Abstract
Preterm birth is a global health issue impacting millions of mothers and babies. However, the etiology of preterm birth is not clearly understood. Our recent finding that premature decidual senescence with terminal differentiation is a cause of preterm birth in mice with uterine Trp53 deletion, encoding p53 protein, led us to explore other potential factors that are related to preterm birth. Using proteomics approaches, here, we show that 183 candidate proteins show significant changes in deciduae with Trp53 deletion as compared with normal deciduae. Functional categorization of these proteins unveiled new pathways that are influenced by p53. In particular, down-regulation of a cluster of antioxidant enzymes in p53-deficient deciduae suggests that increased oxidative stress could be one cause of preterm birth in mice harboring uterine deletion of Trp53.
Pregnancy is a complex process that encompasses major events such as ovulation, fertilization, preimplantation embryo development, implantation, decidualization, placentation, and parturition. Each event is critical for the success of pregnancy. Preterm birth is a pathological state of pregnancy and a global health concern. Prematurity is a direct cause of nearly 30% of all neonatal deaths, and many babies who survive premature birth suffer serious long-term disabilities (1). Although inflammation/infection, stress, uterine overdistension, cervical aberration, and genetic and environmental factors are potential risk factors, the definitive cause of preterm birth remains elusive. Therefore, intense basic research exploring new approaches to combat preterm birth and prematurity is warranted. Animal models that show spontaneous preterm birth are desired tools for studying the underlying mechanism and for developing prevention and treatment strategies. We have recently generated mice with conditional uterine deletion of p53 that show spontaneous preterm birth (2).
Mutation of the Trp53 gene encoding p53 is present in various types of cancers. However, its functions in normal physiological processes, including female reproduction, remain ill understood (3). To examine the role of p53 during pregnancy events, we generated mice with conditional deletion of uterine Trp53 (p53d/d) by crossing floxed Trp53 (Trp53loxP/loxP) mice with progesterone receptor (Pgr)-Cre mice (PgrCre/+) (2, 4, 5). Trp53loxP/loxPPgr+/+ (p53f/f) mice, obtained as littermates of p53d/d mice, were used as controls. Although p53d/d females showed normal implantation, more than 50% of them had preterm birth with neonatal death due to premature decidual senescence resulting from terminal differentiation and senescence-associated growth restriction of decidual cells with up-regulation of p21, phosphorylated (p) protein kinase B (Akt), and phosphorylated S6 levels (2, 6). S6 kinase, which phosphorylates S6, is a downstream player in the mammalian target of rapamycin complex 1 (mTORC1) pathway. mTORC1 activity is activated by pAkt (7, 8). Because either inhibiting mTORC1 or deleting p21 attenuated senescence and prevented preterm birth in p53d/d mice, these molecules are thought to be p53 downstream targets in deciduae (6). In fact, it has been shown in an in vitro model of human decidualization that withdrawal of decidualization stimuli down-regulates p53 but increases the level of pAkt (9, 10). It is also known that p21 participates in decidualization (11). However, the result that deciduae deficient in Trp53 have heightened p21 levels is surprising, because p21 is normally down-regulated in cells with a Trp53 mutation or deletion, such as most cancer cells with the exception of few cases (12, 13). These results prompted us to pursue further studies to explore downstream targets of p53 in deciduae. Because treatment with rapamycin, an inhibitor of mTORC1, attenuates early decidual senescence and preterm labor in p53d/d mice, we sought to explore whether premature senescence and growth restriction affects other pathways that could be associated with adverse pregnancy outcome in p53d/d females.
In the present study, we performed proteomic analyses using p53d/d and p53f/f deciduae on d 8 of pregnancy to identify additional potential downstream targets of p53. Mass spectrometry (MS) characterization of proteins was performed by both a standard commercial MS platform and a new platform coupling an ion mobility separation (IMS) with MS to increase sensitivity and provide greater proteome coverage (14, 15). Because IMS separates ions based on shape, the IMS-MS analysis effectively provides multidimensional separations, greater sensitivity due to efficient ion utilization, and a different view of proteomes of samples without sacrificing measurement throughput (16). These analyses indicate that although decidual Trp53 deficiency positively regulates DNA replication, this deficiency adversely affects antioxidant status, ATP production, and the proteinase system during decidualization. It is an exciting finding to see that Trp53 deletion down-regulates the expression of 10 antioxidant enzymes, including peroxiredoxin-6 (PRDX6), a unique antioxidant enzyme. We previously observed that Prdx6 is robustly expressed in the deciduum surrounding the embryo and acts against reactive oxygen species (ROS) during implantation (17). We also found that 21 proteins in mitochondrial ATP production were down-regulated in p53d/d deciduae, whereas three proteins associated with DNA replication were up-regulated. Moreover, Trp53 deficiency attenuated the levels of five proteinase inhibitors, including α2-macroglobulin-P (A2MP), which is known to be expressed predominantly in mouse deciduae and is thought to regulate trophoblast invasion (18, 19). Collectively, our results show that loss of decidual Trp53 is associated with increased DNA replication and suppressed antioxidant status, ATP production, and proteolytic activity, unveiling new pathways that are influenced by p53 in mouse deciduae during early pregnancy.
Materials and Methods
Mice
Trp53loxP/loxP mice were obtained from the Mouse Models of Human Cancer Consortium (Frederick, MD), whereas PgrCre/+ mice were provided by John B. Lydon and Francesco J. DeMayo (Baylor College of Medicine, Houston, TX) (4, 5). To generate mice with uterine deletion of Trp53, Trp53loxP/loxP mice (FVB/129) were crossed with PgrCre/+ mice (C57BL6/129). To collect d 8 implantation sites (d 1, vaginal plug), littermate Trp53loxP/loxP/Pgr+/+ (p53f/f) and Trp53loxP/loxP/PgrCre/+ (p53d/d) mice on the mixed background were mated with wild-type fertile males to induce pregnancy. All mice used in this investigation were housed in the Cincinnati Children's Hospital Medical Center Animal Care Facility according to National Institutes of Health and institutional guidelines for the use of laboratory animals. All protocols of the present study were reviewed and approved by Cincinnati Children's Hospital Research Foundation Institutional Animal Care and Use Committee.
Preparation of protein samples for MS
Deciduae were dissected from d 8 mouse implantation sites free of embryos and myometria. Tissues were collected from three p53d/d and three p53f/f mice. Multiple deciduae were pooled from each mouse, and they were homogenized in 100 mm ammonium bicarbonate. The initial protein concentration was determined by BCA Protein Assay (Pierce, Rockford, IL). Urea and dithiothreitol (Sigma-Aldrich, St. Louis, MO) were added at a final concentration of 8 m and 10 mm, respectively, and incubated for 1 h at 37 C with shaking at 1000 rpm in Thermomixer R. The samples were diluted 5-fold with 50 mm ammonium bicarbonate (pH 7.8) before adding sequencing grade modified trypsin (Promega, Madison, WI) and 1 mm CaCl2 (to stabilize the trypsin and reduce autolysis) for protein digestion (1:50 wt/wt trypsin-to-protein). The samples were then placed in a Thermomixer R for incubation overnight at 37 C and cleaned via strong cation exchange solid phase extraction (Supelco, Bellefonte, PA). All samples were concentrated down (∼50 μl) in a Speed-Vac SC 250 Express (Thermo Savant, Holbrook, NY). The final peptide concentration was determined by BCA Protein Assay, and peptide samples were stored at −80 C until MS analysis.
