Abstract
Understanding the interactions between small interfering RNAs (siRNAs) and the RNA-induced silencing complex (RISC) – the key protein complex of RNA interference (RNAi) – is of great importance to the development of siRNAs with improved biological, and potentially therapeutic, function. Although various chemically modified siRNAs have been reported, relatively few studies with modified nucleobases exist. Here we describe the synthesis and hybridization properties of siRNAs bearing size-expanded RNA (xRNA) nucleobases, and their use as a novel and systematic set of steric probes in RNAi. xRNA nucleobases are expanded by 2.4 Å using benzo-homologation and retain canonical Watson-Crick base-pairing groups. Our data show that the modified siRNA duplexes display small changes in melting temperature (+1.4 to −5.0 °C); substitutions near the center are somewhat destabilizing to the RNA duplex, while substitutions near the ends are stabilizing. RNAi studies in a dual-reporter luciferase assay in HeLa cells revealed that xRNA nucleobases in the antisense strand reduce activity at some central positions near the seed region, but are generally well tolerated near the ends. Most importantly, we observed that xRNA substitutions near the 3′-end increased activity over wild-type siRNAs. The data are analyzed in terms of site-dependent steric effects in RISC. Circular dichroism experiments show that single xRNA substitutions do not significantly distort the native A-form helical structure of the siRNA duplex, and serum stability studies demonstrated that xRNA substitutions protect siRNAs against nuclease degradation.
INTRODUCTION
Over the past decade, RNA interference (RNAi) has become one of the most powerful and widely used tools for genetic research in various eukaryotes1–3. This post-translational gene silencing mechanism downregulates expression of targeted mRNAs through a process initiated by a small interfering RNA (siRNA) duplex (~21 nt in length with 2-nt overhangs) bound to a protein complex known as the RNA-induced silencing complex (RISC)4,5. RISC is able to unwind the siRNA duplex and uses the strand having sequence complementarity to the target mRNA (‘guide’ or antisense strand) as a template to bind to mRNA for targeted cleavage by the protein complex6. The other strand of the siRNA duplex (‘passenger’ or sense strand) is eventually degraded7.
Beyond its widespread use as a research tool, RNAi is also regarded as a potential therapeutic strategy to silence disease-causing genes using siRNAs8,9; however, the use of unmodified siRNAs presents significant challenges, including short half-lives in serum and undesired immune responses10,11. To address these concerns, various chemical modifications have been made to the sugar and phosphate backbone of siRNAs12,13. However, our understanding of effective nucleobase modifications remains relatively limited14.
In the literature, two broad classes of modified nucleobases in siRNAs have been described: those that retain their canonical Watson-Crick base pairing groups, and those that possess modified/deleted hydrogen bonding groups. In the latter case, altered nucleobases possessing alkylated hydrogen-bonding donors15 and non-polar, non-hydrogen bonding groups16–18 were used to probe the hydrogen bonding requirements of RISC, and have been shown to have a slight or significant reduction in RNAi activity when compared to natural, fully complementary siRNAs. On the other hand, modified nucleobases that retain their Watson-Crick base pairing groups, such as major/minor groove modifiers19–21, 4-thiouridine22, 2-thiouridine23, pseudouridine23, and dihydrouridine23 have shown potent gene silencing activity when used at certain positions in a siRNA duplex. Modified nucleobases can alter biological effects by changing nuclease susceptibility or by imposing hybridization-stabilizing or -destabilizing effects that can affect region-dependent RNAi activity in the siRNA duplex14. In addition, the use of 5-alkylated pyrimidines to test for steric effects in the major groove of the siRNA duplex showed lowered activity at some positions due to apparent steric clashes in RISC19; however, substitutions at other positions stabilized hybridization and increase activity over that of natural RNA.
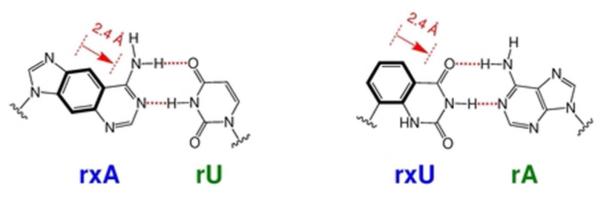
To further explore the effects of sterics in RNAi while retaining canonical Watson-Crick base pairing groups, we considered the use of size-expanded RNA (xRNA) nucleobases, in which natural purines and pyrimidines are expanded linearly by 2.4 Å via benzo-fusion (Figure 1). In the DNA context, it has been previously shown that single, internal size-expanded DNA (xDNA) substitutions in duplexes decrease helix stability as a result of conformationally unfavorable DNA/xDNA junctions. However, xDNA nucleobases stack quite strongly and stabilize duplexes markedly when substituted consecutively within helices24,25. Recently, we reported the synthesis of a four-monomer xRNA nucleoside genetic set and described their favorable fluorescence emission properties26,27. We anticipated that xRNA-containing siRNAs might serve as sequence-selectable probes to investigate the degree of steric tolerance in complex with Argonaute (Ago), the key catalytic component in RISC28. In conjunction with recently published crystal structure data from Thermus thermophilus Ago bound to a series of oligonucleotides29–31, RNAi activity data from this novel class of chemically modified siRNAs might provide new insights into the mechanism by which Ago recognizes both the antisense strand and its mRNA target. Herein we report the synthesis of two xRNA monomer phosphoramidite derivatives, their first successful incorporation into synthetic oligoribonucleotides, and their use as probes of positional steric sensitivity in RNAi. We find that the expanded analogs display strong position-dependent effects, with negative influences on RNAi activity at some central positions, but with favorable effects on gene suppression activity near the ends. We also describe the expanded nucleobases’ influences on siRNA duplex thermal stability, resistance against nuclease degradation, and helical structure.
Figure 1.
Duplexes containing expanded RNAs. Size-expanded versions of adenosine (rxA) and uridine (rxU) nucleotides hydrogen bonded to their natural RNA compliments.
