Abstract
Zhengzhou is the capital of Henan province, where severe HIV prevalence was found in former paid plasma donors. In recent years, the HIV epidemic in men who have sex with men (MSM) increased rapidly in the city. To explore the subtype distribution and genetic characterization of HIV in MSM in Zhengzhou city, phylogenetic analysis was fulfilled based on the full-length gag, pol, and partial env gene. A total of 31 HIV-1-seropositive MSM individuals were enrolled. The full length gag, pol, and partial env gene were amplified and sequenced. Multiple subtypes, including CRF01_AE (45.2%), subtype B (38.7%), and CRF07_BC (16.1%), were identified. Close phylogenetic relationships among our strains with strains from the Henan local area, Hebei MSM population, Beijing area, and Liaoning area were found, suggesting a multiple introduction of HIV into the population. The results will provide clues for prevention and for changes in behavior in the Henan MSM population and also detailed sequence data for vaccine design.
In China, the percentages of newly reported HIV cases attributed to men who have sex with men (MSM) increased dramatically in the past decade. In 2001, only 0.2% of new Chinese HIV cases were estimated to be infected through homosexual contacts; the proportion increased to 12.2% in 2007 and to 32.5% by 2009.1–4 MSM has become the second most vulnerable population for HIV infection in China after injection drug users,5 especially in some metropolitan areas of China, where the estimated infectious rates reach 3.0–4.6% of the population.2,6,7
Henan province, located in central eastern China, is one of the worst affected provinces in terms of HIV/AIDS. Initially, the HIV-1 epidemic had been largely confined to former plasma donors who were infected before 1996 when the laws banning the commercial collection of blood and blood products were introduced and enforced. The original prevalence rate of HIV infection in former plasma donors in Henan province was very high, which caused a severe public health problem.
According to the studies done between 2004 and 2006, the HIV prevalence rate in former plasma donors in some prefectures in Henan province was as high as 10.9%, although more than 10 years has passed since the outbreak of HIV infection in the population.8 Recent studies showed that HIV transmission that originated from former plasma donors had only a limited role in the spread of HIV. HIV transmission between discordant couples and from infected former plasma donor mothers to child was kept at a low level.
It seems that the problem caused by the prevalence of HIV in the former HIV high-epidemic province is being solved. However, in recent years, HIV-1 prevalence among MSM has increased rapidly in Henan and more of an emphasis has been put on it.9 According to the sentinel surveillance site in Zhengzhou monitoring HIV among MSM, the HIV prevalence among MSM surveyed rose from 0.88% in 2005 to 2.67% in 2006.8 Up to now, no information was available on subtype distribution and genetic characterizations of HIV prevalent in the population. To gain a more accurate understanding of HIV prevalence in MSM in Henan province, we analyzed the genetic characterization of HIV-1 strains prevalent in the MSM group in Zhengzhou city, the capital of Henan province in China, based on the HIV-1 full-length gag, pol, and partial env genes.
A total of 31 HIV-positive MSM residing in Zhengzhou City were randomly recruited at the local HIV/AIDS sentinel surveillance site into the study in 2010 with informed consents. The epidemiological background was collected through specific investigation by trained interviewers. The average age of the participants was 33.2 years (ranging from 21 to 59 years). All subjects were of Han ethnicity. Two participants presented at least one symptom, including fever, oral thrush, nausea, debility, diarrhea, and depression. CD4+ T cells were counted using flow cytometry and reagents were provided by Becton Dickinson Biosciences (San Jose, CA) with fresh whole blood samples before plasma was separated and stored in a −80°C freezer. The CD4 T cell counts varied widely, ranging from 80 to 1122 cells/μl.
Viral RNA was extracted from 500 μl of HIV-1-positive plasma specimens after being concentrated using a high pure viral RNA kit (Roche, USA). Viral full length gag gene, pol gene, and partial env gene (the C2-V3 region) were amplified separately using reverse transcriptional nested PCR according to methods previously described.10 Positive PCR products were sequenced by the Huada Genomics Company (China) with a variety of internal specific primers (available on request) after being purified. All of the sequenced fragments were edited and assembled into contiguous sequences as described before.11 A total of 29 full length gag genes (93.5%), 14 full length pol genes (45.2%), and 21 env C2-V3 genes (67.7%) were successfully obtained. A basic local alignment search tool (BLAST) search (http://hiv-web.lanl.gov/content/index) against the HIV-1 sequence database and among themselves showed no evidence of sample contamination. Analysis of sequences showed that all of the gene structures were normal with the proper open reading frames (ORFs).
