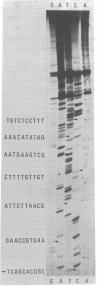
Abstract
We have employed the dideoxynucleotide chain-terminating method to determine the nucleotide sequence of T7 DNA between the physical map positions 18.9% and 19.8%. The most striking features of this sequence are two perfect 21-basepair repeats, each of which appears to contain a promoter for late transcription. In each case the promoter sequence incorporates a putative translational terminator on its left (5'-side of the "sense" strand), and overlaps a potential ribosome-binding site on its right. The region probably lies immediately distal to the early operon, and may contain two short, hitherto unreported protein-coding sequences.
Full text
PDF



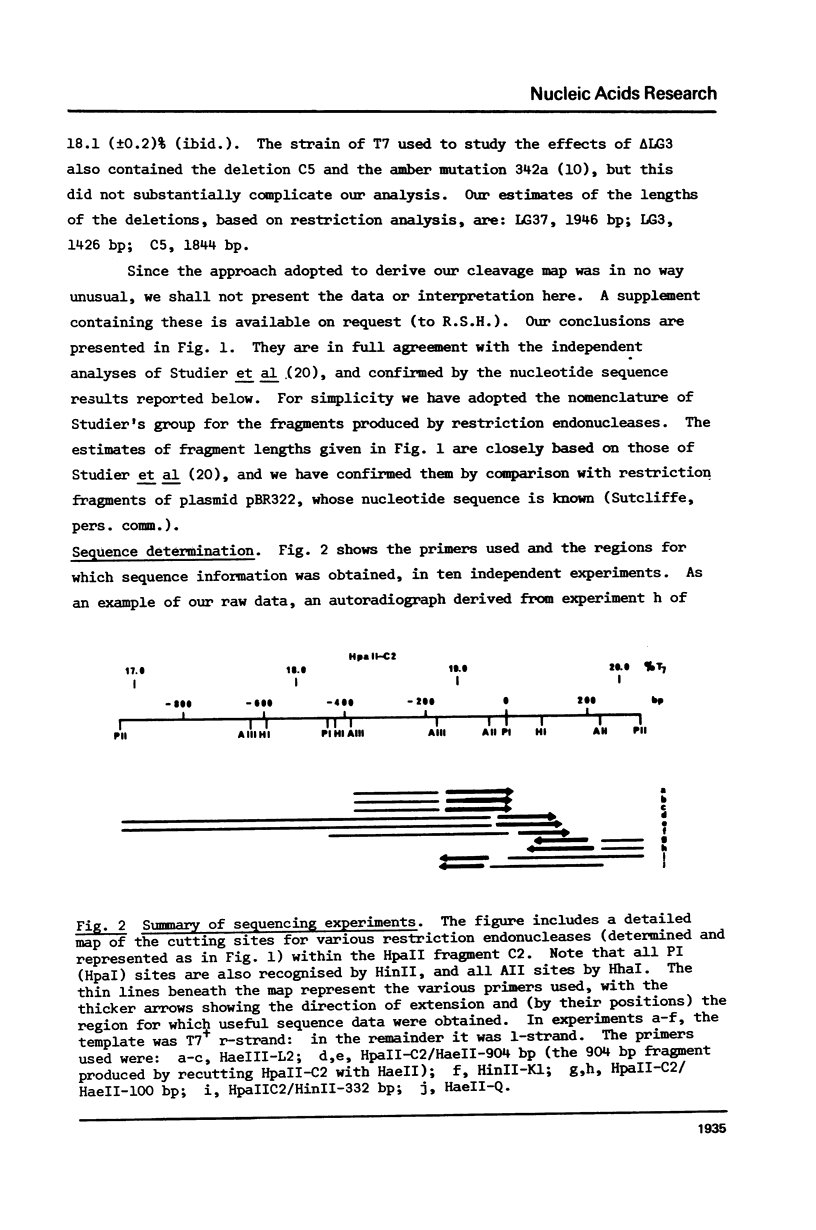


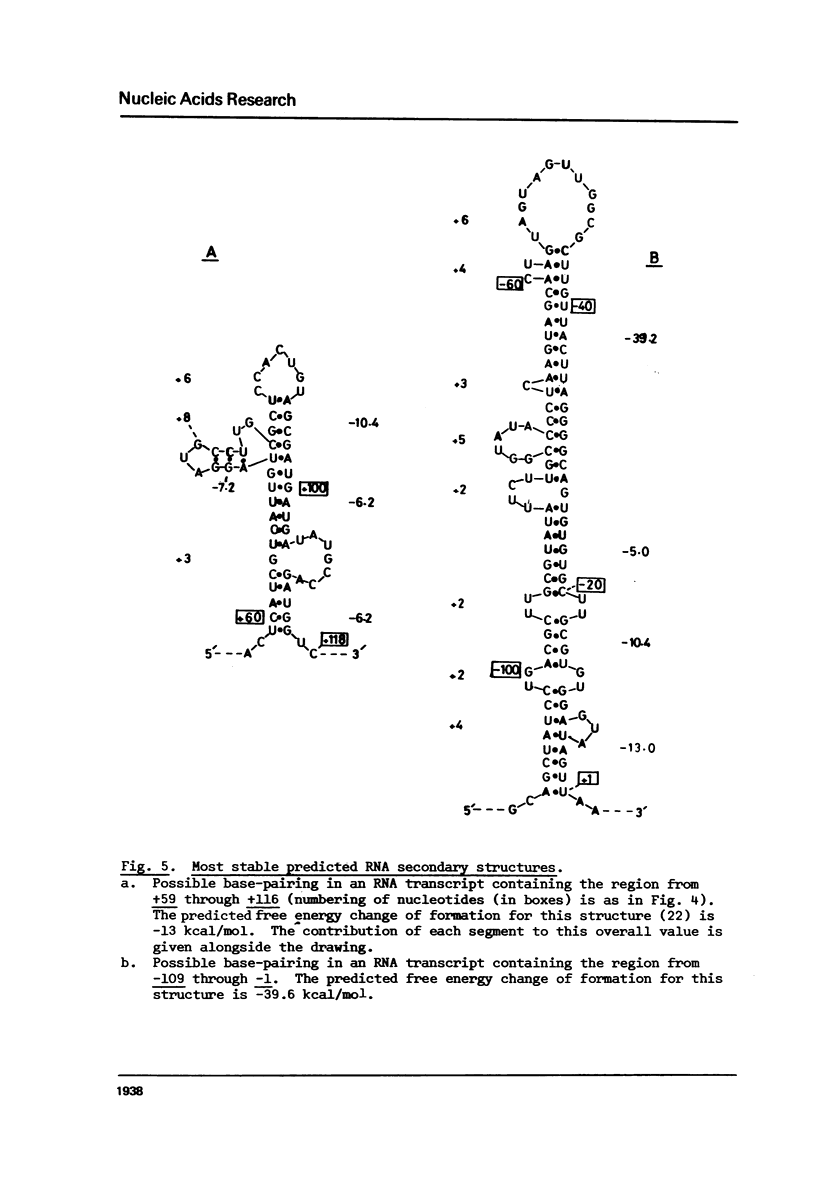





Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Blumberg B. M., Nakamoto T., Kézdy F. J. Kinetics of initiation of bacterial protein synthesis. Proc Natl Acad Sci U S A. 1979 Jan;76(1):251–255. doi: 10.1073/pnas.76.1.251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chamberlin M., McGrath J., Waskell L. New RNA polymerase from Escherichia coli infected with bacteriophage T7. Nature. 1970 Oct 17;228(5268):227–231. doi: 10.1038/228227a0. [DOI] [PubMed] [Google Scholar]
- Chamberlin M., Ring J. Characterization of T7-specific ribonucleic acid polymerase. 1. General properties of the enzymatic reaction and the template specificity of the enzyme. J Biol Chem. 1973 Mar 25;248(6):2235–2244. [PubMed] [Google Scholar]
- Delius H., Westphal H., Axelrod N. Length measurements of RNA synthesized in vitro by Escherichia coli RNA polymerase. J Mol Biol. 1973 Mar 15;74(4):677–687. doi: 10.1016/0022-2836(73)90056-9. [DOI] [PubMed] [Google Scholar]