RESULTS AND DISCUSSION
xRNA monomer phosphoramidites
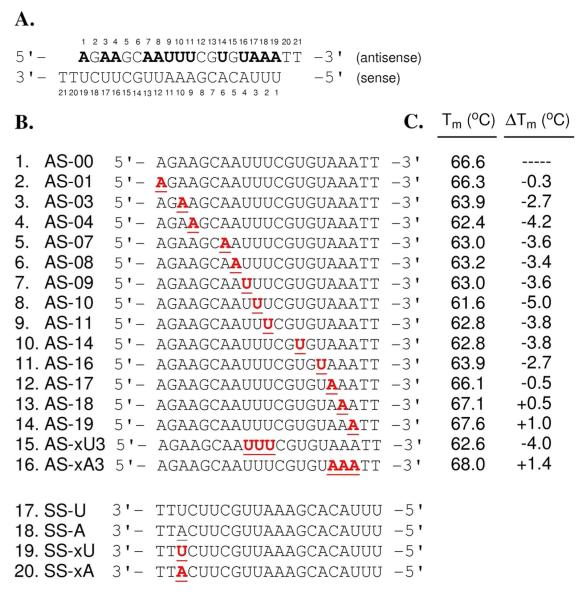
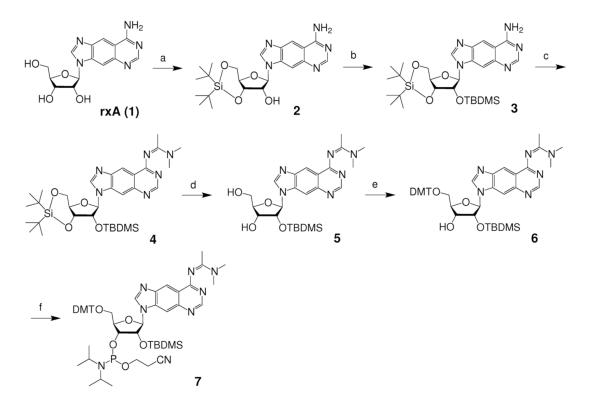
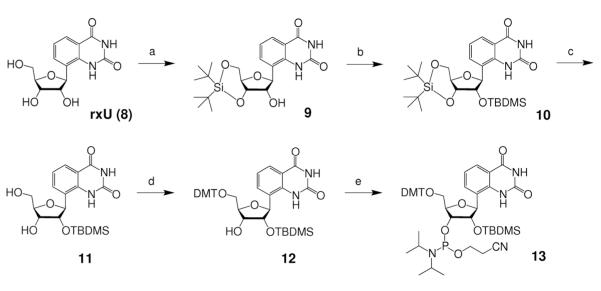
To explore the effects of expanded nucleobases and base pair size on RNAi in a systematic way, we chose a target site in a firefly luceriferase reporter gene whose sequence (Supplementary Figure S1) would allow for expanded versions of adenosine (rxA) and uridine (rxU) to be substituted at thirteen positions along the antisense sequence (Figure 2A). In preparation for automated oligonucleotide synthesis, we investigated various methods to perform the required regiospecific protections of the xRNA monomers. Ultimately, we adopted a strategy previously reported by Piccirilli et al.32, which proved to be successful. In this approach, rxA triol (1)26 was 3′,5′-O-bis-silylene protected (2), followed by 2′-O-TBDMS protection to afford 3 in excellent yield (Scheme 1). After N-protection of the nucleobase (4), selective desilylation of the bis-silylene protecting group generated 5 in very good yield. Successively, compound 5 was 5′-O-4,4′-dimethoxytrytyl (DMT) protected (6), then 3′-O-phosphoramidite protected to give rxA phosphoramidite, 7. Compound 7 was synthesized from 1 in 43.8% yield, and from commercially available starting materials in 2.2% yield (14 steps, longest linear sequence)26. A similar procedure was followed for the synthesis of rxU phosphoramidite, 13, beginning from rxU triol (8)26, except there was no exocyclic amine protection of the nucleobase, and different DMT and phosphitylation protection conditions were required (Scheme 2). Compound 13 was synthesized in 41.0% yield from nucleoside 8, and in 9.3% overall yield (11 steps, longest linear sequence) from commercially available starting materials26.
Figure 2.
siRNA sequence, analogs and melting temperatures. (A) Design of siRNA duplexes with 19-nt sequence and 2-nt overhangs, containing thirteen sites for rxA and rxU substitutions (bold letters), derived from nucleotides 1203-1221 of the luc+ gene sequence in the pGL3 control vector (firefly, Supplementary Figure S1). (B) Analogs of individual antisense (entries 1-16) and sense (entries 17-20) strands containing either natural (underlined) or xRNA (bold and underlined) substitutions. (C) Melting temperature (Tm) data for siR-U series. Change in Tm (ΔTm) compared to Tm of unmodified control duplex, siR-U-00 (i.e. SS-U annealed to AS-00). Triplicate readings taken in 1.0 μM siRNA in 15 mM HEPES-KOH (pH 7.4), 50 mM KOAc and 1 mM MgCl2. Tm errors < ± 0.5 °C.
Scheme 1. Reagents and conditions.
(a) tBu2Si(OTf)2, DMF, 0°C to r.t., 1 h, 85%; (b) TBDMSCI, imidazole, pyridine, r.t., overnight, 88%; (c) N,N-dimethylacetamide dimethyl acetal, MeOH, r.t., 90%; (d) HF-pyridine, THF, 0°C to r.t., 15 min, 87%; (e) DMTCI, DMAP, pyridine, 12 h, 86%; (f) 2-cyanoethyl N,N-diisopropylcholo-rophosphoramidite, 1-methylimidazole, iPr2NEt, CH2CL2, r.t. 6 h, 87%.
Scheme 2. Reagents and conditions.
(a) tBu2Si(OTf)2, DMF, 0°C to r.t., 1 h, 71%; (b) TBDMSCI, imidazole, pyridine, r.t., overnight, 89%; (c) HF-pyridine, THF, 0°C to r.t., 15 min, 86%; (d) DMTCI, AgNO3, 2,6-di-tert-butylpyridine, CH3CN, r.t., 20 min., 82%; (e) 2-cyanoethyl N,N,N’,N’-tetraisopropylphosphordiamidite, pyridinium trifluoroacetate, CH3CN, r.t., 18 h, 92%.
To assure that the bis-silylene protecting group was substituted on the expected 3′-O- and 5′-O-positions in 2 and 9, gCOSY NMR experiments were used to confirm the presence of the expected correlation between the 2′-hydroxyl proton and the 2′-ribose proton (see Supporting Information). The same technique was used to confirm the correlation of the 3′-hydroxyl proton and 3′-ribose proton in 6 and 12 to ensure that 5′-O-DMT protection occurred at the expected position.