All of sequences obtained in the study were submitted to the NCBI viral genotyping tool (http://www.ncbi.nih.gov/projects/genotyping/formpage.cgi) to determine genotype. The results based on different gene regions were combined together to determine the subtype of the strain, which was further confirmed by phylogenetic analysis. For phylogenetic analysis, all of assembled sequences were aligned with the reference sequences representing subtypes A–D, F–H, J, K, CRF01_AE, CRF07_BC, and CRF08_BC (http://www.hiv.lanl.gov) using Clustal_W online software (http://www.ebi.ac.uk/Tools/msa/clustalw2/) and edited manually using BioEdit software (version 7.0.0; T. Hall, North Carolina State University, Raleigh, NC).
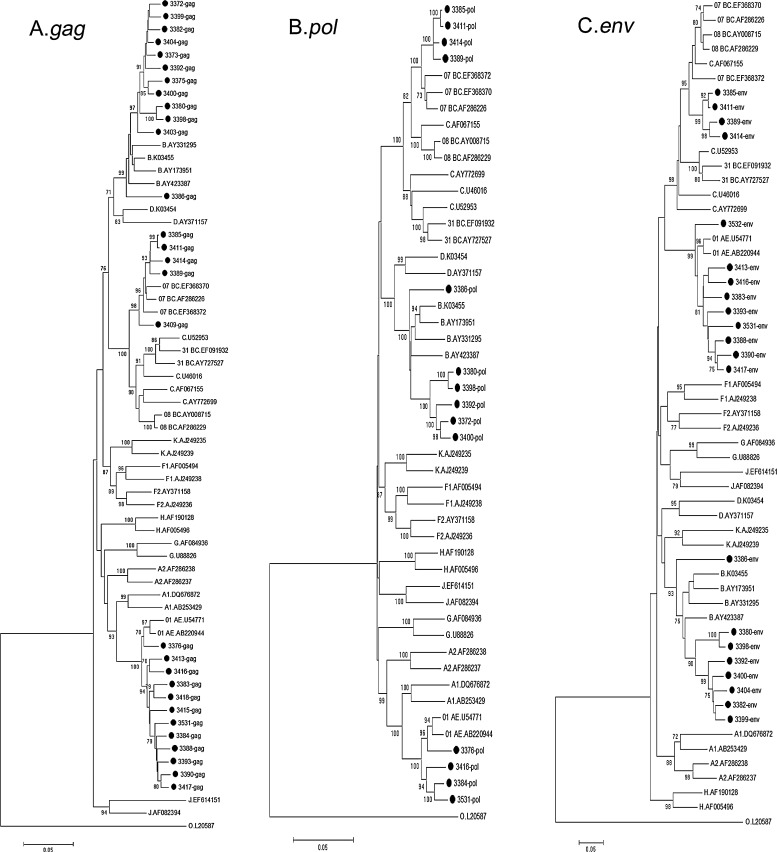
Evolutionary distances were computed by the Kimura two-parameter method including both transitions and transversions and phylogenetic trees were generated using the neighbor-joining method with MEGA 5.0. The reliability of the topologies was estimated by performing bootstrap analysis with 1000 replicates. As shown in Fig. 1, HIV subtypes from 31 cases were successfully determined and the distribution of HIV-1 genotypes in the study population was as follows: 14 cases (45.2%) of subtype CRF01_AE, 5 cases (16.1%) of subtype CRF07_BC, and 12 cases (38.7%) of subtype B. The possible intertype mosaicism was screened with the online Recombination Identification Program (RIP; version 3.0; http://hiv-web.lanl.gov) and further confirmed by Bootscan and SimPlot (version 3.2; S. Ray, Johns Hopkins University, Baltimore, MD), and no recombination was identified (data not shown).
FIG. 1.
Phylogenetic tree analysis. A neighbor-joining tree was created with the full length gag gene (A), pol gene (B), and partial env gene (C) of HIV-1 sequences from men who have sex with men (MSMs) in Zhengzhou (black dots) and the reference sequences of subtypes A–D, F–H, J, K, CRF01_AE, CRF07_BC, CRF08_BC, and group O (http://hiv-web.lanl. gov/). The lengths of the gag, pol, and partial env genes of HIV-1 sequences obtained in the study were 1503, 3056, and 542 base pairs separately by using HXB2 as the genomic calibrator. Each reference sequence is labeled with the HIV-1 subtype, followed by the sequence name. The bootstrap probability (more than 70%, 1000 replica) is indicated at the corresponding nodes of the tree. The scale bar represents 5% genetic distance (0.05 substitutions per site).