- Golomb M., Chamberlin M. A preliminary map of the major transcription units read by T7 RNA polymerase on the T7 and T3 bacteriophage chromosomes. Proc Natl Acad Sci U S A. 1974 Mar;71(3):760–764. doi: 10.1073/pnas.71.3.760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kassavetis G. A., Chamberlin M. J. Mapping of class II promoter sites utilized in vitro by T7-specific RNA polymerase on bacteriophage T7 DNA. J Virol. 1979 Jan;29(1):196–208. doi: 10.1128/jvi.29.1.196-208.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Küpper H., Sekiya T., Rosenberg M., Egan J., Landy A. A rho-dependent termination site in the gene coding for tyrosine tRNA su3 of Escherichia coli. Nature. 1978 Mar 30;272(5652):423–428. doi: 10.1038/272423a0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maniatis T., Jeffrey A., van deSande H. Chain length determination of small double- and single-stranded DNA molecules by polyacrylamide gel electrophoresis. Biochemistry. 1975 Aug 26;14(17):3787–3794. doi: 10.1021/bi00688a010. [DOI] [PubMed] [Google Scholar]
- Maxam A. M., Gilbert W. A new method for sequencing DNA. Proc Natl Acad Sci U S A. 1977 Feb;74(2):560–564. doi: 10.1073/pnas.74.2.560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McConnell D. J. The DNA sequence at the T7 C promoter. Nucleic Acids Res. 1979 Feb;6(2):525–544. doi: 10.1093/nar/6.2.525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDonell M. W., Simon M. N., Studier F. W. Analysis of restriction fragments of T7 DNA and determination of molecular weights by electrophoresis in neutral and alkaline gels. J Mol Biol. 1977 Feb 15;110(1):119–146. doi: 10.1016/s0022-2836(77)80102-2. [DOI] [PubMed] [Google Scholar]
- McMahon J. E., Tinoco I., Jr Sequences and efficiencies of proposed mRNA terminators. Nature. 1978 Jan 19;271(5642):275–277. doi: 10.1038/271275a0. [DOI] [PubMed] [Google Scholar]
- Minkley E. G., Pribnow D. Transcription of the early region of bacteriophage T7: selective initiation with dinucleotides. J Mol Biol. 1973 Jun 25;77(2):255–277. doi: 10.1016/0022-2836(73)90335-5. [DOI] [PubMed] [Google Scholar]
