CTD-phosphatase 1 (Fcp1), have been controversial when both activation with human Pin138 and inhibition with yeast homologue Ess117 have been observed. We reasoned that the discrepancy comes from the different recognition sites for Fcp1 and Pin1. Pin1 binds the Ser5 of the CTD33 whereas Fcp1 highly favors Ser2, and only binds Ser5 weakly.39 When the same recognition motif is being recognized, the effect of the PPIase towards isomer-specific phosphatases should be more consistent between the yeast and human versions of the PPIase.
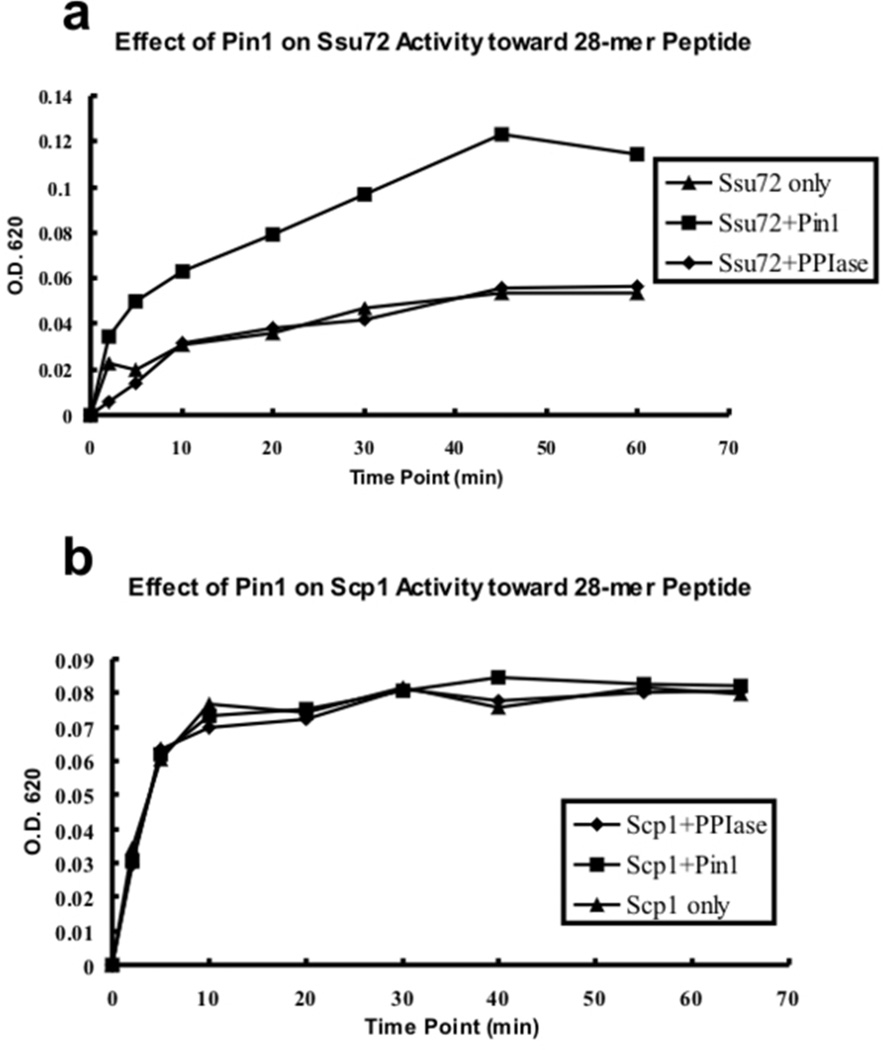
In this study, we used Drosophila Ssu72 to test how Pin1 affects its activity. Drosophila Ssu72 shares 60% identity with human Ssu72, and structural conservation of 0.56 Å in the main chain.24 The active site superimposes perfectly between the Drosophila and human counterparts (Supplementary Figure 3). The Drosophila version of Ssu72 has much higher thermostability, making it a better version to use for the kinetic experiments. Consistent with prior reports,25, 26 Ssu72 is activated upon Pin1 addition (Figure 3a) by about 3-fold. This result is consistent with a scenario in which Pin1 quickly converts the trans-Pro to cis-Pro, and by doing so, makes the cis-trans ratio reach equilibrium much faster than the uncatalyzed auto-conversion. Therefore, the consumable substrate concentration for Ssu72 was increased in the presence of Pin1, and led to the apparent increased activity of Ssu72. Such effect is specifically caused by the prolyl isomerase activity of Pin1, because when we used a truncated version of Pin1, PPIase domain, which cannot target CTD substrate to PPIase active site,33 the activation effect is lost (Figure 3a).
Figure 3.