Modified RNA synthesis
Using compounds 7 and 13, we prepared thirteen single-xRNA-substituted antisense strand analogs along with the unmodified antisense strand (entries 1–14, Figure 2B). To test for effects of consecutive substitutions near the center and end, we also prepared two triply substituted antisense strand analogs (entries 15–16, Figure 2B). Finally, to test for effects of mismatching at the 3′-end of the sense strand, we prepared four variants of the sense strand sequence with U, A, xU, and xA at position 19 (entries 17–20, Figure 2B). In this study, we use the abbreviation siR-X-YY to denote a siRNA duplex containing SS-X as the sense strand and AS-YY as the antisense strand (see strand abbreviations in Figure 2B). After automated oligonucleotide synthesis, cleavage/deprotection and purification, the strands were quantified by absorption at 260 nm and characterized by MALDI-TOF mass spectrometry (Supplementary Table S1).
RNAi activity and structure-activity relationships
A dual reporter firefly/Renilla luciferase assay was used to perform RNAi activity experiments in CCL-2 HeLa cells co-transfected with firefly and Renilla luciferase plasmids, and annealed synthetic siRNAs17. As previously mentioned, the selected antisense strand sequence (Figure 2A) targets firefly luciferase mRNA for degradation. Renilla luciferase mRNA is not complementary to either siRNA sequences in this study and served as a control for transfection efficiency.
A preliminary study using 20 pmoles of the unmodified control duplex, siR-U-00, in prepared HeLa cell culture (33.3 nM) showed acceptable levels of RNAi activity (data not shown). For the full sets of studies, two additional amounts of siRNA duplexes were used in the co-transfection assay experiments to test for dose-dependent behavior: 2 and 100 pmoles (3.33 and 166 nM in the HeLa cell culture, respectively).
We tested the complete series of modified antisense strand RNAs (entries 1–16, Figure 2B) annealed to SS-U (siR-U series), at the three dose ranges – each set measured in triplicate (Supplementary Figure S2). Dose-dependent responses were observed in all cases. Compared to the control siRNA duplex, xRNA substitution at position 1 of the antisense strand gave a modest loss of gene suppression activity, as shown by an increase in normalized luciferase signal relative to siR-U-00 at the same dose. Proceeding along positions 2-10, a steady increase in luciferase signal was also observed, indicating progressively poorer gene suppression. xRNA substitutions at position 11 displayed the lowest RNAi activity in the set. Moving away from the center of the antisense strand toward the 3′-end, however, the luciferase signal began to decrease markedly, indicating progressively stronger RNAi activity. Most significantly, RNAi activity for xRNA substitutions at single positions nearest to the end (i.e. positions 18 and 19) were superior compared to the results with siR-U-00.
The findings from this first set of experiments were used to deduce relationships between the modified guide (or antisense) strand as a steric probe bound to RISC and structural data recently acquired from non-human Ago-oligonucleotide complexes29–31. Beginning at the 5′-end, structural data reveals the nucleobase of position 1 of the guide strand anchored to a binding pocket in the Mid domain29,31. Having a particular preference for adenosine or uridine, the binding pocket also features a nucleotide specificity loop, which can recognize nucleobases via hydrogen bonding33. Moreover, the nucleobase is able to base-stack to a nearby aromatic residue in the binding pocket33. Our data show that a single xRNA substitution at position 1 (i.e. siR-U-01) was reasonably accepted by Ago (see Figure S2). This would indicate that the nucleotide specificity loop, and the binding pocket in general, is flexible enough to accommodate an additional 2.4 Å of size with only a modest penalty.
Structural data also shows residues in Ago making phosphodiester backbone contacts at positions 2-10 of the guide strand (known as the ‘seed region’), as well as hydrogen bonding taking place across the nucleobases of the target-guide duplex at those positions31. It is known that disruption in base pairing by mismatches in the ‘seed region’ greatly reduces RNAi activity30. Similarly, our data show a progressive reduction in activity when single xRNA substitutions are made in the ‘seed region’. Interestingly, position 4 appears to have the best RNAi activity in the entire region.
Moreover, structural evidence indicates that the target strand is cleaved between positions 10 and 11 (opposite of the guide strand) by a divalent-ion, aspartate catalytic triad in the PIWI domain31,34. At position 11, we observe a nearly 2.5-fold increase in luciferase signal (compared to siR-U-00), which suggest that the RNAi mechanism is greatly perturbed at the catalytic site. Not surprisingly, the crystal structure shows both target and guide strands making tight backbone contacts with residues in the PIWI and PAZ domains in the constrained, bilobal compartment of the cleavage site in the region corresponding to positions 10 and 11 of the guide strand31. The fact that complete arrest of RNAi activity was not observed around this site suggests that there is some degree of flexibility, albeit small, that can accommodate an added 2.4 Å of space.
At positions 12-16 of the guide strand, the Ago/guide/target structure reveals further base pairing and backbone contacts between the strands31. Our data shows RNAi activity improving progressively moving from the center towards the 3′ direction. Remarkably, positions 18 and 19 of the siR-U series displayed RNAi activity that is better than wild-type (siR-U-00) activity. However, it is not yet known how the 3′-end of the guide strand interacts with the protein complex because structural data shows duplex migration being blocked at positions 16 by residues in the N domain31. An earlier Ago-guide strand crystal structure, however, revealed that the 3′-end of the guide strand is able to interact with a binding pocket in the PAZ domain containing aromatic residues29. Similarly, others have found a highly conserved, hydrophobic binding pocket within PAZ that can interact well with the 3′-end of an oligonucleotide strand and duplex35,36, although the binding fate of the 3′-end of the guide strand (i.e. once the 5′-end is anchored to RISC) during the dynamic mRNA cleavage process remains under debate37,38. Given our observation of improved activity with 3′-end xRNA substitutions, we speculate that the enhanced RNAi activity observed at the 3′-end of the antisense strand may be due to favorable π-stacking interactions between the expanded nucleobase of xRNAs and aromatic residues within the PAZ domain, which would allow the strand to be better bound to Ago and thus possibly enhance its catalytic activity. Further studies would be needed to test this possibility.
Modified siRNA duplex thermal stability
xRNA nucleobases have not been previously incorporated into oligonucleotides, and so their effects on RNA stability were not known. Since thermal stability of siRNA duplexes has in some cases been correlated with activity17,19, we carried out thermal denaturation experiments to evaluate the effects of single and multiple xRNA substitutions in the siR-U series. Compared to control duplex, siR-U-00, the data show melting temperature (Tm) decreasing as the xRNA nucleobase is moved from the 5′-end of the antisense strand toward the center, and Tm increasing as the nucleobase is moved from the center to the 3′-end (Figure 2C and Supplementary Figures S3–S4). A single xRNA substitution is found to cause similar destabilization from positions 4 to 14 (ΔTm = −3.4 to −5.0 °C); an effect similar in magnitude to that seen with xDNA paired with its natural complement in the center of a DNA duplex24. The destabilization caused by rxA and rxU substitutions were similar in magnitude; for example, the rxA and rxU substitutions at adjacent positions 8 and 9, (entries 6 and 7, Figure 2C) respectively, led to identical RNA duplex stabilities within error. In contrast, single xRNA substitutions at the 3′-end (entries 13 and 14, Figure 2C) show higher Tm than the control (by up to 1.0 °C), likely due to enhanced stacking by the larger, more hydrophobic nucleobases.
