Abstract
The trans-activator of transcription (Tat) of HIV-1 plays an important role in viral infection and pathogenesis. We examined the genetic characteristics of exon 1 of the tat gene derived from 102 seropositive subjects from southern India. Database-derived Indian (n=105) and global (n=413) HIV-1C sequences were also used for viral epidemiological signature pattern analysis in the Tat open reading frame (ORF). We identified HIV-1C as the most predominant genetic subtype (99%) and the presence of a novel A1C recombinant strain in one study participant. After examining all the available HIV-1C Indian sequences from primary clinical isolates and database-derived sequences, we found a high level of sequence conservation (92.6±12%) within Tat amino acid residues. Furthermore, signature pattern analysis identified five amino acid positions in Tat that contained signature residues unique for Indian HIV-1C consisting of 21A, 24N, 29K, 40K, and 60Q. Our data have direct relevance for subunit-based Tat HIV-1 vaccine development.
Human immunodeficiency virus type 1 (HIV-1) trans-activator of transcription (Tat) is an 86–101 residue regulatory protein (9–11 kDa), which plays an important role in viral replication. Apart from its primary role in transcriptional activation, Tat is involved in a number of intracellular and extracellular functions during viral infection, all collectively contributing to viral pathogenesis.1,2 Over the past two decades, the primary focus of vaccine development programs has been the viral structural proteins including env, gag, and pol; however, success was eluded in most of these clinical trials. High genetic diversity of the structural proteins could partly underlie the poor outcome of these studies. The conserved nature of Tat, with its central role in disease pathogenesis, makes this viral antigen a potential candidate for vaccine development against HIV/AIDS. A few clinical trials are presently in progress with Tat as a preventive and therapeutic HIV vaccine (http://www.hiv1tat-vaccines.info).
In India, an estimated 2.5 million people have been infected with HIV-1. The epidemic is predominantly driven by HIV-1 subtype C.3,4 Two different HIV vaccine clinical trials both using the “prime-boost” regimen are presently in progress in India. The components of these vaccines include the synthetic env, gag, reverse transcriptase, rev, tat, and nef antigens of HIV-1 subtype C.5 Unfortunately, data on the genetic characteristics of these viral antigens derived directly from the circulating viral strains are lacking. To fill this gap, we aim to characterize Tat exon-1 derived from a southern Indian clinical cohort, using a cross-sectional study design. Furthermore, using tat, we examine the nature of the viral genetic subtype distribution in southern India. Importantly, we delineate several signature amino acid residues in Tat unique for India and characterized conserved domains and epitopes in Tat with implications for vaccine design. The study was approved by the Institutional Ethical Review Board, St. John's Medical College and Hospital, Bangalore, India.
A single peripheral blood sample was collected from 110 HIV-1-infected patients attending the Infectious Disease Clinic at St. John's Medical College Hospital, Bangalore, between November 2009 and May 2011. Routine CD4 count was performed using a dual-platform flow cytometer (FACSCalibur, BD, USA) and viral load was measured with Abbott m2000rt system (Abbott, Germany). Genomic DNA from whole blood was extracted using a commercial kit (QIAamp Blood DNA kit, Qiagen, Germany). HIV-1 tat exon 1 was amplified using nested polymerase chain reaction (PCR) followed by double pass population sequencing. The first round of PCR was carried out using the following external primers: N1331 (5′–TAGTAGAGGATAGATGGAACAAGSCCCCAG-3′) and N1332 (5′– TCTGTGGGTACACAGGCATGTGTRGCCCA-3′). The cycling conditions were 1 cycle at 95°C for 2 min, 3 cycles at 94°C for 1 min, 55°C for 1 min, and 72°C for 3 min, followed by 32 cycles at 94°C for 1 min, 60°C for 1 min, and 72°C for 3 min with a final extension at 72°C for 10 min. Thereafter, an approximately 450 bp fragment containing tat exon 1 was amplified with internal primers N111 (5′-GGGTGYCARCATAGCAGAATAGGCATT-3′) and N1156 (5′-TCATTGCCACTGTCTTCTGCTCT-3′). The cycling conditions were 1 cycle at 94°C for 2 min, 3 cycles at 94°C for 1 min, 55°C for 30 s, and 72°C for 3 min, followed by 32 cycles at 94°C for 1 min, 60°C for 1 min, and 72°C for 1 min with a final extension at 72°C for 5 min.