Liquid chromatography (LC)-tandem MS (MS/MS), LC-MS, and LC-IMS-MS analyses
Analysis of digested peptide mixtures of samples was performed on both Thermo Fisher Scientific LTQ Orbitrap Velos MS (Velos) (San Jose, CA) operated in MS/MS mode and an in-house built instrument that couples a 1-m IMS (IMS-MS) with an Agilent 6224 time-of-flight MS (Agilent Technologies, Santa Clara, CA) upgraded to a 1.5-m flight tube providing resolution of approximately 25,000 (14). A fully automated in-house built four-column HPLC system equipped with in-house packed capillary columns was used for both instruments with mobile phase A consisting of 0.1% formic acid in water and phase B comprised of 0.1% formic acid in acetonitrile (20). A 100-min LC gradient was performed on the Velos MS (using 60-cm-long columns with an outer diameter of 360 μm, inner diameter of 75 μm, and 3-μm C18 packing material), whereas only a 60-min gradient with shorter columns (30 cm long with same dimensions and packing) was used with the IMS-MS; this additional IMS separation helps address detector suppression. Both gradients linearly increased mobile phase B from 0 to 60% until the final 2 min of the run when B was purged at 95%. Each sample (5 μl) was injected for both analyses, and the HPLC was operated under a constant flow rate of 0.4 μl/min for the 100-min gradient and 1 μl/min for the 60-min gradient. The Velos MS data were collected from 400-2000 m/z at a resolution of 60,000 (automatic gain control target, 1 × 106) followed by data dependent ion trap MS/MS spectra (automatic gain control target, 1 × 104) of the 10 most abundant ions using a collision energy setting of 35%. A dynamic exclusion time of 60 sec was used to discriminate against previously analyzed ions. IMS-MS data were collected from 100-3200 m/z.
Identification and quantification of the detected peptide peaks were performed using the accurate mass and time (AMT) tag approach (21). Briefly, Velos MS/MS data were searched by SEQUEST (Thermo Scientific, Rockford, IL), and the resulting 13,448 peptides that had a MS-GF score greater than or equal to 1E-9 were used to populate our AMT tag database (22, 23). Multiple in-house developed (publicly available http://ncrr.pnnl.gov/software) informatics tools were used to process the LC-MS data and correlate the resulting LC-MS features to an AMT tag database that contained accurate mass and LC separation elution time information for peptide tags generated from decidual proteins. Among the tools used were algorithms for peak-picking and for determining isotopic distributions and charge states (24). Further downstream data analysis incorporated all possible detected peptides into a visualization program, VIPER, to correlate LC-MS features to the peptide identifications in the AMT tag database (25). VIPER provided an intensity report for all detected features, normalized LC elution times via alignment to the database, and featured identification. In DAnTE software, peptide peak intensity values were converted to a log2 scale, normalized with a central tendency algorithm, and assessed at a protein level using rollup parameters; statistical comparisons were done at the peptide and protein level using ANOVA performed as t tests (p53d/d vs. p53f/f) in our analysis (26).
The focus of our analysis was peptide centric, because our MS technologies made possible measurements at the peptide level. Redundant peptide identifications in the case of a single peptide matching multiple proteins (typically protein isoforms) were removed. Therefore, each reported peptide matches back to a single protein. Differences in significance between p53d/d and p53f/f data were determined at the peptide level with a P value less than 0.05. A small subset of these peptides (∼7%) was deemed significant from an “all or none comparison,” where a peptide was detected in more than or equal to five datasets of one sample type and not detected in the other sample type. Tables 1–5 and Supplemental Tables 1 and 2, published on The Endocrine Society's Journals Online web site at http://endo.endojournals.org, depict these significant peptides as rolled up protein values (all peptides, regardless of significance, were rolled up to protein values in Supplemental Tables 3 and 4). Supplemental Table 5 contains the final IMS-MS and Velos-filtered results, their associated quantitative information, and functional categorization of each protein. Tables 1–5 highlight functional categories of interest. Proteins reported in this manuscript were to be identified by at least two peptides. Protein functionality was inferred from UniProtKB (http://www.uniprot.org), and links for each protein can be found in Supplemental Table 5, GO annotation in Supplemental Table 6, and KEGG information in Supplemental Tables 7–9. This section is expanded on in Supplemental methods.
Table 1.
Stress-related proteins
| UniProt Gene ID | UniProt accession no. | Protein name | log2 (p53d/d/p53f/f) | P value |
|---|---|---|---|---|
| Antioxidants | ||||
| Abat | P61922 | 4-Aminobutyrate aminotransferase, mitochondriala | −0.759 | 2.4E-05 |
| Cat | P24270 | Catalase | −0.593 | 7.3E-04 |
| Cryab | P23927 | α-Crystallin B chaina | −0.373 | 9.6E-04 |
| FAM120A | Q6A0A9 | Constitutive coactivator of PPAR-γ-like protein 1 | −0.553 | 2.5E-03 |
| G6pdx | Q00612 | Glucose-6-phosphate 1-dehydrogenase X | −0.511 | 9.6E-06 |
| Park7 | Q99LX0 | Protein DJ-1a | −0.932 | 2.9E-03 |
| Prdx2 | Q61171 | Peroxiredoxin-2a | −0.444 | 6.3E-05 |
| Prdx3 | P20108 | Thioredoxin-dependent peroxide reductase, mitochondriala | −0.444 | 1.0E-03 |
| Prdx6 | O08709 | Peroxiredoxin-6a | −1.174 | 2.3E-08 |
| Sod1 | P08228 | Superoxide dismutase (Cu-Zn) | −0.628 | 1.2E-03 |
| Stress-induced proteins | ||||
| Clu | Q06890 | Clusterin | 0.450 | 3.3E-03 |
| Dnaja2 | Q9QYJ0 | DnaJ homolog subfamily A member 2a | 0.735 | 1.9E-04 |
| Hsp90aa1 | P07901 | Heat shock protein HSP 90-α | 0.663 | 1.3E-03 |
| Hsp90ab1 | P11499 | Heat shock protein HSP 90-βa | 0.822 | 1.5E-03 |
| Hspd1 | P63038 | 60-kDa heat shock protein, mitochondrial | 0.502 | 9.1E-03 |
| Hsph1 | Q61699 | Heat shock protein 105 kDa | 0.533 | 5.0E-04 |
| Myof | Q8R3B4 | Myoferlin | 0.232 | 4.0E-03 |
| Stip1 | Q60864 | Stress-induced-phosphoprotein 1 | 0.192 | 1.5E-03 |
| Trap1 | Q9CQN1 | Heat shock protein 75 kDa, mitochondrial | 4.939 | 2.6E-04 |
| Hspb1 | P14602 | Heat shock protein β-1 | −0.309 | 3.9E-04 |
| Rbm3 | O89086 | Putative RNA-binding protein 3 | p53f/f onlyb | — |
Characterized by IMS-MS and Velos instruments (average values used for fold change and P values).
p53f/f only, p53d/d samples do not show any signal.
Table 2.