This stacking ability likely affects some of the other duplexes as well. The entire thermal stability profile of the siR-U series showed relatively modest changes in Tm (ΔTm = +1.4 to −5.0 °C) compared to ΔTm ranges of other previously reported modified nucleobases14. Interestingly, the xRNA substitutions near the 3′-end of the antisense strand showed improved stability compared to siR-U-00 (Figure 2C). We have seen a similar enhanced stabilization effect in dangling-end studies involving short DNA duplexes containing a single xDNA overhang24. In both cases, we attribute this enhanced stabilization to stronger stacking by the expanded nucleobase. We also observed a rough correlation between RNAi activity and Tm data (Supplementary Figure S5) similar to what was seen in one of our previous RNAi studies17. However, this effect is likely analog-dependent since with some nucleobase analogs, stabilization and destabilization effects in the siRNA duplex were region-specific and were not described by a definitive overall trend14.
To complement these experiments, we also measured J coupling constants from 1H-NMR data of rxA and rxU nucleosides to determine their sugar puckering preferences. Our results show that rxA triol has a sugar conformation that is similar to that of native adenosine (Supplementary Table S2). Notably, rxU triol predominantly prefers a C2′-endo (south) conformation, which differs significantly from that of native uridine, which is more balanced in its north/south preference. This bias of rxU away from the north conformations is likely due to the fact that the structure lacks an electronegative atom at C1 on the nucleobase, thus removing the standard anomeric effect.
Thermodynamic versus steric effects in modified siRNAs
In addition, we investigated the effects of both steric and hydrogen bonding mismatches at the terminus of the duplexes using the dual reporter firefly/Renilla luciferase assay. Previously, it was shown that lowering the thermodynamic stability of either the 3′-end of the sense strand or the 5′-end of the antisense strand will bias the loading of the antisense strand into RISC39,40. It would be of great interest to know if making these substitutions would result in enhanced RNAi activity due to preferential loading of the xRNA-containing antisense strand. Using sense strand analogs 18–20 (Figure 2B), we prepared three new sets of siRNA duplexes with antisense strands 1–14 (Figure 2B) to test the effects of an A-A mismatch (siR-A series, Figure 3), a xU-A size “mismatch” (siR-xU series, Supplementary Figure S6) and a xA-A mismatch (siR-xA series, Supplementary Figure S7). A summary of the overall trends from the experimental results, including that of the siR-U series for comparison, is shown in Figure 4.
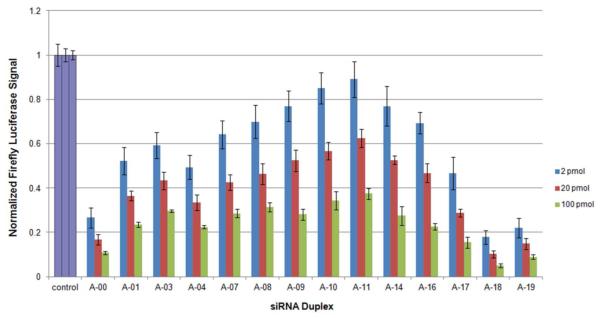
Figure 3.
Normalized RNAi activity data for siR-A series. A-00 corresponds to the unmodified siRNA duplex. Data are averages and s.d. of three independent experiments. HeLa cells were co-transfected with 2 pmol (3.33 nM), 20 pmol (33.3 nM) or 100 pmol (166 nM) of siRNA duplex. Decreased firefly luciferase signal indicates enhanced RNAi activity. All control experiments were conducted in an identical manner (wells contained HeLa cells treated with firefly and Renilla luciferase plasmids in the absence of siRNA).
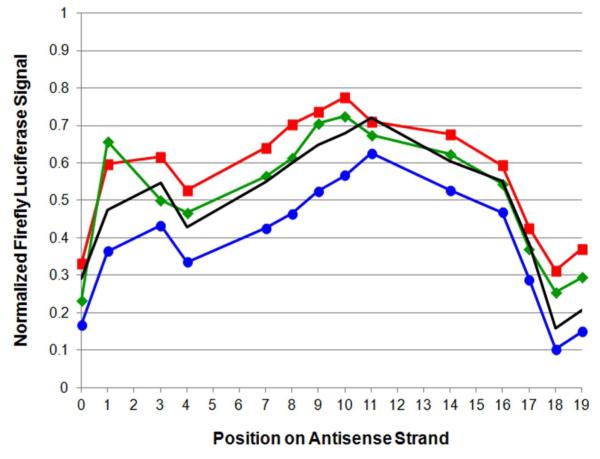
Figure 4.
Comparison of RNAi activities for various sets of siRNA duplexes. Position-dependent normalized RNAi activity data for siR-U (no marker), siR-A ( ), siR-xU (
), siR-xU ( ) and siR-xA (
) and siR-xA ( ) series of modified siRNA duplexes, showing effect of mismatching at 3′-end of sense strand. Activity data are taken from experimental results using 20 pmol (33.3 nM) siRNA duplex as shown in Figure 3 and Supplementary Figures S2, S6 and S7. Data at position 0 corresponds to the fully natural antisense strand (AS-00).
) series of modified siRNA duplexes, showing effect of mismatching at 3′-end of sense strand. Activity data are taken from experimental results using 20 pmol (33.3 nM) siRNA duplex as shown in Figure 3 and Supplementary Figures S2, S6 and S7. Data at position 0 corresponds to the fully natural antisense strand (AS-00).
In general, position-dependent results were very similar for the four series of modified siRNAs. RNAi activity results for the siR-A series (Figure 3) show that the firefly luciferase signal of fully natural siR-A-00 is reduced nearly two-fold compared to the luciferase signal of the natural, fully complementary control (siR-U-00). This confirms the benefit of the terminal mismatch39. The positive benefit of the A-A mismatch also held true for the entire xRNA-containing siR-A series, which showed the best activity of the four series (Figure 4). Results for the siR-xA and siR-xU series showed poorer RNAi activity than either the siR-U or siR-A series, respectively. Interestingly, both the match (siR-xU series) and the mismatch (siR-xA series) showed very similar trends in activity. Overall, our data reveal enhanced RNAi activity with a natural A-A mismatch at position 19 of the sense strand (Figure 4), but reduced activity with matched or mismatched xRNA nucleobases in place of A. This suggests that xRNA nucleobases may inhibit unwinding near their substitution positions, even when mismatched, by their (presumed) superior base stacking propensities (i.e. increased thermodynamic stability).