Effect of human Pin1 on the activities of Drosophila Ssu72 (a) and human Scp1 (b) phosphatases. The activities of both phosphatases toward a 28-mer peptide [sequence: (YSPTpSPS)4] were measured using malachite green assay. The Pin1 is wild-type full length protein and PPIase domain is a truncated version of Pin1 with residues 51–163. (a) The reaction (20 µL total volume) for Ssu72 was carried out in buffer containing 100 ng of Ssu72, 20 µM of peptide, 100 mM MES pH 6.5, and 10 ng of Pin1 or PPIase domain. (b) The reaction of Scp1 was performed in the buffer containing 5 ng of Scp1, 10 µM of peptide, 50 mM Tris-acetate pH 5.5, 10 mM MgCl2 and 10 ng of Pin1 or the PPIase domain. The reactions were quenched by adding 40 µL of malachite green reagent at different time points. The release of inorganic phosphate was detected by measuring the absorbance at 620 nm.
It should be noted that the fraction of peptide that was dephosphorylated by Ssu72 (~1/3) consists of both substrate that was in cis-conformation initially (estimated to be ~20%26), and substrate that was auto-converted to cis-conformation during the process of the reaction. The difference between the reactions with and without Pin1 is caused by the effect of Pin1 'outracing' trans-to-cis auto-conversion.
Combined with a previous experiment that a catalytically impaired mutant of yeast homologue of Pin1, Ess1, cannot activate Ssu72,25 our result shows that it is the isomerization of the CTD that promotes the enhanced phosphatase activity of Ssu72, rather than stabilization of Ssu72 protein or reducing the non-specific adsorption of Ssu72 protein to the test tube. Furthermore, we ruled out the possibility that Pin1 activates Ssu72 by physically interacting with Ssu72. Firstly, Pin1 specifically recognizes a Ser/Thr-Pro motif in its substrates only when the Ser/Thr is phosphorylated.5 However, Ssu72 contains no pSer/Thr-Pro motif in its primary sequence. Secondly, we tested whether Pin1 and Ssu72 can directly interact with each other to form a stable complex using gel filtration chromatography. In this experiment, roughly equal amount of Pin1 and Ssu72 (~300 µM each) were mixed together and incubated at 4 °C for 6 hr. The mixture was then loaded on Superdex 75 column (GE Healthcare). No peak corresponding to a possible Pin1-Ssu72 complex was observed (Supplementary Figure 4). This experiment shows that the enhancement of Ssu72 activity is not due to its physical interaction with Pin1.
In contrast, when we tested human Scp1 in the same assay, the phosphatase activity is not significantly affected by Pin1 (Figure 3b). The insensitivity of Scp1 to Pin1 is consistent with the structural observations for prolyl isomeric states. Both Pro3 and Pro6 of the CTD peptide exhibit only the trans conformation in the complex structure of Scp1 and CTD peptide.27 Unlike Ssu72, Pro3 two residues upstream of pSer5 is a recognition determinant for Scp1 binding. Since the majority of the peptide substrate (estimated to be 80%26) has the proline in the trans conformation, Scp1 recognizes the substrate readily and dephosphorylates the substrate. In this case the addition of Pin1 only marginally improves substrate accessibility of Scp1. The slight improvement is hard to distinguish due to the sensitivity level of malachite green assay and is thus insignificant.
Discussion
Implication of Pin1 mechanism from the structures
Both cis- and trans-prolines are subject to isomerization by Pin1 with cis and trans alkene inhibitors mimicking substrate/product of the Pin1 isomerization reaction. The pSer-Pro dipeptide with either cis or trans conformation is modeled, based on our present complex structures, in the Pin1 structure to illustrate the real substrate binding (Figure 4b and 4c). The complex structures show that the same structural elements are used to recognize the substrate/product and interconvert the two species to reestablish the equilibrium cis:trans ratio.
Figure 4.
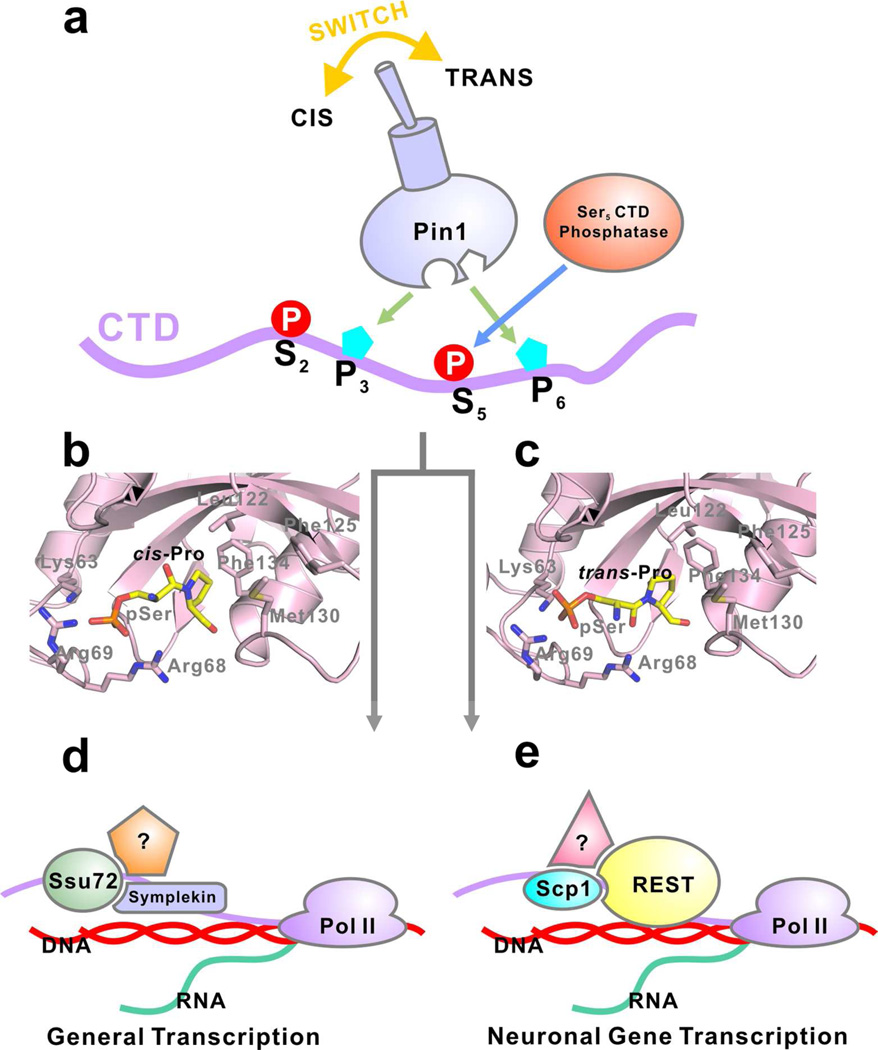