Purified PCR products were sequenced using the primers N111 and N1156. The sequences thus generated were manually edited in Bio-Edit version 7.0.9.0. Country-specific (n=2027) Tat sequences were downloaded from the HIV-1 Los Alamos Database (www.hiv.lanl.gov) taking care to exclude duplicate sequences from the same subject (accessed on July 2011). Phylogenetic trees were constructed using the neighbor-joining method (Kimura two-parameter model) in 500 bootstrapped data sets, using the Molecular Evolutionary Genetics Analysis software ver. 5 (MEGA 5) for subtyping.6 A viral epidemiological signature pattern was identified using the VESPA program available at the Los Alamos Database.7 To assess the presence of recombination events in the Tat sequences, RIP 3.0 was primarily used. For sequences that indicated an event of recombination, bootscan analysis was performed in SimPlot version 3.5.1 using 100 bp window size and 20 bp step size to map the precise breakpoint.8 The recombination event was further confirmed by region-specific phylogenetic analysis in MEGA 5.
Out of the total 110 subjects included in this study, we could successfully amplify tat exon 1 from 102. The clinical characteristics of the 102 study subjects were as follows: the mean age was 38 years (SD±8.2), 60.7% (62/102) were male, the median CD4 count was 184 cells/mm3 (IQR: 32–42), and the median viral load was 5.9 log10 copies/ml (IQR: 5.2–6.0) (data not available for 21 patients), and 14.7% (15/102) were on antiretroviral treatment (ART).
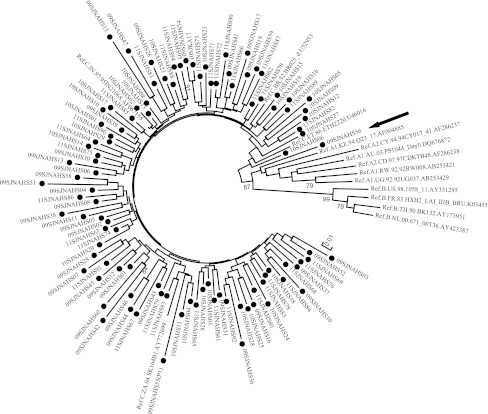
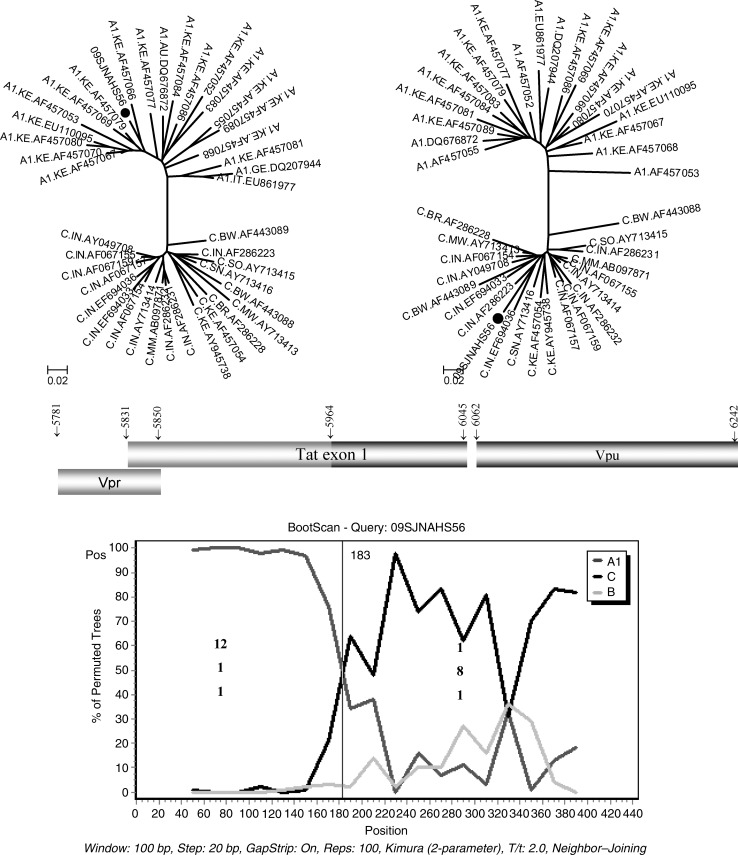
While a large majority of the viral isolates (101/102, 99%) were found to be subtype C in our clinical cohort, the predominant genetic subtype in India, a single viral strain was identified to be an A1C recombinant based on the tat gene (Fig. 1). Bootscan analysis carried out in the vpr/tat/vpu cassette (HXB2 position 5781 to 6242) mapped the recombination breakpoint to position 5964 of HXB2, which correlates with amino acid 44 in Tat exon 1 (Fig. 2). The fragment-specific phylogenetic analysis also confirmed the recombination event. Additional recombination events have been found in the V4 region of the env gene of this viral strain (data not shown). A repeat sampling after 1 year reconfirmed the presence of the A1C recombinant strain with the same breakpoints in tat and the env gene suggesting genetic stability of the recombination event.
FIG. 1.