Studies with triply substituted siRNAs
The RNAi activity results in the siR-U series showed unfavorable effects on gene suppression at positions 4-14 near the seed region, along with lowered thermal stability associated at these substitutions. In contrast, substitutions near the 3′-end gave both favorable increases in RNAi activity and increases in Tm. Given the observed activity-Tm correlation previously mentioned (Figure S5), one might explain the results either by variable acceptance of the added size, or by the change in duplex stabilities. We therefore prepared two sets of triply substituted duplexes by annealing AS-xU3 (siR-xU3 series) and AS-xA3 (siR-xA3 series) to sense strand analogs 17–20 (Figure 2B) and performed both RNAi activity (Supplementary Figure S8) and thermal denaturation experiments (entries 15 and 16, Figure 2C) to observe their effects.
For the siR-xA3 series, the experiments showed similar RNAi activities as the unmodified, control duplex (siR-U-00). In contrast, the triple substitution at the center of the sequence (siR-xU3 series), displayed almost non-existent RNAi activity, indicating a highly unfavorable substitution. Thermal denaturation experiments were consistent with the general observations of the singly substituted siRNAs experiments. Triple rxU substitutions at positions 9–11 (siR-U-xU3) resulted in destabilization similar (although interestingly, not greater than) single rxU substitutions at those positions. In contrast to this, triple rxA substitutions at positions 17-19 (siR-U-xA3) yielded thermal stability that was slightly higher than singly substituted rxAs at the same positions.
xRNA substitution and nuclease degradation susceptibility
Modified nucleobases might effect RNAi activity not only by structural and thermodynamic effects in RISC, but also by affecting siRNA lifetime in the presence of nucleases. To test this possibility, we performed a serum stability assay under native conditions on duplexes in the siR-U series representing single substitutions made at the 5′-end, the middle, the 3′-end, as well as the triply substituted sets. As shown in Supplementary Figure S9, the fully natural siRNA (siR-U-00) is completely digested at 30 min., whereas all singly substituted siRNA duplexes are stabilized slightly, showing substantial amounts of fully intact RNA remaining at 30 min. The results for the triply substituted duplexes show that in both cases these more extensive substitutions confer greater stability, with fully intact siRNA remaining at 60 min.
Our serum stability results show that single xRNA substitutions can inhibit degradation to a small extent, and triple substitutions to a substantial degree. It seems possible that this may enhance activity of the modified siRNAs. However, it is interesting that stability against degradation overall does not correlate with activity. For example, a triple xRNA substitution near the center shows strong stability but poor activity, and one of our most active modified siRNA (siR-U-18) was not the most stable duplex. This may be consistent with other effects (e.g. sterics, duplex stability, etc.) as being dominant in our results.
Helical structure of xRNA-containing siRNAs
An important criterion in siRNA design is the ability for siRNAs to adopt an A-form helical structure, as it is thought that recognition of the A-form major groove is required for RNAi mediation15,41. To investigate this, we used CD spectroscopy to analyze a selected set of modified duplexes in the siR-U series, including cases having a single xRNA substitution at the 5′-end, in the middle, and the 3′-end, as well as the two triply substituted duplexes.
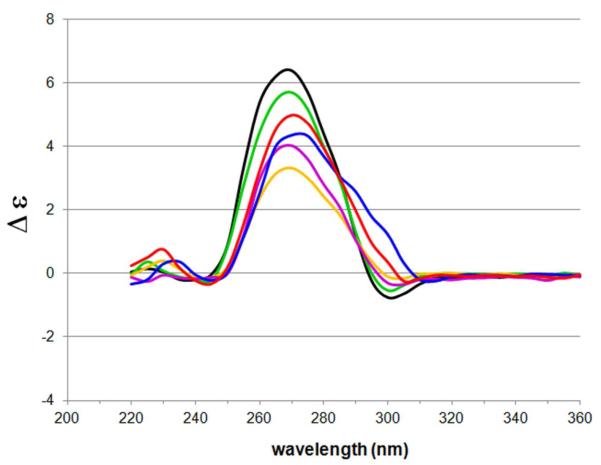
Our findings show that all three of the singly substituted duplexes do not significantly distort the native A-form helical structure of the siRNA duplex (Figure 5). An A-form duplex is characterized by a long-wavelength positive CD band centered at ~260 nm in the CD spectrum42, consistent with our observed spectra. Interestingly, both triply substituted siRNAs showed a red-shifted shoulder band at ~300 nm, which may be due to the known red shifting of absorbance by the xRNA nucleobases26. The findings are consistent with previous findings with xDNA-substituted DNAs, where small substitutions did not greatly perturb the native B-form DNA helix in most cases.24. Overall, it appears that xRNA-containing siRNAs adopt a helical structure that is close to the required native A-form.
Figure 5.
Circular dichroism spectroscopy data of xRNA-containing siRNAs in siR-U series. Duplexes include the fully natural siR-U-00 (black); single-xRNA-containing duplexes siR-U-01 (green), siR-U-10 (yellow) and siR-U-19 (magenta); and triple-xRNA-containing siR-U-xA3 (blue) and siR-U-xU3 (red). siRNA (5 μM) annealed in 15 mM HEPES–KOH (pH 7.4), 50 mM KOAc and 1 mM MgCl2.
In conclusion, we have described the synthesis and use of size-expanded versions of adenosine (rxA) and uridine (rxU) incorporated into biologically active siRNAs as a novel set of steric probes to investigate the RNAi mechanism and use in biophysical studies. These studies demonstrate the utility of xRNA nucleobases as mechanistic tools in biologically functioning siRNAs. Our findings suggest that there exist substantial steric restrictions in the seed region of the duplex, with increasing constriction to approximately position 14. Conversely, there appears to be little steric restriction at positions 16–19. In addition, we find practical advantages to these modifications as well. Substitutions at the 3′ end of the guide strand are found to enhance gene suppression activity over that of natural RNA. Furthermore, the expanded RNA nucleobases confer a measure of nuclease resistance to the siRNAs and they do not impose a significant distortion to the native A-form helical structure required for RNAi activity15,41. Our results also highlight the utility of retaining canonical hydrogen bonding groups when designing effective nucleobase substitutions in siRNAs. Finally, xRNA nucleobases are efficient fluorophores26, and may be useful probes in that regard as well.