Model of the cross-talk between Ser5 dephosphorylation and prolyl isomerization of the CTD. (a) The Pin1 acts as molecular switch that changes the isomeric state of the two prolines, resulting in recruitment of different transcription complex and therefore, different outcomes of transcription. (b) pSer-cis-Pro dipeptide modeled in Pin1 structure. (c) pSer-trans-Pro dipeptide modeled in Pin1 structure. (d) Ssu72 is recruited with specific regulatory factors (colored shapes) to control general gene transcription in response to cis-proline. (e) Scp1 is recruited with a different set of regulatory factors (colored shapes) to control neuronal gene transcription in response to trans-proline.
A challenge of this field has been making substrates locked in only one conformation. Alkenes have a long history as peptide bond isosteres. The carbon-carbon double bond is close to the same length as the amide carbonyl-nitrogen bond, 1.40 Å vs 1.32 Å, and the distance between the α-carbons is identical, 3.8 Å.40 The dynamics of both cis- and trans-locked ligands are dramatically affected by which conformation is bound to Pin1; cis is more rigid than trans, and the rigidity of bound cis results in 23-fold tighter binding.29, 41 The protein dynamics of a conduit between the PPIase and WW domains of Pin1 are also differentially affected by binding of cis- or trans-locked substrate isosteres.35
Previously, we have obtained crystal structures of Pin1 complexed with two high affinity peptide inhibitors; Ac-Phe-(D/L)-pThr-Pip-Nal-Gln-NH2, (D-PEPTIDE or L-PEPTIDE, respectively). Our new structures replace the prolyl-peptide which is subject to isomerization with non-rotatable carbon-carbon double bond, locking the two states of substrate-bound mode of Pin1. When we compared these two pairs of complex structures of Pin1, substrate-mimicking (cis and trans isosteres) versus high affinity inhibitors (D- and L-PEPTIDE), it has been observed that the phosphate positions of different Pin1 inhibitors are highly diversified. The architecture of the triad positive residues allows the rolling of the phosphate group to form electrostatic interactions with Lys63, Arg68 and Arg69. The side chains of these three residues also adopt different conformations with each isomer to accommodate different positions of the phosphate of the substrate, allowing the rotation of the phosphate group, yet still within the pocket. On the contrary, the C-terminus of the signature motif (pSer/Thr-Pro) provides a strong hold for the peptide. The proline pocket does not accommodate free rotation of the proline and the hydrogen bonding between the carbonyl of proline and the amide of Gln131 is highly conserved among all of our Pin1 structures (Supplementary Figure 5). These observations suggest that dynamic interactions of the phosphate group and the protein allows the rotation of the prolyl peptide at the N-terminus of the peptide subject to isomerization, which echoes the discovery in NMR studies on the issue.41, 42 In addition, the flexibility of the interactions between the phosphate group and the positive triad during the rotation permits transition-state stabilization.36
Implications for the regulatory mechanism of CTD
During the progression of the RNA polymerase II-mediated transcription cycle, many CTD-specific kinases and phosphatases are recruited to the CTD. The dynamic phosphorylation/dephosphorylation is a major regulatory mechanism of the CTD, greatly influencing transcription. However, various phosphatases and kinases may have different specificity toward the isomeric states of the prolines adjacent to the major phosphorylation sites. Even though cis and trans forms of proline can reach equilibrium slowly under thermal isomerization called auto-conversion, the rate is too slow to allow efficient signal transduction in cells. Therefore, Pin1-mediated prolyl isomerization of the CTD is necessary to couple with the phosphorylation regulation to generate suitable substrates for both cis- and trans-specific kinases/phosphatases (Figure 4a). The dynamic nature of the CTD phosphorylation states during transcription determines that different outcomes can be reached for different phosphatases when their proline isomeric specificity is different (Figure 4).