Phylogenetic analysis of HIV-1 subtype C Tat exon 1 sequences. A phylogenetic tree was constructed using 102 sequences of Tat exon 1 generated through the present work (marked with filled circles). The neighbor-joining method (Kimura two-parameter model) was employed with 500 bootstrapped data sets, using the Molecular Evolutionary Genetics Analysis software ver. 5 (MEGA 5). The reference subtype A1, A2, B, and C Tat sequences were downloaded from the Los Alamos database. The A1C recombinant viral strain is highlighted with an arrow.
FIG. 2.
Bootscan and region-specific phylogenetic analysis of the A1C recombinant strain. The recombination breakpoint was identified at HXB2 position 5964. Region-specific phylogenetic analysis using the neighbor-joining tree also confirmed the recombination event. Informative sites are shown for both the segments.
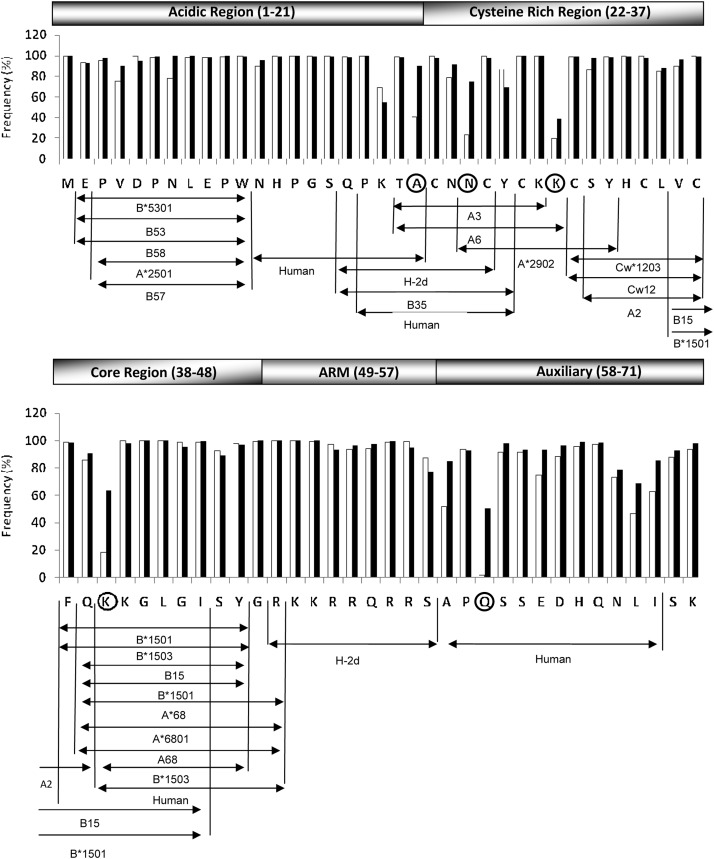
The frequency of conservation of each of the amino acid residues in Tat and the respective experimentally predefined CTL epitopes are presented in Fig. 3. With the exception of five positions, a high level of conservation [mean 92.6% (SD±12)] was observed in Tat amino acid residues in the Indian viral strains. Importantly, we found signature residues 21A, 24N, 29K, 40K, and 60Q in Tat exon 1 in Indian Tat C sequences (Fig. 3). Of note, in a previous analysis, two of these positions contained different signature amino acid residues 29H and 60P in Tat.9 It was interesting to observe changes in subtype C-specific signature residues from 29H and 60P to 29K and 60Q in Indian C sequences after the inclusion of our cohort sequences (n=101) with database sequences (n=105). In Indian C Tat sequences, 9.7% (20/206) and 45.6% (94/206) had conserved 29H and 60P, respectively.
FIG. 3.
Conservation of amino acid residues in the functional domains of Tat. The frequency of amino acid conservation at each position in Tat exon 1 has been evaluated comparing a total of 206 Tat sequences of Indian origin (including 101 sequences from the present study) with 403 global subtype C sequences. Five positions that represent the signature residues of the Indian strains have been encircled. Horizontal lines with arrowheads represent known CTL epitopes in Tat and the HLA classes by which the epitopes are restricted have been indicated.
HIV genetic variability and recombination are important confounding factors in the quest for developing effective intervention strategies. Efforts have been directed toward the development of therapeutic and prophylactic vaccines that are expected to be effective against the diverse circulating HIV-1 strains. Our study for the first time identified the presence of an A1C recombinant strain in southern India with a mosaic pattern different from the A1C recombinants reported previously from western India (AF067156, EU447777, and EU158864).8,10 Recent studies from the north and northeastern parts of India have also identified recombination events within the accessory gene vpr as well as tat.11,12 A previous study identified three different recombinant forms of B/C from southern India based on LTR and env genes.13 Collectively, these reports allude to the possibility of an increasing incidence of viral recombination in India. Our finding of the highly conserved nature of the tat gene among global HIV-1C strains has implications for subunit-based vaccine development strategies targeting structural, accessory, and Tat regulatory proteins.