Protein modification and degradation
| UniProt Gene ID | UniProt accession no. | Protein name | log2 (p53d/d/p53f/f) | P value |
|---|---|---|---|---|
| Posttranslational modification | ||||
| Calu | O35887 | Calumenin | −0.486 | 7.4E-06 |
| Man2a1 | P27046 | α-Mannosidase 2 | p53f/f onlyb | — |
| Padi2 | Q08642 | Protein-arginine deiminase type-2 | p53f/f onlyb | — |
| Pdia6 | Q922R8 | Protein disulfide-isomerase A6a | −1.284 | 2.3E-03 |
| Rpn1 | Q91YQ5 | Dolichyl-diphosphooligosaccharide-protein glycosyltransferase subunit 1 | −0.739 | 1.4E-05 |
| Rpn2 | Q9DBG6 | Dolichyl-diphosphooligosaccharide-protein glycosyltransferase subunit 2a | −0.702 | 2.5E-03 |
| Sptlc2 | P97363 | Serine palmitoyltransferase 2 | −0.741 | 2.9E-03 |
| Tgm2 | P21981 | Protein-glutamine γ-glutamyltransferase 2a | −0.609 | 3.9E-04 |
| Fkbp5 | Q64378 | Peptidyl-prolyl cis-trans isomerase FKBP5 | −0.810 | 3.6E-06 |
| Fkbp1a | P26883 | Peptidyl-prolyl cis-trans isomerase FKBP1Aa | 0.610 | 1.0E-03 |
| Fkbp4 | P30416 | Peptidyl-prolyl cis-trans isomerase FKBP4a | 0.757 | 8.9E-04 |
| Proteasome and related proteins | ||||
| Cand1 | Q6ZQ38 | Cullin-associated NEDD8-dissociated protein 1 | 0.440 | 5.6E-04 |
| Ecm29 | Q6PDI5 | Proteasome-associated protein ECM29 homolog | 0.493 | 3.7E-03 |
| Psmd1 | Q3TXS7 | 26S proteasome non-ATPase regulatory subunit 1 | p53d/d onlyc | — |
| Psmd12 | Q9D8W5 | 26S proteasome non-ATPase regulatory subunit 12a | 0.283 | 1.3E-03 |
| Psmd2 | Q8VDM4 | 26S proteasome non-ATPase regulatory subunit 2 | 0.490 | 2.8E-03 |
| Psmd3 | P14685 | 26S proteasome non-ATPase regulatory subunit 3 | 0.474 | 1.3E-04 |
| Psmd8 | Q9CX56 | 26S proteasome non-ATPase regulatory subunit 8 | 0.483 | 1.0E-03 |
| Psme3 | P61290 | Proteasome activator complex subunit 3 | 0.390 | 8.3E-04 |
| Trim28 | Q62318 | Transcription intermediary factor 1-βa | 0.441 | 1.6E-03 |
| Uba1 | Q02053 | Ubiquitin-like modifier-activating enzyme 1 | 0.474 | 7.6E-04 |
| Ubc | Q9Z0H9 | Ubiquitin | 0.933 | 2.6E-03 |
| Psma1 | Q9R1P4 | Proteasome subunit α type-1 | −1.641 | 4.1E-03 |
| Ubqln2 | Q9QZM0 | Ubiquilin-2 | p53f/f onlyb | — |
| Usp14 | Q9JMA1 | Ubiquitin carboxyl-terminal hydrolase 14 | p53f/f onlyb | — |
| Proteinases | ||||
| Clpp | O88696 | Putative ATP-dependent Clp protease proteolytic subunit, mitochondrial | −0.553 | 4.9E-04 |
| Dpep1 | P31428 | Dipeptidase 1 | −0.939 | 1.6E-03 |
| Gzmb | P04187 | Granzyme B(G,H)a | −0.903 | 1.7E-03 |
| Thop1 | Q8C1A5 | Thimet oligopeptidase | −1.111 | 2.6E-03 |
| Spcs2 | Q9CYN2 | Signal peptidase complex subunit 2 | −0.553 | 8.9E-04 |
| Xpnpep1 | Q6P1B1 | Xaa-Pro aminopeptidase 1 | −0.443 | 1.3E-05 |
| Capns1 | O88456 | Calpain small subunit 1 | 0.703 | 2.4E-03 |
| Npepps | Q11011 | Puromycin-sensitive aminopeptidase | 0.195 | 2.6E-03 |
| Scpep1 | Q920A5 | Retinoid-inducible serine carboxypeptidasea | 0.592 | 6.6E-04 |
| Proteinases inhibitors | ||||
| A2mp | Q6GQT1 | α-2-Macroglobulin-Pa | −1.999 | 7.7E-13 |
| Cstb | Q62426 | Cystatin-Ba | −1.000 | 1.0E-03 |
| Serpina3k | P07759 | Serine protease inhibitor A3K | −1.109 | 4.6E-04 |
| Serpinb6 | Q60854 | Serpin B6a | −0.391 | 7.1E-04 |
| Serpinc1 | P32261 | Antithrombin-III | −0.459 | 2.0E-05 |
Characterized by IMS-MS and Velos instruments (average values used for fold change and P values).
p53f/f only, p53d/d samples do not show any signal.
p53d/d only, p53f/f samples do not show any signal.
Table 3.
Metabolism
| UniProt Gene ID | UniProt accession no. | Protein name | log2 (p53d/d/p53f/f) | P value |
|---|---|---|---|---|
| Lipid metabolism and related proteins | ||||
| Acot11 | Q8VHQ9 | Acyl-coenzyme A thioesterase 11 | −0.701 | 1.8E-04 |
| Ech1 | O35459 | δ(3,5)-δ(2,4)-dienoyl-CoA isomerase, mitochondrial | −0.654 | 2.3E-03 |
| Pafah1b2 | Q61206 | Platelet-activating factor acetylhydrolase IB subunit βa | −0.605 | 2.7E-04 |
| Psap | Q61207 | Sulfated glycoprotein 1a | −0.622 | 5.7E-05 |
| Sgpl1 | Q8R0 × 7 | Sphingosine-1-phosphate lyase 1a | −0.533 | 1.6E-04 |
| Apoa1 | Q00623 | Apolipoprotein A-Ia | 0.672 | 1.4E-03 |
| Apoe | P08226 | Apolipoprotein E | 0.952 | 6.9E-04 |
| Mitochondrial ATP production (fatty acid β-oxidation) | ||||
| Acaa2 | Q8BWT1 | 3-Ketoacyl-CoA thiolase, mitochondriala | −0.698 | 1.8E-06 |
| Acadvl | P50544 | Very long-chain specific acyl-CoA dehydrogenase, mitochondrial | −0.515 | 2.9E-03 |
| Dbi | P31786 | Acyl-CoA-binding proteina | −0.538 | 2.5E-04 |
| Hadh | Q61425 | Hydroxyacyl-coenzyme A dehydrogenase, mitochondrial | −0.725 | 4.6E-03 |
| Hadha | Q8BMS1 | Trifunctional enzyme subunit α, mitochondrial | p53f/f onlyb | — |
| Hadhb | Q99JY0 | Trifunctional enzyme subunit β, mitochondrial | −0.724 | 2.3E-03 |
| Mitochondrial ATP production (electron transport chain) | ||||
| Atp5c1 | Q91VR2 | ATP synthase subunit γ, mitochondrial | −0.568 | 2.4E-04 |
| Atp5i | Q06185 | ATP synthase subunit e, mitochondrial | −0.417 | 1.0E-03 |
| Cox5b | P19536 | Cytochrome c oxidase subunit 5B, mitochondriala | −1.105 | 2.6E-03 |
| Cox7a2 | P48771 | Cytochrome c oxidase subunit 7A2, mitochondrial | −0.609 | 3.8E-05 |
| Cyb5a | P56395 | Cytochrome b5 | −0.931 | 3.9E-05 |
| Etfa | Q99LC5 | Electron transfer flavoprotein subunit α, mitochondrial | −0.327 | 5.1E-04 |
| Mtco2 | P00405 | Cytochrome c oxidase subunit 2 | p53f/f onlyb | — |
| Mtnd5 | P03921 | NADH-ubiquinone oxidoreductase chain 5 | −0.837 | 1.9E-03 |
| Cisd1 | Q91WS0 | CDGSH iron sulfur domain-containing protein 1 | 0.622 | 4.7E-03 |
| Mitochondrial ATP production (tricarboxylic acid cycle) | ||||
| Aco2 | Q99KI0 | Aconitate hydratase, mitochondrial | −0.597 | 1.7E-03 |
| Dld | O08749 | Dihydrolipoyl dehydrogenase, mitochondrial | −0.857 | 2.3E-03 |
| Idh1 | O88844 | Isocitrate dehydrogenase (NADP) cytoplasmica | −0.635 | 9.4E-04 |
| Idh2 | P54071 | Isocitrate dehydrogenase (NADP), mitochondriala | −0.716 | 2.7E-03 |
| Idh3g | P70404 | Isocitrate dehydrogenase (NAD) subunit-γ, mitochondrial | −0.809 | 2.7E-03 |
| Mdh2 | P08249 | Malate dehydrogenase, mitochondrial | −0.532 | 4.7E-05 |
| Suclg1 | Q9WUM5 | Succinyl-CoA ligase (GDP-forming) subunit α, mitochondrial | −1.231 | 1.6E-03 |
| Other metabolism | ||||
| Acat1 | Q8QZT1 | Acetyl-CoA acetyltransferase, mitochondrial | −0.609 | 2.3E-03 |
| Adh5 | P28474 | Alcohol dehydrogenase class-3 | −0.398 | 1.3E-04 |
| Akr1b1 | P45376 | Aldose reductase | −1.016 | 3.0E-03 |
| Aldh9a1 | Q9JLJ2 | 4-Trimethylaminobutyraldehyde dehydrogenase | −0.661 | 1.6E-03 |
| Asl | Q91YI0 | Argininosuccinate lyasea | −0.693 | 3.4E-05 |
| Ckb | Q04447 | Creatine kinase B-type | −0.713 | 2.9E-03 |
| Cyb5r3 | Q9DCN2 | NADH-cytochrome b5 reductase 3 | −0.378 | 2.8E-03 |