In such a scenario, the prolyl isomerase activity can greatly affect the outcome when the pool of one species of the substrate is rapidly depleted. For a cis-specific enzyme, such as Ssu72, the isomerase activity of Pin1 guarantees the availability of cis-form substrate that is rapidly depleted. On the other hand, since Scp1 utilizes the trans-form CTD as substrate, which is the major species, the apparent dephosphorylation will not be affected much by Pin1. So even though both Scp1 and Ssu72 recognize the pSer5 of CTD as substrate, their response towards Pin1 PPIase activity in cells will differ dramatically (Figure 4d and 4e). Their different responses will in turn affect the regulatory factors recruited to the vicinity of genes during transcription. Specifically, Ssu72 together with its binding partner, the scaffold protein symplekin26 and other regulatory factors from the cleavage/polyadenylation specificity factor (CPSF) complex,43 will be recruited in response to pSer-cis-Pro, thus regulate general transcription. On the other hand, Scp1, together with REST complex,21 will be recruited in response to pSer-trans-Pro to turn off neuronal gene expressions. To a certain extent, the phosphorylation state of the CTD is governed not only by the phosphorylation/dephosphorylation mechanism, but also the prolyl isomerization mechanism (Figure 4). The combination of the various post-translational modifications on CTD can lead to different transcription outcome and therefore, various fates for the cell. The recognition of both phosphorylation and isomerization states of CTD by partner proteins are very likely to be a general mechanism adopted by other CTD-binding proteins in transcription regulation, indicating a “combinatorial” CTD code.
Conclusions
In this study, we determined the complex structures of human Pin1 with two isomer-locked peptidomimetics that mimic the substrates in the cis or trans form of a pSer-Pro peptide bond. The recognition by Pin1 has an impact on the downstream regulatory phosphatases of CTD to a different extent based on their specificity towards proline isomeric states. The existence of Pin1 isomerase activity can greatly stimulate the activity of a cis-proline specific phosphatase by increasing the potential substrate pool. However, the Pin1 effect is more limited on trans-proline specific phosphatases. Therefore, the up-regulation of Pin1 activity can alter the signal transduction pathway in CTD-mediated transcriptional regulation. The cross-talk between prolyl-isomerase and CTD phosphatases can differentially lead to various transcriptional outcomes in cells.
Methods
Synthesis of the cis and trans peptide mimetic inhibitors
The cis and trans isosteres, Boc–Ser–Ψ[(Z/E)CH=C]-Pro–OH, where (Z) is the cis mimic, and (E) is the trans mimic, were synthesized as previously reported.44 Both peptidomimetics (Figure 1), Ac–Phe–Phe–pSer–Ψ[(Z/E)CH=C]-Pro–Arg–NH2, were synthesized using solid-phase peptide synthesis with the Fmoc-protected, block-phosphorylated isosteres as described previously.29
Purification of human Pin1 and human Scp1
The human Pin1 or Scp1 gene was sub-cloned in a pHIS8 vector, a derivative of pET28a vector (Novagene).45 The Pin1 R14A mutant was produced using the QuikChange Site-Directed Mutagenesis Kit (Stratagene, CA). The purification of Pin1 R14A mutant or Pin1 PPIase domain (residue 51–163) was identical to the procedure previous reported.30 Concisely, the protein was overexpressed using E. coli BL21(DE3) strain with isopropyl-β-D-thiogalactopyranoside (IPTG) induction at 16 °C overnight. The cells were pelleted and lysed with subsequent nickel affinity chromatography purification. After imidazole elution of the HIS-tagged recombinant protein, the N-terminal polyhistidine-tag was truncated with thrombin protease during dialysis at 4 °C, and subsequently purified on ion-exchange and size exclusion chromatography columns. The purified protein was homogenous on SDS-PAGE gel.
Purification of Drosophila Ssu72
A pET28b derivative vector, pETHIS8–SUMO, encoding Drosophila Ssu72 proceeded by an N-terminal 8×HIS-SUMO tag was constructed previously.24 The protein was overexpressed in E. coli BL21 (DE3). The cells were grown at 37 °C in Luria-Bertani medium supplemented with 50 µg/mL kanamycin, then induced with 0.5 mM IPTG at 16 °C when the O.D. at 600 nm reached 0.8. After overnight incubation, the cells were harvested by centrifugation and disrupted by sonication. Recombinant protein was initially purified with Ni-NTA column (Qiagen, Switzerland). The N-terminal 8×HIS-SUMO tag was then removed by PreScission protease (GE Healthcare). The protein was further purified by a size exclusion column Superdex-75 (GE Healthcare), equilibrated with 25 mM Tris-HCl (pH8.0) and 200 mM NaCl buffer. The collected Ssu72 protein was passed through the Ni-NTA column again to remove any heterogeneous proteins, as evaluated by SDS-PAGE gels. The pure Ssu72 protein was finally flash frozen in liquid nitrogen and stored at −80 °C.