Acknowledgments
The authors would like to thank the staff at the Infectious Disease Clinic for their generous help with field work and sample and data collections. We would also like to thank the study participants. U.N. was supported by the Einstein AIDS International Training and Research Program grant (D43 TW001403). Sequences generated from this study have been submitted to GenBank (accession numbers JQ241040–JQ241141).
Author Disclosure Statement
No competing financial interests exist.
References
- 1.Jeang KT. Xiao H. Rich EA. Multifaceted activities of the HIV-1 transactivator of transcription, Tat. J Biol Chem. 1999;274:28837–28840. doi: 10.1074/jbc.274.41.28837. [DOI] [PubMed] [Google Scholar]
- 2.Campbell GR. Loret EP. What does the structure-function relationship of the HIV-1 Tat protein teach us about developing an AIDS vaccine? Retrovirology. 2009;6:50. doi: 10.1186/1742-4690-6-50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Neogi U. Prarthana BS. Gupta S, et al. Naturally occurring polymorphisms and primary drug resistance profile among antiretroviral-naïve individuals in Bangalore, India. AIDS Res Hum Retroviruses. 2010;26:1097–1101. doi: 10.1089/aid.2010.0092. [DOI] [PubMed] [Google Scholar]
- 4.Neogi U. Sood V. Banerjee S, et al. Global HIV-1 molecular epidemiology with special reference to genetic analysis of HIV-1 subtypes circulating in North India: Functional and pathogenic implications of genetic variation. Indian J Exp Biol. 2009;47:424–431. [PubMed] [Google Scholar]
- 5.Ramanathan VD. Kumar M. Mahalingam J, et al. A phase 1 study to evaluate the safety and immunogenicity of a recombinant HIV type 1 subtype C-modified vaccinia Ankara virus vaccine candidate in Indian volunteers. AIDS Res Hum Retroviruses. 2009;25:1107–1116. doi: 10.1089/aid.2009.0096. [DOI] [PubMed] [Google Scholar]
- 6.Tamura K. Peterson D. Peterson N. Stecher G. Nei M. Kumar S. MEGA5: Molecular Evolutionary Genetics Analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol. 2011;28:2731–2739. doi: 10.1093/molbev/msr121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Korber B. Myers G. Signature pattern analysis: A method for assessing viral sequence relatedness. AIDS Res Hum Retroviruses. 1992;8:1549–1560. doi: 10.1089/aid.1992.8.1549. [DOI] [PubMed] [Google Scholar]
- 8.Lole KS. Bollinger RC. Paranjape RS, et al. Full-length human immunodeficiency virus type 1 genomes from subtype C-infected seroconverters in India, with evidence of intersubtype recombination. J Virol. 1999;73:152–160. doi: 10.1128/jvi.73.1.152-160.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ranga U. Shankarappa R. Siddappa NB, et al. Tat protein of human immunodeficiency virus type 1 subtype C strains is a defective chemokine. J Virol. 2004;78:2586–2590. doi: 10.1128/JVI.78.5.2586-2590.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lall M. Gupta RM. Sen S. Kapila K. Tripathy SP. Paranjape RS. Profile of primary resistance in HIV-1 infected treatment naïve individuals from western India. AIDS Res Hum Retroviruses. 2008;24:987–990. doi: 10.1089/aid.2008.0079. [DOI] [PubMed] [Google Scholar]
- 11.Mullick R. Sengupta S. Sarkar K. Chakrabarti S. Molecular characterization of tat gene and long terminal repeat region of human immunodeficiency virus type-1 detected among the injecting drug users (IDUs) of Manipur, India: Identification of BC recombinants. Virus Res. 2010;147:195–201. doi: 10.1016/j.virusres.2009.10.024. [DOI] [PubMed] [Google Scholar]
- 12.Bano AS. Sood V. Neogi U, et al. Genetic and functional characterization of human immunodeficiency virus type 1 VprC variants from north India: Presence of unique recombinants with mosaic genomes from B, C and D subtypes within the open reading frame of Vpr. J Gen Virol. 2009;90:2768–2776. doi: 10.1099/vir.0.011080-0. [DOI] [PubMed] [Google Scholar]
- 13.Siddappa NB. Dash PK. Mahadevan A, et al. Identification of unique B/C recombinant strains of HIV-1 in the southern state of Karnataka, India. AIDS. 2005;19:1426–1429. doi: 10.1097/01.aids.0000180795.49016.89. [DOI] [PubMed] [Google Scholar]