| Dck | P43346 | Deoxycytidine kinase | −0.836 | 3.6E-03 |
| Dhcr24 | Q8VCH6 | 24-Dehydrocholesterol reductasea | −0.888 | 1.9E-05 |
| Es1 | P23953 | Liver carboxylesterase Na | −0.781 | 1.0E-04 |
| Gatm | Q9D964 | Glycine amidinotransferase, mitochondriala | −0.499 | 5.1E-04 |
| Gda | Q9R111 | Guanine deaminase | −0.961 | 4.1E-04 |
| Glul | P15105 | Glutamine synthetasea | −0.953 | 2.9E-04 |
| Mpi | Q924M7 | Mannose-6-phosphate isomerasea | −0.495 | 3.3E-04 |
| Nampt | Q99KQ4 | Nicotinamide phosphoribosyltransferase | −0.300 | 2.4E-04 |
| Nudt5 | Q9JKX6 | ADP-sugar pyrophosphatasea | −0.404 | 4.4E-04 |
| Oat | P29758 | Ornithine aminotransferase, mitochondrial | −1.362 | 3.3E-03 |
| Pik3c3 | Q6PF93 | Phosphatidylinositol 3-kinase catalytic subunit type 3 | −0.754 | 2.4E-03 |
| Ahcy | P50247 | Adenosylhomocysteinase | 0.220 | 1.8E-04 |
| Ak1 | Q9R0Y5 | Adenylate kinase isoenzyme 1a | 0.687 | 1.1E-04 |
| Aldh1a2 | Q62148 | Retinal dehydrogenase 2 | 0.601 | 2.0E-05 |
| Fmo1 | P50285 | Dimethylaniline monooxygenase (N-oxide-forming) 1 | 1.227 | 3.8E-03 |
| Gc | P21614 | Vitamin D-binding protein | 0.872 | 1.7E-03 |
| Gpd1l | Q3ULJ0 | Glycerol-3-phosphate dehydrogenase 1-like protein | 0.373 | 3.6E-03 |
| Hdlbp | Q8VDJ3 | Vigilin | 0.374 | 2.5E-03 |
| Prpsap1 | Q9D0M1 | Phosphoribosyl pyrophosphate synthase-associated protein 1 | 0.463 | 1.5E-03 |
| Sqrdl | Q9R112 | Sulfide:quinone oxidoreductase, mitochondriala | 0.800 | 5.0E-07 |
| Tdo2 | P48776 | Tryptophan 2,3-dioxygenasea | 0.455 | 2.6E-03 |
| Umps | P13439 | Uridine 5′-monophosphate synthase | 0.532 | 1.3E-04 |
Characterized by IMS-MS and Velos instruments (average values used for fold change and P values).
p53f/f only, p53d/d samples do not show any signal.
Table 4.
Nuclear proteins
| UniProt Gene ID | UniProt accession no. | Protein name | log2 (p53d/d/p53f/f) | P value |
|---|---|---|---|---|
| Association to DNA | ||||
| H2afy | Q9QZQ8 | Core histone macro-H2A.1 | −1.232 | 6.3E-04 |
| Hist1h1b | P43276 | Histone H1.5a | −0.485 | 2.3E-03 |
| Hist1h4a | Q5T006 | Histone H4 | −0.892 | 1.1E-03 |
| Hmgn2 | P09602 | Nonhistone chromosomal protein HMG-17 | −0.703 | 1.5E-03 |
| Nap1l4 | Q78ZA7 | Nucleosome assembly protein 1-like 4 | 0.343 | 2.1E-03 |
| DNA replication | ||||
| Mcm5 | P49718 | DNA replication licensing factor MCM5 | 0.357 | 1.8E-04 |
| Mcm6 | P97311 | DNA replication licensing factor MCM6 | 0.376 | 1.1E-03 |
| Pcna | P17918 | Proliferating cell nuclear antigen | 0.531 | 3.7E-03 |
| Nuclear import | ||||
| Kpna2 | P52293 | Importin subunit α-2 | 0.352 | 7.6E-04 |
| Kpnb1 | P70168 | Importin subunit β-1 | 0.476 | 1.9E-03 |
| Ipo9 | Q91YE6 | Importin-9 | 0.630 | 1.0E-05 |
| Ipo7 | Q9EPL8 | Importin-7 | 0.776 | 4.7E-04 |
| Mvp | Q9EQK5 | Major vault protein | 0.605 | 2.7E-04 |
Characterized by IMS-MS and Velos instruments (average values used for fold change and P values).
Table 5.
Other proteins
| UniProt Gene ID | UniProt accession no. | Protein name | log2 (p53d/d/p53f/f) | P value |
|---|---|---|---|---|
| Expressed exclusively in decidua | ||||
| Prl3c1 | Q9QUN5 | Prolactin-3C1a | −1.974 | 9.2E-09 |
| Transport | ||||
| Anxa1 | P10107 | Annexin A1 | −1.227 | 1.2E-03 |
| Ap1b1 | O35643 | AP-1 complex subunit β-1 | −0.459 | 2.3E-03 |
| Atl3 | Q91YH5 | Atlastin-3a | −0.984 | 2.2E-03 |
| Cp | Q61147 | Ceruloplasmina | −0.653 | 1.3E-03 |
| Fabp4 | P04117 | Fatty acid-binding protein, adipocyte | −1.090 | 8.3E-07 |
| Lman2 | Q9DBH5 | Vesicular integral-membrane protein VIP36 | −0.362 | 3.0E-03 |
| Rab8a | P55258 | Ras-related protein Rab-8A | −0.789 | 3.5E-03 |
| Sec23b | Q9D662 | Protein transport protein Sec23B | −1.085 | 1.8E-05 |
| Sec61a1 | P61620 | Protein transport protein Sec61 subunit α isoform 1 | −0.521 | 3.5E-03 |
| Sec61b | Q9CQS8 | Protein transport protein Sec61 subunit β | −0.731 | 1.1E-04 |
| Tom1 | O88746 | Target of Myb protein 1 | −0.375 | 3.4E-03 |
| Tspo | P50637 | Translocator protein | −0.653 | 2.8E-03 |
| Vapa | Q9WV55 | Vesicle-associated membrane protein-associated protein A | −1.014 | 1.2E-04 |
| Copb1 | Q9JIF7 | Coatomer subunit β | 0.611 | 4.4E-03 |
| Timm44 | O35857 | Mitochondrial import inner membrane translocase subunit TIM44 | 1.502 | 1.6E-03 |
| Uso1 | Q9Z1Z0 | General vesicular transport factor p115 | 0.291 | 4.0E-03 |
| Cell adhesion | ||||
| Emilin1 | Q99K41 | EMILIN-1 | 0.748 | 3.8E-03 |
| Fermt2 | Q8CIB5 | Fermitin family homolog 2 | 0.435 | 9.4E-06 |
| Fn1 | P11276 | Fibronectina | 0.539 | 1.3E-03 |
| Ilk | O55222 | Integrin-linked protein kinase | 0.371 | 1.0E-03 |
| Lama5 | Q61001 | Laminin subunit α-5a | 0.604 | 5.0E-04 |
| Lamb2 | Q61292 | Laminin subunit β-2a | 0.601 | 4.4E-04 |
| Tgfbi | P82198 | Transforming growth factor-β-induced protein ig-h3 | 1.006 | 2.7E-03 |
| Tjp1 | P39447 | Tight junction protein ZO-1 | 0.341 | 3.7E-03 |
| Cdh3 | P10287 | Cadherin-3 | −1.201 | 3.4E-03 |
| Pecam1 | Q08481 | Platelet endothelial cell adhesion molecule | −0.517 | 9.5E-04 |
| Cytoskeleton | ||||
| Actn1 | Q7TPR4 | α-Actinin-1 | 0.580 | 3.5E-07 |
| Actn4 | P57780 | α-Actinin-4a | 0.480 | 1.4E-03 |
| Add3 | Q9QYB5 | γ-Adducin | 0.455 | 2.3E-03 |
| Anxa3 | O35639 | Annexin A3 | 0.265 | 3.3E-03 |
| Cap1 | P40124 | Adenylyl cyclase-associated protein 1a | 0.571 | 4.9E-05 |
| Col14a1 | Q80 × 19 | Collagen α-1(XIV) chaina | 0.491 | 1.1E-03 |
| Col1a1 | P11087 | Collagen α-1(I) chaina | 0.787 | 1.4E-05 |
| Csrp1 | P97315 | Cysteine and glycine-rich protein 1 | 0.427 | 1.2E-04 |
| Dctn2 | Q99KJ8 | Dynactin subunit 2 | 0.362 | 4.1E-03 |
| Dstn | Q9R0P5 | Destrin | 0.594 | 1.0E-03 |
| Krt18 | P05784 | Keratin, type I cytoskeletal 18 | 0.773 | 1.1E-03 |
| Lima1 | Q9ERG0 | LIM domain and actin-binding protein 1 | 0.492 | 5.9E-04 |
| Map4 | Q8CFP5 | Microtubule-associated protein 4 | 0.396 | 5.0E-04 |
| Myh10 | Q61879 | Myosin-10a | 0.598 | 1.3E-04 |
| Myh9 | Q8VDD5 | Myosin-9a | 0.429 | 4.3E-04 |
| P4ha1 | Q60715 | Prolyl 4-hydroxylase subunit α-1a | 0.971 | 5.7E-04 |
| Parva | Q9EPC1 | α-Parvin | 0.702 | 3.3E-04 |
| Plod2 | Q9R0B9 | Procollagen-lysine,2-oxoglutarate 5-dioxygenase 2a | 0.750 | 3.1E-04 |
| Serpinh1 | P19324 | Serpin H1a | 0.442 | 1.1E-04 |
| Tpm4 | Q6IRU2 | Tropomyosin α-4 chaina | 0.579 | 5.1E-05 |
| Ugdh | O70475 | UDP-glucose 6-dehydrogenase | 0.791 | 1.2E-03 |
| Cnn3 | Q9DAW9 | Calponin-3 | −0.291 | 4.3E-03 |
| Hspg2 | Q05793 | Basement membrane-specific heparan sulfate proteoglycan core proteina | −0.812 | 1.5E-03 |
| Pfn1 | P62962 | Profilin-1a | −0.515 | 2.2E-03 |
| Thy1 | P01831 | Thy-1 membrane glycoprotein | −0.929 | 3.3E-03 |
Characterized by IMS-MS and Velos instruments (average values used for fold change and P values).