Crystallization, soaking and data collection
The R14A variant of human Pin1 was crystallized by vapor diffusion using a hanging drop of 1 µL protein plus 1 µL well solution. The crystals were obtained at 1.9–2.2 M ammonium sulfate, 1% PEG400 at pH 7.5 in 50 mM HEPES buffer. The crystals were then transferred to mother liquor containing 40% PEG400 and 50 mM HEPES pH 7.5 with 0.2 mM of peptide mimetic inhibitor. The crystals were soaked for 4 weeks with buffer exchange using fresh mother-liquor containing peptide mimetic inhibitor every week. The crystals were then frozen in liquid nitrogen and subjected to in-house X-ray beam using DIP100 imaging plate (MacScience, CO) with data collection. Diffraction data were processed using HKL2000. The statistics of the data are summarized in Table 1.
Table 1.
Crystallographic data statistics
| Pin1 with cis compound | Pin1 with trans compound | |
|---|---|---|
| Data collection | ||
| Space group | P3121 | P3121 |
| Cell dimensions: a, b, c (Å) | 69.4, 69.4, 79.6 | 69.3, 69.3, 79.7 |
| α, β, γ (°) | 90.0, 90.0, 120.0 | 90.0, 90.0, 120.0 |
| Resolution (Å) | 50.00 – 2.10 (2.18 – 2.10)* | 50.00– 2.26 (2.34 – 2.26)* |
| No. of unique reflections | 12458 (1106) | 10488 (908) |
| Rsym or Rmerge (%) | 5.4 (47.6) | 4.3 (30.1) |
| I/σ(I) | 26.4 (2.5) | 33.2 (5.4) |
| Completeness (%) | 93.2 (84.8) | 97.5 (85.9) |
| Redundancy | 5.5 (5.0) | 4.6 (4.3) |
| Refinement | ||
| Resolution (Å) | 33.19 – 2.10 | 47.95 – 2.27 |
| No. of reflections (test set) | 10431 (1187) | 9168 (1037) |
| Rwork / Rfree (%)# | 22.3 / 26.5 | 21.7 / 25.6 |
| No. of atoms: Protein | 1164 | 1164 |
| Ligand | 30 | 39 |
| PEG | 24 | 24 |
| Water | 87 | 64 |
| B-factors (Å2): Protein | 32.6 | 29.0 |
| Ligand | 47.0 | 55.6 |
| PEG | 31.7 | 30.2 |
| Water | 39.1 | 32.5 |
| R.m.s deviations: Bond lengths (Å) | 0.011 | 0.020 |
| Bond angles (°) | 1.411 | 1.977 |
| Ramachandran plot (%): Most favored | 92.8 | 92 |
| Additionally allowed | 6.4 | 7.2 |
| Generally allowed | 0.0 | 0.0 |
| Disalloweda | 0.8 | 0.8 |
Highest resolution shell is shown in parenthesis.
Rfree is calculated with 10% of the data randomly omitted from refinement.
Leu7 (chain A) in both structures is close to the N-terminus of Pin1.
Structure determination and analysis
The complex structure of human Pin1 R14A with cis or trans peptide mimetic inhibitors were determined using molecular replacement with Pin1 complex structure with a high affinity inhibitor (PDB code: 2itk) as a search model. The solution of the structure was identified using AMoRe, a program in the CCP4 program suite.46 The refinement of the complex structures was performed using the program refmac in CCP4.47 Electron density maps (sigmaA weighted 2Fo-Fc and Fo-Fc maps) were calculated after each cycle of refinement, and inspected to guide model rebuilding using Coot.48 The quality of the final model was evaluated using Procheck.49 The statistics of the final model for both structures are summarized in Table 1.
Malachite green assay for Scp1 and Ssu72
The activity of the CTD phosphatases Scp1 and Ssu72 in the presence or absence of Pin1 toward 28-mer CTD peptide was measured in this assay.50 The 28-mer peptide contains 4 repeats of the consensus sequence with each Ser5 phosphorylated: (YSPTpSPS)4. The reaction (20 µL total volume) for human Scp1 was carried out in buffer containing 5 ng of Scp1, 10 µM of peptide, 50 mM Tris-acetate pH 5.5, 10 mM MgCl2 and 10 ng of Pin1 or the PPIase domain, and was incubated at 37 °C. The reaction (20 µL total volume) of Drosophila Ssu72 was performed in the buffer containing 100 ng of Ssu72, 20 µM of peptide, 100 mM MES pH 6.5, and 10 ng of Pin1 or PPIase domain, and was incubated at 28 °C. The reactions were quenched by adding 40 µL of malachite green reagent at different time points. The release of inorganic phosphate was detected by measuring the absorbance at 620 nm.
Accession code
Coordinates of the Pin1-cis compound and Pin1-trans compound complex structures have been deposited in the Protein Data Bank with the accession numbers 3tcz and 3tdb.