- Oakley J. L., Coleman J. E. Structure of a promoter for T7 RNA polymerase. Proc Natl Acad Sci U S A. 1977 Oct;74(10):4266–4270. doi: 10.1073/pnas.74.10.4266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oakley J. L., Strothkamp R. E., Sarris A. H., Coleman J. E. T7 RNA polymerase: promoter structure and polymerase binding. Biochemistry. 1979 Feb 6;18(3):528–537. doi: 10.1021/bi00570a023. [DOI] [PubMed] [Google Scholar]
- Pachl C. A., Yound E. T. The size and messenger RNA activity of bacteriophage T7 late transcripts synthesized in vivo. J Mol Biol. 1978 Jun 15;122(1):69–101. doi: 10.1016/0022-2836(78)90109-2. [DOI] [PubMed] [Google Scholar]
- Panayotatos N., Wells R. D. Recognition and initiation site for four late promoters of phage T7 is a 22-base pair DNA sequence. Nature. 1979 Jul 5;280(5717):35–39. doi: 10.1038/280035a0. [DOI] [PubMed] [Google Scholar]
- Peters G. G., Hayward R. S. Dinucleotide sequences in the regions of T7 DNA coding for termination of early transcription. Eur J Biochem. 1974 Oct 1;48(1):199–208. doi: 10.1111/j.1432-1033.1974.tb03757.x. [DOI] [PubMed] [Google Scholar]
- Peters G. G., Hayward R. S. Transcriptional termination in vitro: the 3'-terminal sequence of coliphage T7 "early" RNA. Biochem Biophys Res Commun. 1974 Nov 27;61(2):809–816. doi: 10.1016/0006-291x(74)91029-8. [DOI] [PubMed] [Google Scholar]
- Roberts R. J. Restriction and modification enzymes and their recognition sequences. Gene. 1978 Nov;4(3):183–194. doi: 10.1016/0378-1119(78)90017-3. [DOI] [PubMed] [Google Scholar]
- Rosa M. D. Four T7 RNA polymerase promoters contain an identical 23 bp sequence. Cell. 1979 Apr;16(4):815–825. doi: 10.1016/0092-8674(79)90097-7. [DOI] [PubMed] [Google Scholar]
- Rosenberg M., Court D., Shimatake H., Brady C., Wulff D. L. The relationship between function and DNA sequence in an intercistronic regulatory region in phage lambda. Nature. 1978 Mar 30;272(5652):414–423. doi: 10.1038/272414a0. [DOI] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharp P. A., Sugden B., Sambrook J. Detection of two restriction endonuclease activities in Haemophilus parainfluenzae using analytical agarose--ethidium bromide electrophoresis. Biochemistry. 1973 Jul 31;12(16):3055–3063. doi: 10.1021/bi00740a018. [DOI] [PubMed] [Google Scholar]