Western blotting
Protein extraction and Western blotting were performed as described (27). Antibodies to PRDX6 (LabFrontier Co., Seoul, Korea) and ACTIN (Santa Cruz Biotechnology, Inc., Santa Cruz, CA) were used. ACTIN served as a loading control.
Reverse transcription-polymerase chain reaction
RT-PCR was performed as described using primers 5′-TCATGGGGCATTCTCTTTTC-3′ and 5′-AAGGTCCCTGCCCTTATCAT-3′ for Prdx6 (product size 237 bps), 5′-GACCTTGTTGTTGACAAGGACTTA-3′ and 5′-CCTGAATGTACAGTAGAGGAATCA-3′ for A2mp (product size 277 bps), and 5′-GTGGGCCGCCCTAGGCACCAG-3′ and 5′- CTCTTTGATGTCACGCACGATTTC-3′ for β-Actin (product size 540 bps) (27). The housekeeping gene β-Actin served as an internal control.
Results
MS characterizes p53-regulated proteins in deciduae
The heterogeneous population of decidual cell types, presumably with different functions, made quantitation sensitivity very important. Because different cell types are difficult to distinguish, we homogenized the whole decidua and diluted protein expression changes that represented only few cells. For increased protein coverage and quantitative information, MS analysis of decidual tissues was performed with both a LTQ Orbitrap Velos MS (Velos) and new IMS-MS platform. These analyses identified a total of 283 proteins (Supplemental Table 5) that showed different levels between d 8 deciduae of p53d/d and p53f/f females; 183 of these proteins are represented in the functional categories of Tables 1–5. A reliable overlap was observed between platforms; 76 proteins were found significant by both Velos and IMS-MS (Supplemental Table 5). As expected, IMS-MS illustrated more significant proteins than the Velos alone (15). Figure 1 shows the intensity changes of IMS-MS spectra for selected significant peptides that are down-regulated (Fig. 1, A and B) and up-regulated (Fig. 1C) in p53d/d deciduae compared with those in p53f/f deciduae. Tables 1–5 show 103 down-regulated proteins and 80 up-regulated proteins in the p53d/d deciduae. The P values shown in these tables were calculated using all instrument runs and biological variability among six mice (three each of p53d/d and p53f/f) and are depicted by average protein intensities for the IMS-MS subset of the data (Fig. 2). We analyzed protein functions using UniProtKB, GO annotation, and KEGG information (Supplemental Tables 5–9) and categorized proteins into 18 functional pathways (Tables 1–5 and Supplemental Tables 5–9). In the p53d/d deciduae, most protein species were down-regulated in the following 12 pathways: antioxidant enzymes, posttranscriptional modifications, proteinases, proteinase inhibitors, lipid metabolism, fatty acid β-oxidation, electron transport chain, tricarboxylic acid cycle, metabolism, association to DNA, transporter, and known molecules previously found to be associated with decidualization. Proteins mostly up-regulated in the p53d/d deciduae are associated with six pathways, including stress-induced proteins, proteasome and related proteins, DNA replication, nuclear import, cell adhesion, and cytoskeleton. Of note, we found that prolactin-3C1, a known decidual marker (28), and platelet endothelial cell adhesion molecule 1 (29), an endothelial cell marker, are down-regulated in p53d/d deciduae.
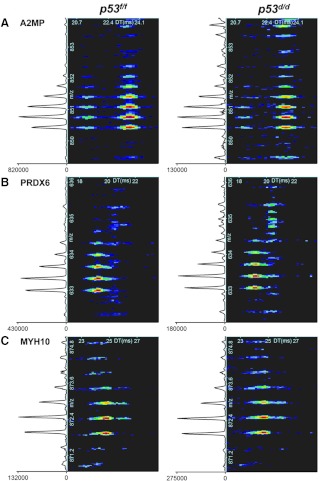
Fig. 1.
IMS-MS spectra of A2MP, PRDX6, and myosin-10 (MYH10) peptides. A, YIFIDESHITQALTWLSQQQK from A2MP. B, LIALSIDSVEDHLAWSK from PRDX6. The signal intensity located at the bottom left of each spectra denotes an increase in peptide intensity in the p53f/f samples by 6.3- and 2.4-fold change for these A2MP and PRDX6 peptides, respectively. C, IMS-MS spectra for the significant peptide QLLQANPILESFGNAK from MYH10 showed a 2.1-fold intensity increase in the p53d/d sample. Mouse identifiers WT_8718 and KO_7988 (see Supplemental Tables 1–9) were used as examples for this figure.
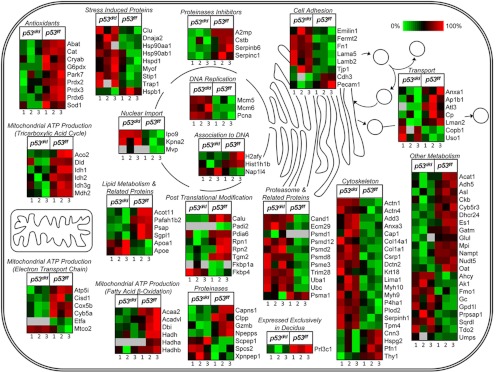
Fig. 2.
Disruption of p53 signaling adversely affects many cellular processes. Heat maps depict average protein intensities, calculated from multiple IMS-MS analyses, for each of the total six mice (three of p53d/d and p53f/f each).
DNA replication is up-regulated in p53d/d deciduae
Decidualization is a process by which endometrial stromal cells undergo proliferation and differentiation after embryo implantation. We record weights of implantation sites as an index of the extent of decidualization. During decidualization, there are massive increases in cell numbers with heightened DNA synthesis in many cells leading to endoreduplication and polyploidy (11). In p53d/d females, the number of decidual cells undergoing polyploidy with terminal differentiation and cellular senescence activity substantially increases. These changes are reflected in growth restriction, smaller decidual size, and preterm birth (2). In the present study, we found three proteins related to DNA replication- DNA replication licensing factor minichromosome maintenance complex components 5 and 6 and proliferating cell nuclear antigen are all up-regulated in p53d/d deciduae compare with those in p53f/f dams (Table 4).