Supplementary Material
Acknowledgement
We thank M. Hackert for his reading and comments about the manuscript. This work was supported by Welch Foundation Grant (F-1778 to Y. Z.), NIH R03DA030556 (Y.Z.), and R01 CA110940 (F.A.E.). Instrumentation and technical assistance for this work were provided by the Macromolecular Crystallography Facility, with financial support from the College of Natural Sciences, the Office of the Executive Vice President and Provost, and the Institute for Cellular and Molecular Biology, the University of Texas at Austin.
Footnotes
Supporting information
Five figures as described in the text are included in the supporting material. This material is available free of charge via the Internet at http://pubs.acs.org.
The authors declare no competing financial interests.
References
- 1.Brandts JF, Halvorson HR, Brennan M. Consideration of the Possibility that the slow step in protein denaturation reactions is due to cis-trans isomerism of proline residues. Biochemistry. 1975;14:4953–4963. doi: 10.1021/bi00693a026. [DOI] [PubMed] [Google Scholar]
- 2.Schutkowski M, Bernhardt A, Zhou XZ, Shen M, Reimer U, Rahfeld JU, Lu KP, Fischer G. Role of phosphorylation in determining the backbone dynamics of the serine/threonine-proline motif and Pin1 substrate recognition. Biochemistry. 1998;37:5566–5575. doi: 10.1021/bi973060z. [DOI] [PubMed] [Google Scholar]
- 3.Lu KP, Liou YC, Zhou XZ. Pinning down proline-directed phosphorylation signaling. Trends Cell Biol. 2002;12:164–172. doi: 10.1016/s0962-8924(02)02253-5. [DOI] [PubMed] [Google Scholar]
- 4.Lu KP. Pinning down cell signaling, cancer and Alzheimer's disease. Trends Biochem. Sci. 2004;29:200–209. doi: 10.1016/j.tibs.2004.02.002. [DOI] [PubMed] [Google Scholar]
- 5.Lu KP, Hanes SD, Hunter T. A human peptidyl-prolyl isomerase essential for regulation of mitosis. Nature. 1996;380:544–547. doi: 10.1038/380544a0. [DOI] [PubMed] [Google Scholar]
- 6.Yaffe MB, Schutkowski M, Shen M, Zhou XZ, Stukenberg PT, Rahfeld JU, Xu J, Kuang J, Kirschner MW, Fischer G, Cantley LC, Lu KP. Sequence-specific and phosphorylation-dependent proline isomerization: a potential mitotic regulatory mechanism. Science. 1997;278:1957–1960. doi: 10.1126/science.278.5345.1957. [DOI] [PubMed] [Google Scholar]
- 7.Corden JL. Tails of RNA polymerase II. Trends Biochem. Sci. 1990;15:383–387. doi: 10.1016/0968-0004(90)90236-5. [DOI] [PubMed] [Google Scholar]
- 8.Dahmus ME. Phosphorylation of mammalian RNA polymerase II. Methods Enzymol. 1996;273:185–193. doi: 10.1016/s0076-6879(96)73019-7. [DOI] [PubMed] [Google Scholar]
- 9.Dahmus ME. Reversible phosphorylation of the C-terminal domain of RNA polymerase II. J. Biol. Chem. 1996;271:19009–19012. doi: 10.1074/jbc.271.32.19009. [DOI] [PubMed] [Google Scholar]
- 10.Meinhart A, Kamenski T, Hoeppner S, Baumli S, Cramer P. A structural perspective of CTD function. Genes Dev. 2005;19:1401–1415. doi: 10.1101/gad.1318105. [DOI] [PubMed] [Google Scholar]
- 11.Palancade B, Bensaude O. Investigating RNA polymerase II carboxyl-terminal domain (CTD) phosphorylation. Eur. J. Biochem. 2003;270:3859–3870. doi: 10.1046/j.1432-1033.2003.03794.x. [DOI] [PubMed] [Google Scholar]
- 12.Morris DP, Phatnani HP, Greenleaf AL. Phospho-carboxyl-terminal domain binding and the role of a prolyl isomerase in pre-mRNA 3'-End formation. J. Biol. Chem. 1999;274:31583–31587. doi: 10.1074/jbc.274.44.31583. [DOI] [PubMed] [Google Scholar]
- 13.Wu X, Wilcox CB, Devasahayam G, Hackett RL, Arevalo-Rodriguez M, Cardenas ME, Heitman J, Hanes SD. The Ess1 prolyl isomerase is linked to chromatin remodeling complexes and the general transcription machinery. EMBO J. 2000;19:3727–3738. doi: 10.1093/emboj/19.14.3727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhang M, Gill GN, Zhang Y. Bio-molecular architects: a scaffold provided by the C-terminal domain of eukaryotic RNA polymerase II. Nano Rev. 2010;1 doi: 10.3402/nano.v1i0.5502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Meinhart A, Cramer P. Recognition of RNA polymerase II carboxy-terminal domain by 3'-RNA-processing factors. Nature. 2004;430:223–226. doi: 10.1038/nature02679. [DOI] [PubMed] [Google Scholar]