METHODS
Oligonucleotide synthesis
All 21-nt natural and unnatural oligoribonucleotides were synthesized on an Applied Biosystems 394 DNA/RNA synthesizer (1 μmol scale), in “DMT On” mode, using commercially available 2′-O-TBDMS rA, rG, rC, rU and dT phosphoramidites (Glen Research) as well as the prepared xRNA phosphoramidites. 5-Benzylthio-1H-tetrazole was used as activator (Glen Research). Coupling time for rA, rG, rC, rU and xRNA phosphoramidites was 8 minutes, whereas coupling time for dT phosphoramidite was 4 minutes. After synthesis, CPG solid support was transferred to screw-cap vial, suspended in 1 ml of 1:1 NH4OH:CH3NH2 solution, and placed on a heat block for 10 min. (65 °C). After cooling to room temperature, supernatant was transferred to sterile 1.5-mL centrifuge tube. Support was rinsed with 250 μL sterile water and added to sterile tube. Collected fractions were evaporated to dryness in a centrifugal evaporator.
Oligonucleotide purification and characterization
Deprotection of 2′-O-TBDMS group and crude purification of oligoribonucleotides were performed using a Glen-PakTM RNA purification cartridge (Glen Research) following the manufacturer’s procedure. The dried oligonucleotide was further purified by 20% denaturing polyacrylamide gel electrophoresis. Buffers and gel mix were prepared using sterile conditions and reagents. Desired oligoribonucleotide strand was detected by UV lamp, excised from gel, extracted in 0.5 M aq. NaCl using the crush-and-soak method. Supernatant was dialyzed and evaporated to dryness in a centrifugal evaporator. Purified oligoribonucleotide was quantified by absorption at 260 nm and molecular weight (Supplementary Table S1) was confirmed by MALDI-TOF mass spectrometry (Stanford University Protein and Nucleic Acid Facility). Molar extinction coefficients of xRNA-containing oligonucleotides were estimated by subtracting ε260 of rxA and rxU triol nucleosides (pH 7)26 from ε260 of rA and rU triol nucleosides (pH 7)43, respectively, and adding the difference to the calculated ε260 of the corresponding fully natural oligonucleotide (Supplementary Table S1). Previous xDNA studies show that the expanded nucleobases have very low hypochromicity in xDNA-containing oligonucleotides44.
Thermal denaturation assay
Temperature-dependent absorbance curves of siRNA duplexes (1 μM) were measured in triplicate in 15 mM HEPES–KOH (pH 7.4), 50 mM KOAc and 1 mM MgCl2 on a Varian Cary 100 Bio UV-visible spectrophotometer equipped with thermoprogrammer. Oligonucleotides were initially annealed on a heat block set to 95 °C then allowed to slowly cool to room temperature. Samples were warmed at a rate of 1 °C/min from 20 °C to 90 °C while monitoring absorbance at 260 nm. Resulting denaturing data was analyzed on MeltWin v.3.0 software in order to determine the melting temperature (Tm) by computer-fit of the first derivative of absorbance with respect to 1/T.
RNAi activity assay
CCL-2 HeLa cells (ATCC, Mannasas, VA) were grown at 37 °C (5% CO2) in Dulbecco’s modified Eagles’ medium (DMEM, GIBCO) – supplemented with 10% fetal bovine serum (FBS, GIBCO), 100 μg/mL streptomycin (GIBCO) and 100 U/mL penicillin (GIBCO) – until 60–80% confluency was reached. In preparation for co-transfection, cells were trypsinized, suspended in DMEM + 10% FBS, and transferred to 24-well plates (500 μL cell suspension per well). Firefly lucuferase plasmid (pGL3 control vector, Promega) and Renilla luciferase plasmid (pRL-CMV vector, Promega) served as a reporter and control vectors, respectively, and were amplified in E. coli. After 24 hours of incubation, 100 μL of the following substrates were combined into a co-transfection cocktail and added to each well in the 24-well plate: 100 ng firefly luciferase plasmid, 10 ng Renilla luciferase plasmid, a varying amount of annealed siRNA duplex (2 pmol, 20 pmol or 100 pmol), 13.3 μL of LipofectamineTM 2000 (Invitrogen), and the remaining volume consisted of OPTI-MEM® I media (GIBCO). Samples were co-transfected in triplicate. A set of wells containing co-transfection cocktail without siRNA (control) per plate was also prepared. After co-transfection, the final volume was 600 μL per well. Six hour later, media was changed to DMEM + 10% FBS. After additional 18 hours, media was removed and cells were lysed using 100 μL of 1× passive lysis buffer (Promega). Plates were gently shaken for 45 min. at room temperature and 30 μL of cell lysate from each well was added to 96-well MicroliteTM 1 plates (Thermo Scientific). Luciferase activity experiments were measured on a Fluoroskan Ascent FL luminometer (Thermo Scientific) with a 2.0 sec. lag time and a 10.0 sec. integration time. Injectors were set to dispense 40 μL of luciferase assay reagent II (Promega) and 40 μL of Stop & Glo® reagent (Promega) per well. Readings were performed in triplicate. Firefly/Renilla luciferase ratios were calculated by the instrument and RNAi activity was expressed as the averaged luciferase ratio in a triplicate set normalized to the averaged luciferase ratio of the control set.
Serum stability assay
Annealed siRNA duplexes (40 μM) were incubated on a heat block (37 °C) with a mixture of human serum (Sigma Aldrich) diluted in 1× phosphate buffered saline to 10% for various time lengths (0 min., 15 min., 30 min., 60 min., 120 min., 240 min., and 480 min.). At the desired time, a 5-μL aliquot was removed, mixed with 5 μL of quenching mix (10% glycerol solution in 1× TBE buffer) and immediately frozen at −80 °C. Samples were analyzed by 18% native polyacrylamide gel electrophoresis. Additionally, samples of siR-U-00 (10 μM) and SS-U (10 μM) in quenching buffer were loaded onto PAGE gel to serve as reference markers after staining. RNA bands were visualized by Stains-All staining (Sigma Aldrich).