Antioxidant enzymes are down-regulated in p53d/d deciduae
Generation of ROS plays important roles during pregnancy. However, ROS must be balanced with antioxidant enzymes, because excessive ROS causes susceptibility to oxidative stress (OS), which results in cellular damage and senescence (17, 30). We have previously observed that increased polyploidy of stromal cells with and growth restriction in p53d/d deciduae are associated with increased senescence (2), and OS is associated with preterm birth (31). We found that 10 antioxidant enzymes were down-regulated in p53d/d deciduae compared with p53f/f deciduae. Using Western blot and RT-PCR analyses, we confirmed that PRDX6 expression is lower in p53d/d deciduae than p53f/f deciduae (Fig. 3, A and B). These results suggest that p53d/d deciduae with decreased expression of antioxidant enzymes are susceptible to OS, which in turn induces compromised decidualization with restricted decidual growth and premature senescence, ultimately triggering preterm birth in p53d/d deciduae.
Fig. 3.
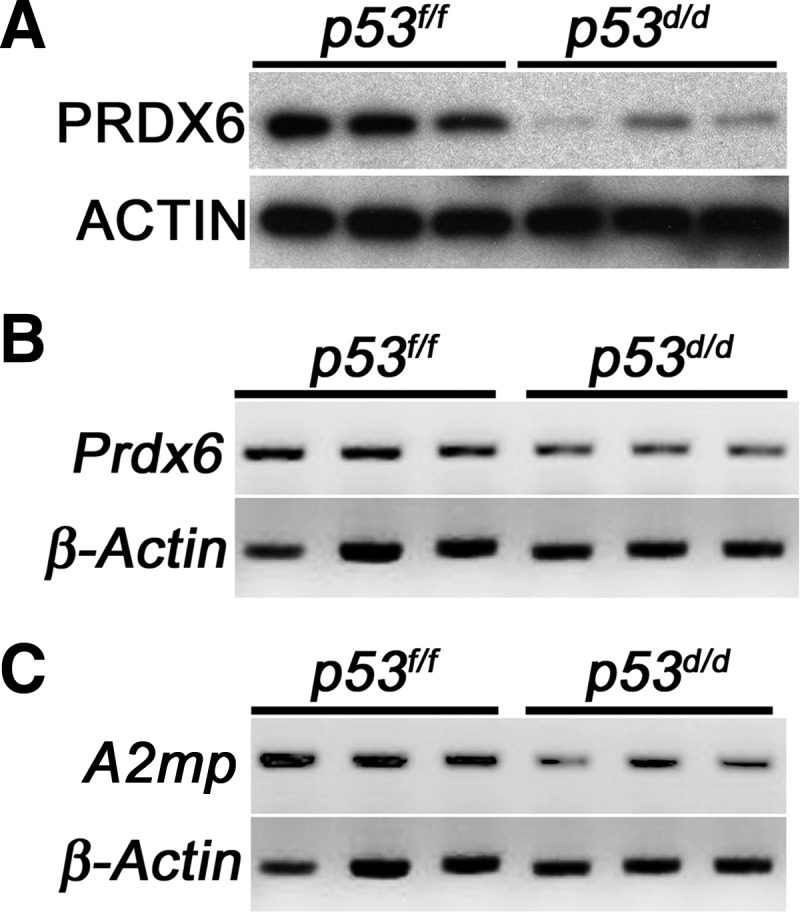
PRDX6 and A2MP are down-regulated in d 8 p53d/d deciduae. A and B, Protein and mRNA levels of PRDX6 in d 8 deciduae of p53d/d and p53f/f mice were examined by Western blotting (A) and RT-PCR (B). C, mRNA level of A2mp, in d 8 deciduae in p53d/d and p53f/f mice was examined by RT-PCR. ACTIN and β-Actin served as loading controls.
Mitochondrial ATP production is down-regulated in p53d/d deciduae
It is interesting to note that expression levels of 21 proteins in the mitochondrial ATP generation pathways were compromised in p53d/d deciduae compared with those in p53f/f females (Table 3). One major function of mitochondria is to generate ATP, and dysfunction of mitochondria accompanied by reduction in ATP production promotes senescence (32–38). Collectively, these findings suggest that reduced ATP generation resulting from mitochondrial dysfunction in p53d/d deciduae induces premature senescence.
Proteinase inhibitors are down-regulated in p53d/d deciduae
Some proteinases and proteinase inhibitors are known to play roles in pregnancy, such as trophoblast invasion (39). By proteomic analysis, we found that five proteinase inhibitors were down-regulated in p53d/d deciduae compared with those in p53f/f females (Table 2). Using RT-PCR analysis, we confirmed that the level of A2MP, induced in deciduae (18), was lower in p53d/d deciduae compared with p53f/f deciduae (Fig. 3C). Mouse A2mp, was cloned from mesometrial deciduae and is thought to be up-regulated by uterine natural killer cells to promote remodeling of the spiral artery to support decidualization (18, 19).
Discussion
Our recent observations have shown that uterine deletion of Trp53 increases the incidence of preterm birth resulting from premature decidual senescence with enhanced terminal differentiation of stromal cells and heightened mTORC1 signaling (2, 6). Mouse models for studying spontaneous preterm labor are very limited, because a drop in progesterone levels, unlike humans, is seen in most mouse models of preterm birth. p53d/d mice serve as a novel model to study the underlying mechanism of spontaneous preterm birth without progesterone withdrawal, a model seems relevant in the pursuit for the cause of premature birth in humans. In addition, our observations of senescence-associated decidual growth restriction and preterm birth with conditional deletion of uterine Trp53 have discovered a new critical role of p53 in uterine biology and parturition involving uterine cyclooxygenase 2-prostagrandin F2α signaling pathway, which is well known to be associated with preterm delivery. This is relevant to the observation that women at ages 35 or older are at higher risk for preterm birth (40), and p53 activity diminishes with ageing of mice (41).
Because many factors, including genetic alterations, OS, and inflammation/infection, can give rise to cellular senescence and preterm birth, we were interested in exploring other potential factors that are associated with these events by comparing decidual protein profiles in p53f/f and p53d/d females. Our findings of down-regulation of a cluster of antioxidant enzymes, including PRDX6, a unique member in the family, and a group of 21 proteins involved in ATP generation, are provocative and add new information to our repertoire. Because down-regulation of antioxidant enzymes provoking OS and mitochondrial dysfunction with reduced ATP generation are contributors to cellular senescence, we believe that preterm birth in p53d/d females arise from several deregulated pathways converging to the senescence pathway. Our results also show that proteins involved in the DNA replication pathway are up-regulated in p53d/d deciduae. This finding corroborates with our previous observation of decidual growth restriction with heightened stromal cell polyploidy and increased DNA content without cytokinesis in the absence of p53 (2).
Antioxidant enzymes help balance ROS generated under normal physiological conditions. Pregnancy is an example of a physiological system in which this balance is tightly regulated. Collapse of the balance due to the excessive generation of ROS imposing OS induces adverse effects on embryo development and pregnancy outcome. However, the mechanism by which a balance between ROS and antioxidant enzymes are achieved during pregnancy is not yet clearly understood (42–45). We have previously shown that FK506-binding protein 4 (FKBP52) mutant females with reduced uterine PRDX6 are susceptible to overt OS and show implantation failure. This failure can be overcome by providing mutant females with excess progesterone and antioxidant supplements (17). We also showed that induced expression of PRDX6 rescues H2O2-induced cell death in Fkbp52-deficient mouse embryonic fibroblasts with reduced PRDX6 levels. These results suggest that Fkbp52 deficiency diminishes the threshold against OS by reducing PRDX6 levels. In the current study, we again found that many antioxidant enzymes, including PRDX6, have levels that are lower in p53d/d deciduae, suggesting higher OS levels. Because it was shown that OS activates mTORC1 signaling, one possibility is that loss of Trp53 induces higher OS by reducing antioxidant levels, thus leading to higher mTORC1 activity (46). Moreover, p53 transcriptionally induces the expression of antioxidant enzymes (46), implying that a decidual Trp53 deficiency might reduce antioxidant activity.