To estimate the diversity of sequences in this study belonging to different subtypes, the genetic distances of different subtype sequences were analyzed using the Kimura two-parameter model (Table 1). A larger mean genetic distance was found among subtype B and CRF01_AE. Relatively smaller mean genetic distances were found in CRF07_BC. The high genetic divergences suggested multiple introductions of HIV strains into the MSM population instead of a founder effect.
Table 1.
Genetic Distances Among Sequences Belonging to Different Subtypes
| |
Genetic distances (mean±SD) |
||
|---|---|---|---|
| Gene (cases) | CRF01_AE (cases) | Subtype B | CRF07_BC |
| gag (n=29) | 0.040±0.003 (n=12) | 0.044±0.003 (n=12) | 0.030±0.003 (n=5) |
| pol (n=14) | 0.040±0.003 (n=4) | 0.045±0.002 (n=6) | 0.012±0.001 (n=4) |
| env (C2-V3) (n=21) | 0.097±0.009 (n=9) | 0.130±0.010 (n=8) | 0.052±0.010 (n=4) |
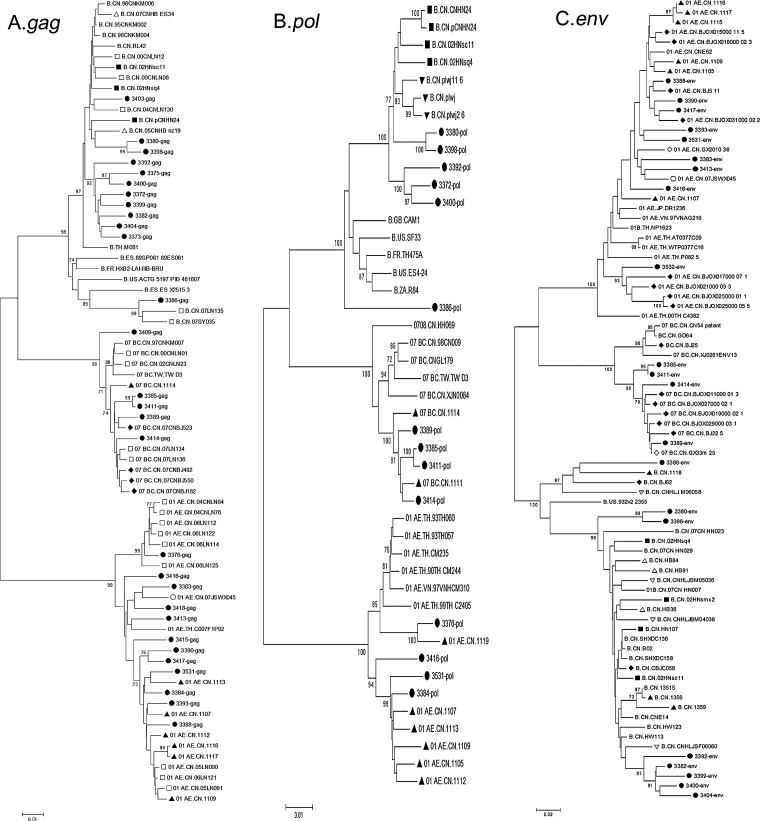
Genetic analysis based on sequence proved to be effective for exploring the relationship among HIV strains from different areas or populations. In a previous study on MSM residing in Shijiazhuang city of Hebei Province, which is close to Henan province, a close relationship was found between HIV strains from the same MSM population in Shijiazhuang city and Beijing city, but not between those prevalent in different populations in Shijiazhuang city, suggesting the intrapopulation transmission of HIV in Shijiazhuang city.10 In this study, we determined whether it was the same in Zhengzhou city as in Shijiazhuang city. To explore strains with the highest similarity to our strains, BLAST was used to search and select the HIV-1 reference sequences with the highest similarity to our strains. At least five sequences with the greatest similarity to each of our strains were downloaded from the Los Alamos National Laboratory (http://www.hiv.lanl.gov) and aligned together with our strains using the online CLUSTAL_W software (http://www.ebi.ac.uk/Tools/msa/clustalw2/).
Phylogenetic trees were constructed by the neighbor-joining (NJ) method listed in the MEGA software. The reliability of the branching patterns was tested by bootstrap analysis with 1000 replicates. For the NJ tree based on gag genes, 29 of our strains were aligned with 47 reference sequences (Fig. 2A). As shown in the phylogenetic tree, sequences of different subtypes grouped together and were supported by a high bootstrap value; 11 of 12 subtype B sequences clustered with Thai B strains and only one strain (3386 gag) showed a closer relationship to European and American subtype B strains. It was reported that subtype B strains prevalent in the Chinese MSM population mainly consisted of European and American subtype B strains; however, in Zhengzhou city, we found that Thai B, which was reported to be prevalent in former plasma donors, was the main subtype B strains prevalent in the MSM population. This observation supposed that MSM in Zhengzhou city had a close relationship with other local high-risk populations.