CD spectroscopy studies
CD spectra were measured on a Jasco-810 spectropolarimeter between 220 and 360 nm. siRNA duplexes (5 μM) were annealed in 15 mM HEPES–KOH (pH 7.4), 50 mM KOAc and 1 mM MgCl2. Spectra were acquired continuously every 1 nm with a 1-nm bandwidth at a scanning speed of 50 nm/min and a response time of 1 sec at 15 °C. Raw data were acquired over 6 scans, wave amplitude was converted from mdeg to Δε using established methods42, and processed using a smoothing function (Δλ = 5 nm) on Microsoft Excel software.
Supplementary Material
ACKNOWLEGEMENTS
We thank the National Institutes of Health (GM063587) for support. We also acknowledge the National Institutes of Health for support of A.R.H. as a Ruth L. Kirschstein NRSA predoctoral fellow (GM084680). We further thank A. Somoza (IMDEA Nanoscience Institute, Spain) for help with the RNAi assay, M. Terrazas (University of Barcelona, Spain) for helpful discussions, and S. R. Lynch (Stanford University) for assistance with NMR experiments.
Footnotes
Supporting Information Available. MALDI-TOF mass spectrometry data, sugar conformation analysis, additional RNAi activity data, serum stability assay results, organic synthesis procedures and associated NMR spectra. This material is available free of charge via the Internet at http://www.pubs.acs.org.
REFERENCES
- (1).Fire A, Xu S, Montgomery MK, Kostas SA, Driver SE, Mello CC. Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature. 1998;391:806–811. doi: 10.1038/35888. [DOI] [PubMed] [Google Scholar]
- (2).Bantounas I, Phylactou LA, Uney JB. RNA interference and the use of small interfering RNA to study gene function in mammalian systems. J. Mol. Endocrinol. 2004;33:545–557. doi: 10.1677/jme.1.01582. [DOI] [PubMed] [Google Scholar]
- (3).Hannon GJ. RNA interference. Nature. 2002;418:244–251. doi: 10.1038/418244a. [DOI] [PubMed] [Google Scholar]
- (4).Meister G, Tuschl T. Mechanisms of gene silencing by double-stranded RNA. Nature. 2004;431:343–349. doi: 10.1038/nature02873. [DOI] [PubMed] [Google Scholar]
- (5).Agrawal N, Dasaradhi PVN, Mohmmed A, Malhotra P, Bhatnagar RK, Mukherjee SK. RNA interference: Biology, mechanism, and applications. Microbiol. Molec. Biol. Rev. 2003;67:657–685. doi: 10.1128/MMBR.67.4.657-685.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (6).Jinek M, Doudna JA. A three-dimensional view of the molecular machinery of RNA interference. Nature. 2009;457:405–412. doi: 10.1038/nature07755. [DOI] [PubMed] [Google Scholar]
- (7).Leuschner PJF, Ameres SL, Kueng S, Martinez J. Cleavage of the siRNA passenger strand during RISC assembly in human cells. EMBO Rep. 2006;7:314–320. doi: 10.1038/sj.embor.7400637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (8).Aagaard L, Rossi JJ. RNAi therapeutics: Principles, prospects and challenges. Adv. Drug Deliv. Rev. 2007;59:75–86. doi: 10.1016/j.addr.2007.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (9).Dykxhoorn DM, Palliser D, Lieberman J. The silent treatment: siRNAs as small molecule drugs. Gene Ther. 2006;13:541–552. doi: 10.1038/sj.gt.3302703. [DOI] [PubMed] [Google Scholar]
- (10).Watts JK, Deleavey GF, Damha MJ. Chemically modified siRNA: Tools and applications. Drug Disc. Today. 2008;13:842–855. doi: 10.1016/j.drudis.2008.05.007. [DOI] [PubMed] [Google Scholar]
- (11).Judge AD, Sood V, Shaw JR, Fang D, McClintock K, MacLachlan I. Sequence-dependent stimulation of the mammalian innate immune response by synthetic siRNA. Nat. Biotech. 2005;23:457–462. doi: 10.1038/nbt1081. [DOI] [PubMed] [Google Scholar]
- (12).Shukla S, Sumaria CS, Pradeepkumar PI. Exploring chemical modifications for siRNA therapeutics: A structural and functional outlook. ChemMedChem. 2010;5:328–349. doi: 10.1002/cmdc.200900444. [DOI] [PubMed] [Google Scholar]
- (13).Manoharan M. RNA interference and chemically modified small interfering RNAs. Curr. Opin. Chem. Biol. 2004;8:570–579. doi: 10.1016/j.cbpa.2004.10.007. [DOI] [PubMed] [Google Scholar]
- (14).Peacock H, Kannan A, Beal PA, Burrows CJ. Chemical modification of siRNA bases to probe and enhance RNA interference. J. Org. Chem. 2011;76:7295–7300. doi: 10.1021/jo2012225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (15).Chiu Y-L, Rana TM. siRNA function in RNAi: A chemical modification analysis. RNA. 2003;9:1034–1048. doi: 10.1261/rna.5103703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (16).Xia J, Noronha A, Toudjarska I, Li F, Akinc A, Braich R, Frank-Kamenetsky M, Rajeev KG, Egli M, Manoharan M. Gene silencing activity of siRNAs with a ribo-difluorotoluyl nucleotide. ACS Chem. Biol. 2006;1:176–183. doi: 10.1021/cb600063p. [DOI] [PubMed] [Google Scholar]
- (17).Somoza A, Chelliserrykattil J, Kool ET. The roles of hydrogen bonding and sterics in RNA interference. Angew. Chem. Int. Ed. 2006;45:4994–4997. doi: 10.1002/anie.200601311. [DOI] [PubMed] [Google Scholar]
- (18).Somoza A, Silverman AP, Miller RM, Chelliserrykattil J, Kool ET. Steric effects in RNA interference: Probing the influence of nucleobase size and shape. Chem. Eur. J. 2008;14:7978–7987. doi: 10.1002/chem.200800837. [DOI] [PubMed] [Google Scholar]
- (19).Terrazas M, Kool ET. RNA major groove modifications improve siRNA stability and biological activity. Nucleic Acids Res. 2009;37:346–353. doi: 10.1093/nar/gkn958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (20).Wahba AS, Azizi F, Deleavey GF, Brown C, Robert F, Carrier M, Kalota A, Gewirtz AM, Pelletier J, Hudson RHE, Damha MJ. Phenylpyrrolocytosine as an unobtrusive base modification for monitoring activity and cellular trafficking of siRNA. ACS Chem. Biol. 2011;6:912–919. doi: 10.1021/cb200070k. [DOI] [PubMed] [Google Scholar]