- 16.Fabrega C, Shen V, Shuman S, Lima CD. Structure of an mRNA capping enzyme bound to the phosphorylated carboxy-terminal domain of RNA polymerase II. Mol. Cell. 2003;11:1549–1561. doi: 10.1016/s1097-2765(03)00187-4. [DOI] [PubMed] [Google Scholar]
- 17.Xu YX, Hirose Y, Zhou XZ, Lu KP, Manley JL. Pin1 modulates the structure and function of human RNA polymerase II. Genes Dev. 2003;17:2765–2776. doi: 10.1101/gad.1135503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Xu YX, Manley JL. Pin1 modulates RNA polymerase II activity during the transcription cycle. Genes Dev. 2007;21:2950–2962. doi: 10.1101/gad.1592807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lu KP, Zhou XZ. The prolyl isomerase PIN1: a pivotal new twist in phosphorylation signalling and disease. Nature reviews. 2007;8:904–916. doi: 10.1038/nrm2261. [DOI] [PubMed] [Google Scholar]
- 20.Prelich G. RNA polymerase II carboxy-terminal domain kinases: emerging clues to their function. Eukaryot. Cell. 2002;1:153–162. doi: 10.1128/EC.1.2.153-162.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yeo M, Lee SK, Lee B, Ruiz EC, Pfaff SL, Gill GN. Small CTD phosphatases function in silencing neuronal gene expression. Science. 2005;307:596–600. doi: 10.1126/science.1100801. [DOI] [PubMed] [Google Scholar]
- 22.Yeo M, Lin PS, Dahmus ME, Gill GN. A novel RNA polymerase II C-terminal domain phosphatase that preferentially dephosphorylates serine 5. J. Biol. Chem. 2003;278:26078–26085. doi: 10.1074/jbc.M301791200. [DOI] [PubMed] [Google Scholar]
- 23.Zhang M, Liu J, Kim Y, Dixon JE, Pfaff SL, Gill GN, Noel JP, Zhang Y. Structural and functional analysis of the phosphoryl transfer reaction mediated by the human small C-terminal domain phosphatase, Scp1. Protein Sci. 2010;19:974–986. doi: 10.1002/pro.375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhang Y, Zhang M, Zhang Y. Crystal structure of Ssu72, an essential eukaryotic phosphatase specific for the C-terminal domain of RNA polymerase II, in complex with a transition state analogue. Biochem. J. 2011;434:435–444. doi: 10.1042/BJ20101471. [DOI] [PubMed] [Google Scholar]
- 25.Werner-Allen JW, Lee CJ, Liu P, Nicely NI, Wang S, Greenleaf AL, Zhou P. cis-Proline-mediated Ser(P)5 dephosphorylation by the RNA polymerase II C-terminal domain phosphatase Ssu72. J. Biol. Chem. 2011;286:5717–5726. doi: 10.1074/jbc.M110.197129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Xiang K, Nagaike T, Xiang S, Kilic T, Beh MM, Manley JL, Tong L. Crystal structure of the human symplekin-Ssu72-CTD phosphopeptide complex. Nature. 2010;467:729–733. doi: 10.1038/nature09391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhang Y, Kim Y, Genoud N, Gao J, Kelly JW, Pfaff SL, Gill GN, Dixon JE, Noel JP. Determinants for dephosphorylation of the RNA polymerase II C-terminal domain by Scp1. Mol. Cell. 2006;24:759–770. doi: 10.1016/j.molcel.2006.10.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Etzkorn FA. Pin1 flips Alzheimer's switch. ACS Chem. Biol. 2006;1:214–216. doi: 10.1021/cb600171g. [DOI] [PubMed] [Google Scholar]
- 29.Wang XJ, Xu B, Mullins AB, Neiler FK, Etzkorn FA. Conformationally locked isostere of phosphoSer-cis-Pro inhibits Pin1 23-fold better than phosphoSer-trans-Pro isostere. J. Am. Chem. Soc. 2004;126:15533–15542. doi: 10.1021/ja046396m. [DOI] [PubMed] [Google Scholar]
- 30.Zhang Y, Daum S, Wildemann D, Zhou XZ, Verdecia MA, Bowman ME, Lucke C, Hunter T, Lu KP, Fischer G, Noel JP. Structural basis for high-affinity peptide inhibition of human Pin1. ACS Chem. Biol. 2007;2:320–328. doi: 10.1021/cb7000044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lu PJ, Zhou XZ, Shen M, Lu KP. Function of WW domains as phosphoserine- or phosphothreonine-binding modules. Science. 1999;283:1325–1328. doi: 10.1126/science.283.5406.1325. [DOI] [PubMed] [Google Scholar]
- 32.Ranganathan R, Lu KP, Hunter T, Noel JP. Structural and functional analysis of the mitotic rotamase Pin1 suggests substrate recognition is phosphorylation dependent. Cell. 1997;89:875–886. doi: 10.1016/s0092-8674(00)80273-1. [DOI] [PubMed] [Google Scholar]
- 33.Verdecia MA, Bowman ME, Lu KP, Hunter T, Noel JP. Structural basis for phosphoserine-proline recognition by group IV WW domains. Nat. Struct. Biol. 2000;7:639–643. doi: 10.1038/77929. [DOI] [PubMed] [Google Scholar]