FIG. 2.
Evolutionary relationship among HIV-1 sequences from Zhengzhou MSMs and other areas. Phylogenetic trees were constructed with the full length gag gene (A), pol gene (B), and partial env gene (C) of HIV-1 sequences from MSMs in Zhengzhou (●) and the reference sequences download from the Los Alamos National Laboratory (http://hiv-web.lanl.gov/), which showed the highest similarity to our strains. Each reference sequence is labeled with the HIV-1 subtype, followed by the isolated country and sequence name. The bootstrap probability (more than 70%, 1000 replica) was also indicated at the corresponding nodes of the tree. The scale bar representing unit genetic distance was labeled below. Reference strains from different areas, including Hubei province (▵), Hebei provinces (▴), Henan local area (▪), Liaoning province (□), Jiangsu province (○), Beijing (♦), Guangxi province (⋄), Heilongjiang province (▿), and Fujian province (▾), were labeled with different symbols.
Of all 12 subtype B sequences, eight strains grouped into one cluster with a high bootstrap value (92%), indicating there was a closely related transmission network in the population. The other four subtype B strains distributed among the reference sequences and showed a close relationship to strains from Hubei province, Liaoning province, and those from ES, suggesting their different origin. Multiple introduction of HIV was also observed in CRF01_AE strains in the same NJ tree based on a gag gene fragment. One CRF01_AE strain (3376 gag) clustered with those from Liaoning province, suggesting that it was a close relative of Liaoning province strains. A separate cluster were constructed with six our CRF01_AE strains, six strains from the Hebei MSM population and three from Liaoning province, providing proof of a close relationship between MSM residing in Henan and Hebei province. The other five CRF01_AE strains were distributed among the reference sequences. Five gag genes of subtype CRF07_BC were obtained from the Zhengzhou MSM population and were closely related to strains from Shijiazhuang and Beijing MSM, and also some strains from Liaoning province.
The high diversity of HIV strains prevalent in the MSM population in Zhengzhou can also be observed in the NJ tree constructed based on pol and env genes (Fig. 2). It has been estimated that there are 17.82 million MSM in China.12 Because of the large size of the population, the recent rapid increase in HIV prevalence in MSM could mean a new emerging HIV epidemic in China. Due to the social discrimination and cultural stigma associated with homosexual behaviors in China, most of the Chinese MSM engaged in sex with both men and women within and out of marriage,13,14 which facilitates the transmission of HIV from high-risk populations to the general population, and may contribute to the growing number of HIV-positive women infected through unprotected sex. The special characterization of the MSM population makes them into the focus of HIV prevention and behavior change. Analysis of the genetic background of HIV in MSM will provide clues for prevention and changes in behavior.
In this study, the genetic characterization of the HIV-1 full-length gag genes, pol genes, and partial env genes in the MSM population in Zhengzhou city was illustrated. High genetic diversity and the multiple introduction of HIV into the MSM population in Zhengzhou city were observed. Furthermore, associations of strains from the MSM population and former plasma donors were found. Those results will facilitate epidemiological investigations and also provide public health strategies for the prevention of the spread of the virus in Henan province, China.15–18
Sequence Data
The gene sequences were deposited in the GenBank with the following accession numbers: JQ234979–JQ235042.
Acknowledgments
This work was supported by the National Key S&T Special Projects on Major Infectious Diseases (Grants 2008ZX10001-004, 2008ZX10001-002, and 2008ZX10001-012) and the National Natural Science Foundation of China (Nos. 30700706 and 81072348). We also want to express our thanks to Dr. Feng Gao at Duke Human Vaccine Institute, Duke University Medical Center for providing the technical support in reverse transcription and nested PCR. Lin Li, Guoqing Sun, and Tianyi Li contributed equally to this work.
Author Disclosure Statement
No competing financial interests exist.