Mitochondria's one major function is to generate ATP, and it is thought that its dysfunction plays a role in senescence (32, 35, 36). It has also been shown that decreased F1-ATPase activity might be responsible for age-dependent mitochondrial dysfunction (35, 36). There is also evidence that ADP/ATP ratio is in parallel with declining mitochondrial functions during cellular senescence of human diploid fibroblasts, which exhibit a limited replicative life span (32). Our findings of down-regulation of mitochondrial proteins related to ATP production are consistent with their role in premature decidual senescence in p53d/d mice. Notably, p53 localizes in mitochondria and directly implies positive effects on mitochondrial biogenesis and function (47, 48).
The family of A2M genes is evolutionarily conserved from lower to higher organisms and categorized as proteinase inhibitors. They are known to play a role in the immune system (49). Two isoforms of A2M, pregnancy zone proteins and A2MP, are expressed in mouse deciduae and are involved in decidualization (18, 50). We found that A2MP is down-regulated in p53d/d deciduae compared with p53f/f deciduae. Although most A2M are produced in the liver and are abundant in the blood, A2MP is instead locally found in the uterus and heart (18). Deciduae of Rag2/γC mutant mice without NK or T cells have reduced levels of A2MP and fail to remodel spiral arteries, but A2M supplementation rescues spiral artery remodeling (18). These data suggest that reduced levels of A2MP caused by Trp53 deletion may affect spiral artery remodeling in deciduae.
In conclusion, proteins related to DNA replication, antioxidation, mitochondrial ATP production, and proteinase inhibitors are potential downstream targets of p53 during decidualization. These findings add to our knowledge of repertoire of factors that could be involved in preterm birth and may open up new avenues to design better approaches to combat preterm birth and prematurity.
Supplementary Material
Acknowledgments
We thank Serenity Curtis for editing the manuscript and Jessica Martin for her assistance preparing samples for MS analysis.
This work was supported by grants from the National Center for Research Resources (5P41RR018522-10) and the National Institute of General Medical Sciences (8 P41 GM103493-10) from the National Institutes of Health for Proteomics, a grant from the Bill and Melinda Gates Foundation through the Grand Challenges Explorations Initiative, NIH Grant HD12304, HD068524 (to S.K.D.), and a Cincinnati Children's Hospital Medical Center Perinatal Institute Pilot/Feasibility grant (T.D.). Y.H. is supported by Precursory Research for Embryonic Science and Technology, Grant-in-Aid for Scientific Research from Japan Society for the Promotion of Science, Mochida Memorial Foundation for Medical and Pharmaceutical Research, and Kanae Foundation for the Promotion of Medical Science. Proteomic work was performed in the Environmental Molecular Sciences Laboratory, a U.S. Department of Energy (DOE) Office of Biological and Environmental Research national scientific user facility on the Pacific Northwest National Laboratory (PNNL) campus. PNNL is multiprogram national laboratory operated by Battelle for the DOE under Contract DE- AC05-76RL01830.
Disclosure Summary: The authors have nothing to disclose.
Footnotes
- Akt
- Protein kinase B
- A2MP
- α2-macroglobulin-P
- AMT
- accurate mass and time
- FKBP52
- FK506-binding protein 4
- IMS
- ion mobility separation
- LC
- liquid chromatography
- MS
- mass spectrometry
- MS/MS
- tandem MS
- mTORC1
- mammalian target of rapamycin complex 1
- OS
- oxidative stress
- p
- phosphorylated
- Pgr
- progesterone receptor
- PRDX6
- peroxiredoxin-6
- ROS
- reactive oxygen species.
References
- 1. Beck S, Wojdyla D, Say L, Betran AP, Merialdi M, Requejo JH, Rubens C, Menon R, Van Look PF. 2010. The worldwide incidence of preterm birth: a systematic review of maternal mortality and morbidity. Bull World Health Organ 88:31–38 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Hirota Y, Daikoku T, Tranguch S, Xie H, Bradshaw HB, Dey SK. 2010. Uterine-specific p53 deficiency confers premature uterine senescence and promotes preterm birth in mice. J Clin Invest 120:803–815 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Vogelstein B, Lane D, Levine AJ. 2000. Surfing the p53 network. Nature 408:307–310 [DOI] [PubMed] [Google Scholar]
- 4. Jonkers J, Meuwissen R, van der Gulden H, Peterse H, van der Valk M, Berns A. 2001. Synergistic tumor suppressor activity of BRCA2 and p53 in a conditional mouse model for breast cancer. Nat Genet 29:418–425 [DOI] [PubMed] [Google Scholar]
- 5. Soyal SM, Mukherjee A, Lee KY, Li J, Li H, DeMayo FJ, Lydon JP. 2005. Cre-mediated recombination in cell lineages that express the progesterone receptor. Genesis 41:58–66 [DOI] [PubMed] [Google Scholar]
- 6. Hirota Y, Cha J, Yoshie M, Daikoku T, Dey SK. 2011. Heightened uterine mammalian target of rapamycin complex 1 (mTORC1) signaling provokes preterm birth in mice. Proc Natl Acad Sci USA 108:18073–18078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Selman C, Tullet JM, Wieser D, Irvine E, Lingard SJ, Choudhury AI, Claret M, Al-Qassab H, Carmignac D, Ramadani F, Woods A, Robinson IC, Schuster E, Batterham RL, Kozma SC, Thomas G, Carling D, Okkenhaug K, Thornton JM, Partridge L, Gems D, Withers DJ. 2009. Ribosomal protein S6 kinase 1 signaling regulates mammalian life span. Science 326:140–144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Harrison DE, Strong R, Sharp ZD, Nelson JF, Astle CM, Flurkey K, Nadon NL, Wilkinson JE, Frenkel K, Carter CS, Pahor M, Javors MA, Fernandez E, Miller RA. 2009. Rapamycin fed late in life extends lifespan in genetically heterogeneous mice. Nature 460:392–395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Pohnke Y, Schneider-Merck T, Fahnenstich J, Kempf R, Christian M, Milde-Langosch K, Brosens JJ, Gellersen B. 2004. Wild-type p53 protein is up-regulated upon cyclic adenosine monophosphate-induced differentiation of human endometrial stromal cells. J Clin Endocrinol Metab 89:5233–5244 [DOI] [PubMed] [Google Scholar]
- 10. Yoshino O, Osuga Y, Hirota Y, Koga K, Yano T, Tsutsumi O, Taketani Y. 2003. Akt as a possible intracellular mediator for decidualization in human endometrial stromal cells. Mol Hum Reprod 9:265–269 [DOI] [PubMed] [Google Scholar]
- 11. Tan J, Raja S, Davis MK, Tawfik O, Dey SK, Das SK. 2002. Evidence for coordinated interaction of cyclin D3 with p21 and cdk6 in directing the development of uterine stromal cell decidualization and polyploidy during implantation. Mech Dev 111:99–113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Parker SB, Eichele G, Zhang P, Rawls A, Sands AT, Bradley A, Olson EN, Harper JW, Elledge SJ. 1995. p53-independent expression of p21Cip1 in muscle and other terminally differentiating cells. Science 267:1024–1027 [DOI] [PubMed] [Google Scholar]
- 13. Macleod KF, Sherry N, Hannon G, Beach D, Tokino T, Kinzler K, Vogelstein B, Jacks T. 1995. p53-Dependent and independent expression of p21 during cell growth, differentiation, and DNA damage. Genes Dev 9:935–944 [DOI] [PubMed] [Google Scholar]
- 14. Baker ES, Clowers BH, Li F, Tang K, Tolmachev AV, Prior DC, Belov ME, Smith RD. 2007. Ion mobility spectrometry-mass spectrometry performance using electrodynamic ion funnels and elevated drift gas pressures. J Am Soc Mass Spectrom 18:1176–1187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Baker ES, Livesay EA, Orton DJ, Moore RJ, Danielson WF, 3rd, Prior DC, Ibrahim YM, LaMarche BL, Mayampurath AM, Schepmoes AA, Hopkins DF, Tang K, Smith RD, Belov ME. 2010. An LC-IMS-MS platform providing increased dynamic range for high-throughput proteomic studies. J Proteome Res 9:997–1006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Mason EA, McDaniel EW. 1988. Transport properties of ions in gases. New York: John Wiley, Sons, Inc.; 1–560 [Google Scholar]