- 34.Li Z, Li H, Devasahayam G, Gemmill T, Chaturvedi V, Hanes SD, Van Roey P. The structure of the Candida albicans Ess1 prolyl isomerase reveals a well-ordered linker that restricts domain mobility. Biochemistry. 2005;44:6180–6189. doi: 10.1021/bi050115l. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Namanja AT, Wang XJ, Xu B, Mercedes-Camacho AY, Wilson KA, Etzkorn FA, Peng JW. Stereospecific gating of functional motions in Pin1. Proc. Natl. Acad. Sci. U. S. A. 2011;108:12289–12294. doi: 10.1073/pnas.1019382108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Xu GG, Zhang Y, Mercedes-Camacho AY, Etzkorn FA. A reduced-amide inhibitor of pin1 binds in a conformation resembling a twisted-amide transition state. Biochemistry. 2011;50:9545–9550. doi: 10.1021/bi201055c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Daum S, Lucke C, Wildemann D, Schiene-Fischer C. On the benefit of bivalency in peptide ligand/pin1 interactions. J. Mol. Biol. 2007;374:147–161. doi: 10.1016/j.jmb.2007.09.019. [DOI] [PubMed] [Google Scholar]
- 38.Kops O, Zhou XZ, Lu KP. Pin1 modulates the dephosphorylation of the RNA polymerase II C-terminal domain by yeast Fcp1. FEBS Lett. 2002;513:305–311. doi: 10.1016/s0014-5793(02)02288-3. [DOI] [PubMed] [Google Scholar]
- 39.Hausmann S, Shuman S. Characterization of the CTD phosphatase Fcp1 from fission yeast. Preferential dephosphorylation of serine 2 versus serine 5. J. Biol. Chem. 2002;277:21213–21220. doi: 10.1074/jbc.M202056200. [DOI] [PubMed] [Google Scholar]
- 40.Shue YK, Tufano MD, Carrera GM, Jr, Kopecka H, Kuyper SL, Holladay MW, Lin CW, Witte DG, Miller TR, Stashko M, Nadzan AM. Double bond isosteres of the peptide bond: synthesis and biological activity of cholecystokinin (CCK) C-terminal hexapeptide analogs. Bioorg. Med. Chem. 1993;1:161–171. doi: 10.1016/s0968-0896(00)82117-3. [DOI] [PubMed] [Google Scholar]
- 41.Namanja AT, Wang XJ, Xu B, Mercedes-Camacho AY, Wilson BD, Wilson KA, Etzkorn FA, Peng JW. Toward flexibility-activity relationships by NMR spectroscopy: dynamics of Pin1 ligands. J. Am. Chem. Soc. 2010;132:5607–5609. doi: 10.1021/ja9096779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Labeikovsky W, Eisenmesser EZ, Bosco DA, Kern D. Structure and dynamics of pin1 during catalysis by NMR. J. Mol. Biol. 2007;367:1370–1381. doi: 10.1016/j.jmb.2007.01.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Krishnamurthy S, He X, Reyes-Reyes M, Moore C, Hampsey M. Ssu72 Is an RNA polymerase II CTD phosphatase. Mol. Cell. 2004;14:387–394. doi: 10.1016/s1097-2765(04)00235-7. [DOI] [PubMed] [Google Scholar]
- 44.Wang XJ, Hart SA, Xu B, Mason MD, Goodell JR, Etzkorn FA. Serine-cis-proline and serine-trans-proline isosteres: stereoselective synthesis of (Z)- and (E)-alkene mimics by Still-Wittig and Ireland-Claisen rearrangements. J. Org. Chem. 2003;68:2343–2349. doi: 10.1021/jo026663b. [DOI] [PubMed] [Google Scholar]
- 45.Jez JM, Ferrer JL, Bowman ME, Dixon RA, Noel JP. Dissection of malonyl-coenzyme A decarboxylation from polyketide formation in the reaction mechanism of a plant polyketide synthase. Biochemistry. 2000;39:890–902. doi: 10.1021/bi991489f. [DOI] [PubMed] [Google Scholar]
- 46.Navaza J. AMoRe: an automated package for molecular replacement. Acta Cryst. 1994;A50:157–163. [Google Scholar]
- 47.Vagin AA, Steiner RA, Lebedev AA, Potterton L, McNicholas S, Long F, Murshudov GN. REFMAC5 dictionary: organization of prior chemical knowledge and guidelines for its use. Acta Cryst. 2004;D60:2184–2195. doi: 10.1107/S0907444904023510. [DOI] [PubMed] [Google Scholar]
- 48.Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr. D. Biol. Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 49.CCP4. Collaborative Computational Project, Number 4. The CCP4 Suite: Programs for Protein Crystallography. Acta Cryst. 1994;D50:760–763. doi: 10.1107/S0907444994003112. [DOI] [PubMed] [Google Scholar]
- 50.Martin B, Pallen CJ, Wang JH, Graves DJ. Use of fluorinated tyrosine phosphates to probe the substrate specificity of the low molecular weight phosphatase activity of calcineurin. J. Biol. Chem. 1985;260:14932–14937. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.