References
- 1.China Ministry of Health, UNAIDS. Beijing, China: China Ministry of Health; 2010. World Health Organization (WHO): 2009 estimates for the HIV/AIDS epidemic in China. [Google Scholar]
- 2.China Ministry of Health. Beijing: State Council AIDS Working Committee Office, UN Theme Group on AIDS in China; 2007. UN Theme Group on HIV/AIDS in China: A joint assessment of HIV/AIDS prevention, treatment, care in China. [Google Scholar]
- 3.China Ministry of Health, UNAIDS. Beijing: National Center for AIDS/STD Prevention and Control; 2006. WHO: 2005 update on the HIV/AIDS epidemic, response in China. [Google Scholar]
- 4.China Ministry of Health, UN Theme Group on HIV/AIDS in China. A joint assessment of HIV/AIDS prevention, treatment, care in China. Beijing, China: Ministry of Health, UNAIDS China Office. 2003. http://data.unaids.org/UNA-docs/china_joint_assessment_2003_en.pdf http://data.unaids.org/UNA-docs/china_joint_assessment_2003_en.pdf
- 5.Zhang BC. Chu QS. MSM and HIV/AIDS in China. Cell Res. 2005;15:858–864. doi: 10.1038/sj.cr.7290359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Choi KH. Lui H. Guo Y, et al. Lack of HIV testing and awareness of HIV infection among men who have sex with men, Beijing, China. AIDS Educ Prev. 2006;18:33–43. doi: 10.1521/aeap.2006.18.1.33. [DOI] [PubMed] [Google Scholar]
- 7.UNAIDS/WHO. AIDS epidemic update. Dec, 2006. 2007. http://www.unaids.org/ http://www.unaids.org/
- 8.Li N. Wang Z. Sun D, et al. HIV among plasma donors and other high-risk groups in Henan, China. J Acquir Immune Defic Syndr. 2010;53S1:S41–47. doi: 10.1097/QAI.0b013e3181c7d717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cui Z. Wang Z. Liu G, et al. An analysis of epidemic and trend of HIV/AIDS in Henan province [in Chinese] Chin J AIDS/STD. 2006;12:324–326. [Google Scholar]
- 10.Li L. Lu X. Li H, et al. High genetic diversity of HIV-1 was found in men who have sex with men in Shijiazhuang, China. Infect Genet Evol. 2011;11:1487–1492. doi: 10.1016/j.meegid.2011.05.017. [DOI] [PubMed] [Google Scholar]
- 11.Li L. Liang S. Chen L, et al. Genetic characterization of 13 subtype CRF01_AE near full-length genomes in Guangxi, China. AIDS Res. Hum. Retroviruses. 2010;26:699–704. doi: 10.1089/aid.2010.0026. [DOI] [PubMed] [Google Scholar]
- 12.Zhang BC. Li XF. Shi TX, et al. Survey on the high risk behaviors and other AIDS/STI related factors among men who have sex with men (MSM) in mainland China (2001) Chin J Dermatol. 2002;35:214–216. [Google Scholar]
- 13.Zhang D. Bi P. Lv F, et al. Changes in HIV prevalence and sexual behavior among men who have sex with men in a northern Chinese city: 2002–2006. J Infect. 2007;55:456–463. doi: 10.1016/j.jinf.2007.06.015. [DOI] [PubMed] [Google Scholar]
- 14.Zhang X. Li S. Li X, et al. Characterization of HIV-1 subtypes and viral antiretroviral drug resistance in men who have sex with men in Beijing, China. AIDS. 2007;21:S59–S65. doi: 10.1097/01.aids.0000304698.47261.b1. [DOI] [PubMed] [Google Scholar]
- 15.de Oliveira T. Pybus OG. Rambaut A, et al. Molecular epidemiology: HIV-1 and HCV sequences from Libyan outbreak. Nature. 2006;444:836–837. doi: 10.1038/444836a. [DOI] [PubMed] [Google Scholar]
- 16.Hue S. Pillay D. Clewley JP, et al. Genetic analysis reveals the complex structure of HIV-1 transmission within defined risk groups. Proc Natl Acad Sci USA. 2005;102:4425–4429. doi: 10.1073/pnas.0407534102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hue S. Clewley JP. Cane PA, et al. HIV-1 pol gene variation is sufficient for reconstruction of transmissions in the era of antiretroviral therapy. AIDS. 2004;18:719–728. doi: 10.1097/00002030-200403260-00002. [DOI] [PubMed] [Google Scholar]
- 18.Yahi N. Fantini J. Tourres C, et al. Use of drug resistance sequence data for the systematic detection of non-B human immunodeficiency virus type 1 (HIV-1) subtypes: How to create a sentinel site for monitoring the genetic diversity of HIV-1 at a country scale. J Infect Dis. 2001;183:1311–1317. doi: 10.1086/319859. [DOI] [PubMed] [Google Scholar]