- 17. Hirota Y, Acar N, Tranguch S, Burnum KE, Xie H, Kodama A, Osuga Y, Ustunel I, Friedman DB, Caprioli RM, Daikoku T, Dey SK. 2010. Uterine FK506-binding protein 52 (FKBP52)-peroxiredoxin-6 (PRDX6) signaling protects pregnancy from overt oxidative stress. Proc Natl Acad Sci USA 107:15577–15582 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. He H, McCartney DJ, Wei Q, Esadeg S, Zhang J, Foster RA, Hayes MA, Tayade C, Van Leuven F, Croy BA. 2005. Characterization of a murine α2 macroglobulin gene expressed in reproductive and cardiovascular tissue. Biol Reprod 72:266–275 [DOI] [PubMed] [Google Scholar]
- 19. Tayade C, Esadeg S, Fang Y, Croy BA. 2005. Functions of α2 macroglobulins in pregnancy. Mol Cell Endocrinol 245:60–66 [DOI] [PubMed] [Google Scholar]
- 20. Livesay EA, Tang K, Taylor BK, Buschbach MA, Hopkins DF, LaMarche BL, Zhao R, Shen Y, Orton DJ, Moore RJ, Kelly RT, Udseth HR, Smith RD. 2008. Fully automated four-column capillary LC-MS system for maximizing throughput in proteomic analyses. Anal Chem 80:294–302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Zimmer JS, Monroe ME, Qian WJ, Smith RD. 2006. Advances in proteomics data analysis and display using an accurate mass and time tag approach. Mass Spectrom Rev 25:450–482 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Eng JK, Mccormack AL, Yates JR. 1994. An approach to correlate tandem mass-spectral data of peptides with amino-acid-sequences in a protein database. J Am Soc Mass Spectrom 5:976–989 [DOI] [PubMed] [Google Scholar]
- 23. Kim S, Gupta N, Pevzner PA. 2008. Spectral probabilities and generating functions of tandem mass spectra: a strike against decoy databases. J Proteome Res 7:3354–3363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Jaitly N, Mayampurath A, Littlefield K, Adkins JN, Anderson GA, Smith RD. 2009. Decon2LS: an open-source software package for automated processing and visualization of high resolution mass spectrometry data. BMC Bioinformatics 10:87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Monroe ME, Toli N, Jaitly N, Shaw JL, Adkins JN, Smith RD. 2007. VIPER: an advanced software package to support high-throughput LC-MS peptide identification. Bioinformatics 23:2021–2023 [DOI] [PubMed] [Google Scholar]
- 26. Polpitiya AD, Qian WJ, Jaitly N, Petyuk VA, Adkins JN, Camp DG, 2nd, Anderson GA, Smith RD. 2008. DAnTE: a statistical tool for quantitative analysis of -omics data. Bioinformatics 24:1556–1558 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Daikoku T, Tranguch S, Friedman DB, Das SK, Smith DF, Dey SK. 2005. Proteomic analysis identifies immunophilin FK506 binding protein 4 (FKBP52) as a downstream target of Hoxa10 in the periimplantation mouse uterus. Mol Endocrinol 19:683–697 [DOI] [PubMed] [Google Scholar]
- 28. Alam SM, Konno T, Sahgal N, Lu L, Soares MJ. 2008. Decidual cells produce a heparin-binding prolactin family cytokine with putative intrauterine regulatory actions. J Biol Chem 283:18957–18968 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Parums DV, Cordell JL, Micklem K, Heryet AR, Gatter KC, Mason DY. 1990. JC70: a new monoclonal antibody that detects vascular endothelium associated antigen on routinely processed tissue sections. J Clin Pathol 43:752–757 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Ramsey MR, Sharpless NE. 2006. ROS as a tumour suppressor? Nat Cell Biol 8:1213–1215 [DOI] [PubMed] [Google Scholar]
- 31. Davis JM, Auten RL. 2010. Maturation of the antioxidant system and the effects on preterm birth. Semin Fetal Neonatal Med 15:191–195 [DOI] [PubMed] [Google Scholar]
- 32. Fan W, Kou H, Shen D, LeRoy EC. 1998. Identification of altered expression of ADP/ATP translocase during cellular senescence in vitro. Exp Gerontol 33:457–465 [DOI] [PubMed] [Google Scholar]
- 33. Ames BN, Shigenaga MK, Hagen TM. 1995. Mitochondrial decay in aging. Biochim Biophys Acta 1271:165–170 [DOI] [PubMed] [Google Scholar]
- 34. Wallace DC, Shoffner JM, Trounce I, Brown MD, Ballinger SW, Corral-Debrinski M, Horton T, Jun AS, Lott MT. 1995. Mitochondrial DNA mutations in human degenerative diseases and aging. Biochim Biophys Acta 1271:141–151 [DOI] [PubMed] [Google Scholar]
- 35. Kröll J. 1994. The mitochondrial F1-ATPase and the aging process. Med Hypotheses 42:395–396 [DOI] [PubMed] [Google Scholar]
- 36. Shigenaga MK, Hagen TM, Ames BN. 1994. Oxidative damage and mitochondrial decay in aging. Proc Natl Acad Sci USA 91:10771–10778 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Ozawa T. 1995. Mechanism of somatic mitochondrial DNA mutations associated with age and diseases. Biochim Biophys Acta 1271:177–189 [DOI] [PubMed] [Google Scholar]
- 38. Pall ML. 1990. Very low ATP/ADP ratios with aging of the natural death senescence mutant of Neurospora crassa. Mech Ageing Dev 52:287–294 [DOI] [PubMed] [Google Scholar]
- 39. Salamonsen LA. 1999. Role of proteases in implantation. Rev Reprod 4:11–22 [DOI] [PubMed] [Google Scholar]
- 40. Goldenberg RL, Culhane JF, Iams JD, Romero R. 2008. Epidemiology and causes of preterm birth. Lancet 371:75–84 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Feng Z, Hu W, Teresky AK, Hernando E, Cordon-Cardo C, Levine AJ. 2007. Declining p53 function in the aging process: a possible mechanism for the increased tumor incidence in older populations. Proc Natl Acad Sci USA 104:16633–16638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Jauniaux E, Poston L, Burton GJ. 2006. Placental-related diseases of pregnancy: involvement of oxidative stress and implications in human evolution. Hum Reprod Update 12:747–755 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Sharma JB, Sharma A, Bahadur A, Vimala N, Satyam A, Mittal S. 2006. Oxidative stress markers and antioxidant levels in normal pregnancy and pre-eclampsia. Int J Gynaecol Obstet 94:23–27 [DOI] [PubMed] [Google Scholar]
- 44. Hausburg MA, Dekrey GK, Salmen JJ, Palic MR, Gardiner CS. 2005. Effects of paraquat on development of preimplantation embryos in vivo and in vitro. Reprod Toxicol 20:239–246 [DOI] [PubMed] [Google Scholar]
- 45. Leyens G, Verhaeghe B, Landtmeters M, Marchandise J, Knoops B, Donnay I. 2004. Peroxiredoxin 6 is upregulated in bovine oocytes and cumulus cells during in vitro maturation: role of intercellular communication. Biol Reprod 71:1646–1651 [DOI] [PubMed] [Google Scholar]
- 46. Vigneron A, Vousden KH. 2010. p53, ROS and senescence in the control of aging. Aging 2:471–474 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Zhou S, Kachhap S, Singh KK. 2003. Mitochondrial impairment in p53-deficient human cancer cells. Mutagenesis 18:287–292 [DOI] [PubMed] [Google Scholar]
- 48. Donahue RJ, Razmara M, Hoek JB, Knudsen TB. 2001. Direct influence of the p53 tumor suppressor on mitochondrial biogenesis and function. FASEB J 15:635–644 [DOI] [PubMed] [Google Scholar]
- 49. Armstrong PB, Quigley JP. 1999. α2-Macroglobulin: an evolutionarily conserved arm of the innate immune system. Dev Comp Immunol 23:375–390 [DOI] [PubMed] [Google Scholar]
- 50. Bao L, Devi S, Bowen-Shauver J, Ferguson-Gottschall S, Robb L, Gibori G. 2006. The role of interleukin-11 in pregnancy involves up-regulation of α2-macroglobulin gene through janus kinase 2-signal transducer and activator of transcription 3 pathway in the decidua. Mol Endocrinol 20:3240–3250 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.