- Shine J., Dalgarno L. The 3'-terminal sequence of Escherichia coli 16S ribosomal RNA: complementarity to nonsense triplets and ribosome binding sites. Proc Natl Acad Sci U S A. 1974 Apr;71(4):1342–1346. doi: 10.1073/pnas.71.4.1342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siebenlist U. Nucleotide sequence of the three major early promoters of bacteriophage T7. Nucleic Acids Res. 1979;6(5):1895–1907. doi: 10.1093/nar/6.5.1895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simon M. N., Studier F. W. Physical mapping of the early region of bacteriophage T7 DNA. J Mol Biol. 1973 Sep 15;79(2):249–265. doi: 10.1016/0022-2836(73)90004-1. [DOI] [PubMed] [Google Scholar]
- Stahl S. J., Chamberlin M. J. An expanded transcriptional map of T7 bacteriophage. Reading of minor T7 promoter sites in vitro by Escherichia coli RNA polymerase. J Mol Biol. 1977 Jun 5;112(4):577–601. doi: 10.1016/s0022-2836(77)80165-4. [DOI] [PubMed] [Google Scholar]
- Steitz J. A., Bryan R. A. Two ribosome binding sites from the gene 0-3 messenger RNA of bacteriophages T7. J Mol Biol. 1977 Aug 25;114(4):527–543. doi: 10.1016/0022-2836(77)90176-0. [DOI] [PubMed] [Google Scholar]
- Steitz J. A., Jakes K. How ribosomes select initiator regions in mRNA: base pair formation between the 3' terminus of 16S rRNA and the mRNA during initiation of protein synthesis in Escherichia coli. Proc Natl Acad Sci U S A. 1975 Dec;72(12):4734–4738. doi: 10.1073/pnas.72.12.4734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Studier F. W. Bacteriophage T7. Science. 1972 Apr 28;176(4033):367–376. doi: 10.1126/science.176.4033.367. [DOI] [PubMed] [Google Scholar]
- Tinoco I., Jr, Borer P. N., Dengler B., Levin M. D., Uhlenbeck O. C., Crothers D. M., Bralla J. Improved estimation of secondary structure in ribonucleic acids. Nat New Biol. 1973 Nov 14;246(150):40–41. doi: 10.1038/newbio246040a0. [DOI] [PubMed] [Google Scholar]