- (21).Kannan A, Fostvedt E, Beal PA, Burrows CJ. 8-Oxoguanosine switches modulate the activity of alkylated siRNAs by controlling steric effects in the major versus minor grooves. J. Am. Chem. Soc. 2011;133:6343–6351. doi: 10.1021/ja2003878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (22).Parrish S, Fleenor J, Xu S, Mello C, Fire A. Functional anatomy of a dsRNA trigger: Differential requirement for the two trigger strands in RNA interference. Mol. Cell. 2000;6:1077–1087. doi: 10.1016/s1097-2765(00)00106-4. [DOI] [PubMed] [Google Scholar]
- (23).Sipa K, Sochacka E, Kazmierczak-Baranska J, Maszewska M, Janicka M, Nowak G, Nawrot B. Effect of base modifications on structure, thermodynamic stability, and gene silencing activity of short interfering RNA. RNA. 2007;13:1301–1316. doi: 10.1261/rna.538907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (24).Gao J, Liu H, Kool ET. Expanded-size bases in naturally sized DNA: Evaluation of steric effects in Watson–Crick pairing. J. Am. Chem. Soc. 2004;126:11826–11831. doi: 10.1021/ja048499a. [DOI] [PubMed] [Google Scholar]
- (25).Krueger AT, Lu H, Lee AHF, Kool ET. Synthesis and properties of size-expanded DNAs: Toward designed, functional genetic systems. Acc. Chem. Res. 2007;40:141–150. doi: 10.1021/ar068200o. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (26).Hernández AR, Kool ET. The components of xRNA: Synthesis and fluorescence of a full genetic set of size-expanded ribonucleosides. Org. Lett. 2011;13:676–679. doi: 10.1021/ol102915f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (27).Leonard NJ, Sprecker MA, Morrice AG. Defined dimensional changes in enzyme substrates and cofactors. Synthesis of lin-benzoadenosine and enzymatic evaluation of derivatives of the benzopurines. J. Am. Chem. Soc. 1976;98:3987–3994. doi: 10.1021/ja00429a040. [DOI] [PubMed] [Google Scholar]
- (28).Liu J, Carmell MA, Rivas FV, Marsden CG, Thomson JM, Song J-J, Hammond SM, Joshua-Tor L, Hannon GJ. Argonaute2 is the catalytic engine of mammalian RNAi. Science. 2004;305:1437–1441. doi: 10.1126/science.1102513. [DOI] [PubMed] [Google Scholar]
- (29).Wang Y, Sheng G, Juranek S, Tuschl T, Patel DJ. Structure of the guide-strand-containing argonaute silencing complex. Nature. 2008;456:209–213. doi: 10.1038/nature07315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (30).Wang Y, Juranek S, Li H, Sheng G, Tuschl T, Patel DJ. Structure of an argonaute silencing complex with a seed-containing guide DNA and target RNA duplex. Nature. 2008;456:921–926. doi: 10.1038/nature07666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (31).Wang Y, Juranek S, Li H, Sheng G, Wardle GS, Tuschl T, Patel DJ. Nucleation, propagation and cleavage of target RNAs in Ago silencing complexes. Nature. 2009;461:754–761. doi: 10.1038/nature08434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (32).Lu J, Li N-S, Koo SC, Piccirilli JA. Synthesis of pyridine, pyrimidine and pyridinone C-nucleoside phosphoramidites for probing cytosine function in RNA. J. Org. Chem. 2009;74:8021–8030. doi: 10.1021/jo9016919. [DOI] [PubMed] [Google Scholar]
- (33).Frank F, Sonenberg N, Nagar B. Structural basis for 5′-nucleotide base-specific recognition of guide RNA by human AGO2. Nature. 2010;465:818–822. doi: 10.1038/nature09039. [DOI] [PubMed] [Google Scholar]
- (34).Yuan Y-R, Pei Y, Ma J-B, Kuryavyi Y, Zhadina M, Meister G, Chen H-Y, Dauter Z, Tuschl T, Patel DJ. Crystal structure of A. aeolicus Argonaute, a site-specific DNA-guided endoribonuclease, provides insights into RISC-mediated mRNA cleavage. Mol. Cell. 2005;19:405–419. doi: 10.1016/j.molcel.2005.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (35).Lingel A, Simon B, Izaurralde E, Sattler M. Nucleic acid 3′-end recognition by the Argonaute2 PAZ domain. Nat. Struct. Mol. Biol. 2004;11:576–577. doi: 10.1038/nsmb777. [DOI] [PubMed] [Google Scholar]
- (36).Ma J-B, Ye K, Patel DJ. Structural basis for overhang-specific small interfering RNA recognition by the PAZ domain. Nature. 2004;429:318–322. doi: 10.1038/nature02519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (37).Filipowicz W. RNAi: The nuts and bolts of the RISC machine. Cell. 2005;122:17–20. doi: 10.1016/j.cell.2005.06.023. [DOI] [PubMed] [Google Scholar]
- (38).Tomari Y, Zamore PD. Perspectives: Machines for RNAi. Genes Dev. 2005;19:517–529. doi: 10.1101/gad.1284105. [DOI] [PubMed] [Google Scholar]
- (39).Schwarz DS, Hutvágner G, Du T, Xu Z, Aronin N, Zamore PD. Asymmetry in the assembly of the RNAi enzyme complex. Cell. 2003;115:199–208. doi: 10.1016/s0092-8674(03)00759-1. [DOI] [PubMed] [Google Scholar]
- (40).Khvorova A, Reynolds A, Jayasena SD. Functional siRNAs and miRNAs exhibit strand bias. Cell. 2003;115:209–216. doi: 10.1016/s0092-8674(03)00801-8. [DOI] [PubMed] [Google Scholar]
- (41).Chiu Y-L, Rana TM. RNAi in human cells: Basic structural and functional features of small interfering RNA. Mol. Cell. 2002;10:549–561. doi: 10.1016/s1097-2765(02)00652-4. [DOI] [PubMed] [Google Scholar]
- (42).Berova N, Nakanishi K, Woody RW, editors. Circular Dichroism: Principles and Applications. 2nd ed Wiley-VCH Publishers, Inc.; New York, NY: 2000. [Google Scholar]
- (43).Fasman GD, editor. CRC Handbook of Biochemistry and Molecular Biology – Nucleic Acids. 3rd ed Vol. 1. CRC Press, Inc.; Boca Raton, FL: 1975. [Google Scholar]
- (44).Liu H, Gao J, Kool ET. Helix-forming properties of size-expanded DNA, an alternative four-base genetic form. J. Am. Chem. Soc. 2005;127:1396–1402. doi: 10.1021/ja046305l. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.