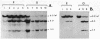Abstract
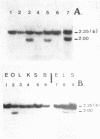
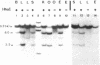
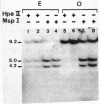
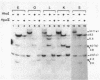
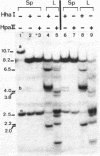
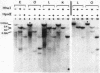
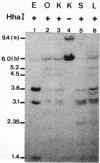
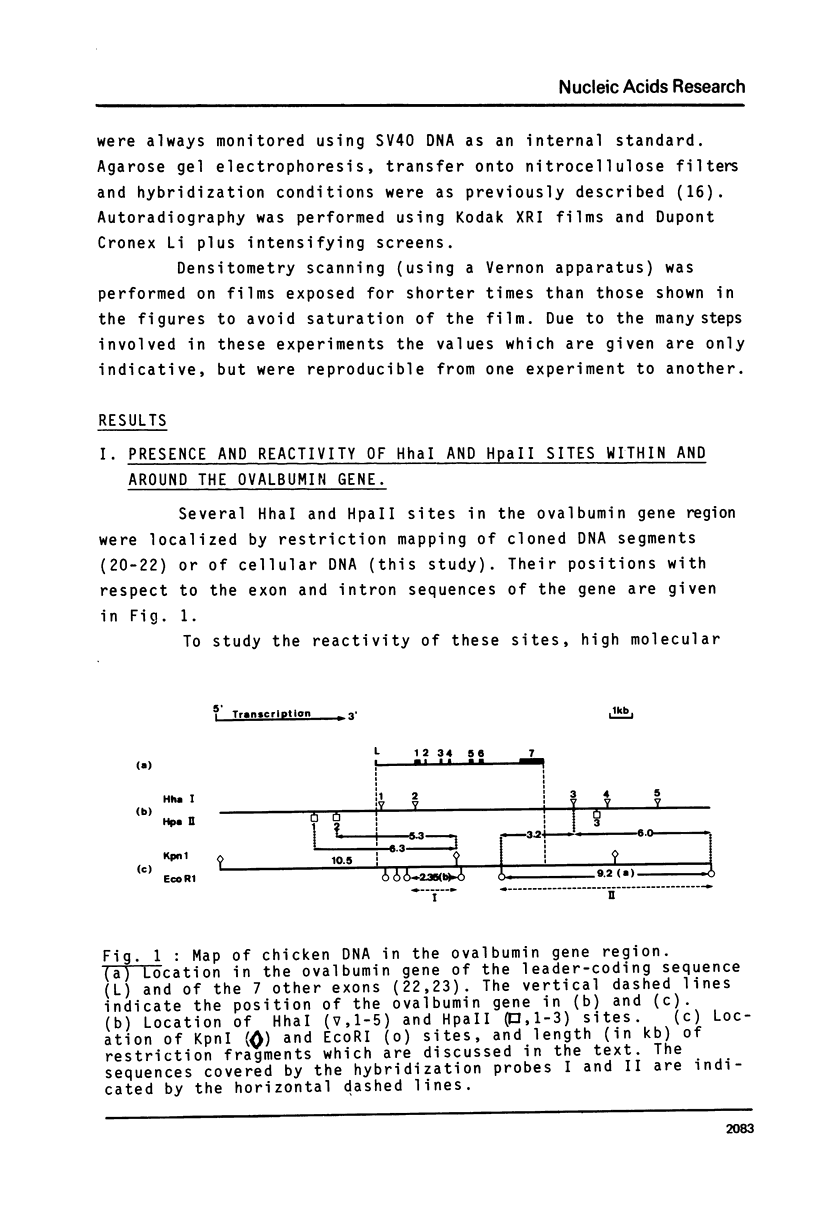
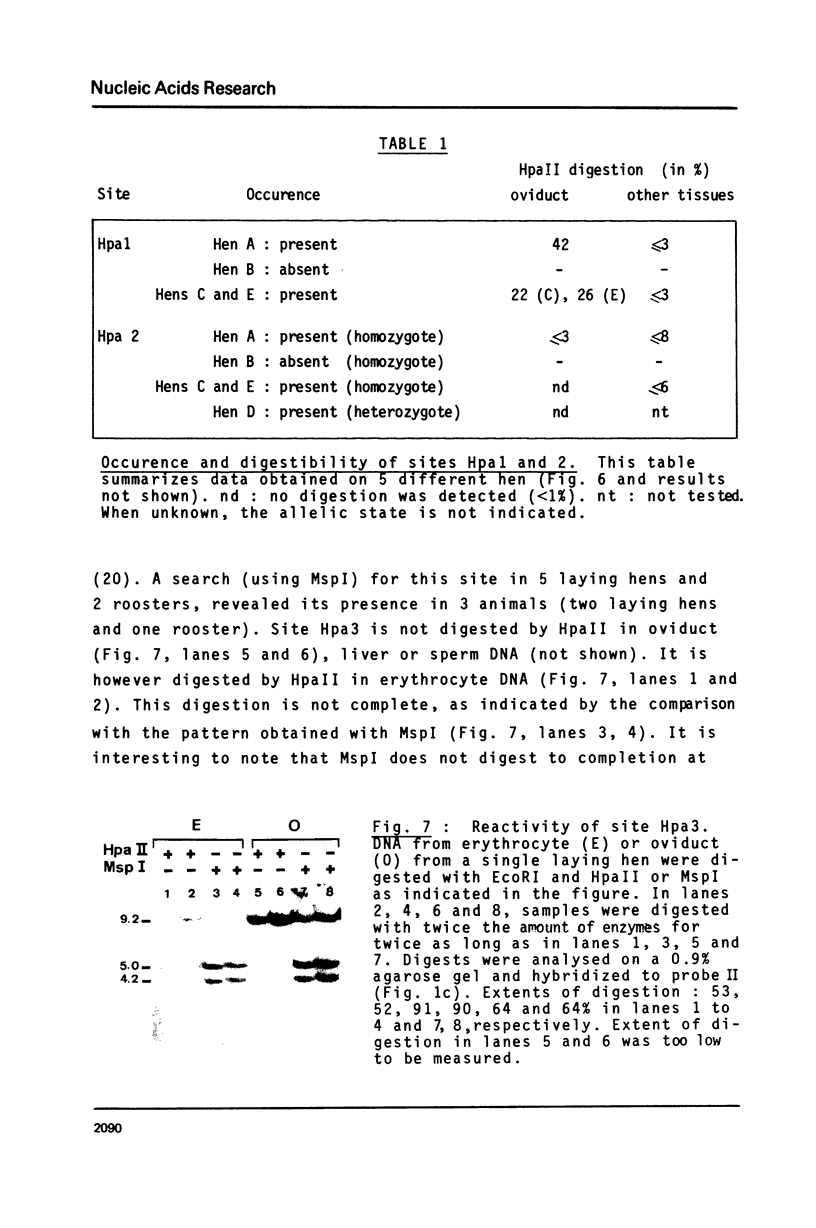
The restriction enzymes HhaI and HpaII, whose activity is inhibited by cytosine methylation within their recognition sites, have been utilised as probes to study methylation in the vicinity of the ovalbumin gene in DNA from various chicken tissues. This was complemented by a preliminary study of methylation in the regions of chicken ovotransferrin (conalbumin), ovomucoid and beta-globin genes. From our data we conclude that HaI or HpaII sites can be divided in 3 classes according to their pattern of methylation in different tissues. In the first class of sites (mV class) the extent of methylation varies in different tissues. The patterns obtained show that methylation at the sites located within and around the 3 genes which code for egg white proteins is in general lowest in oviduct of laying hen, where these genes are expressed. However some sites are not methylated (m- class) and others are 95 to 100% resistant (m+ class) to digestion by HhaI or HpaII in the DNAs of all the tissues which were tested. Our study has also revealed a remarkable number of allelic variants for the presence of HhaI or HpaII sites in the region of the ovalbumin gene.
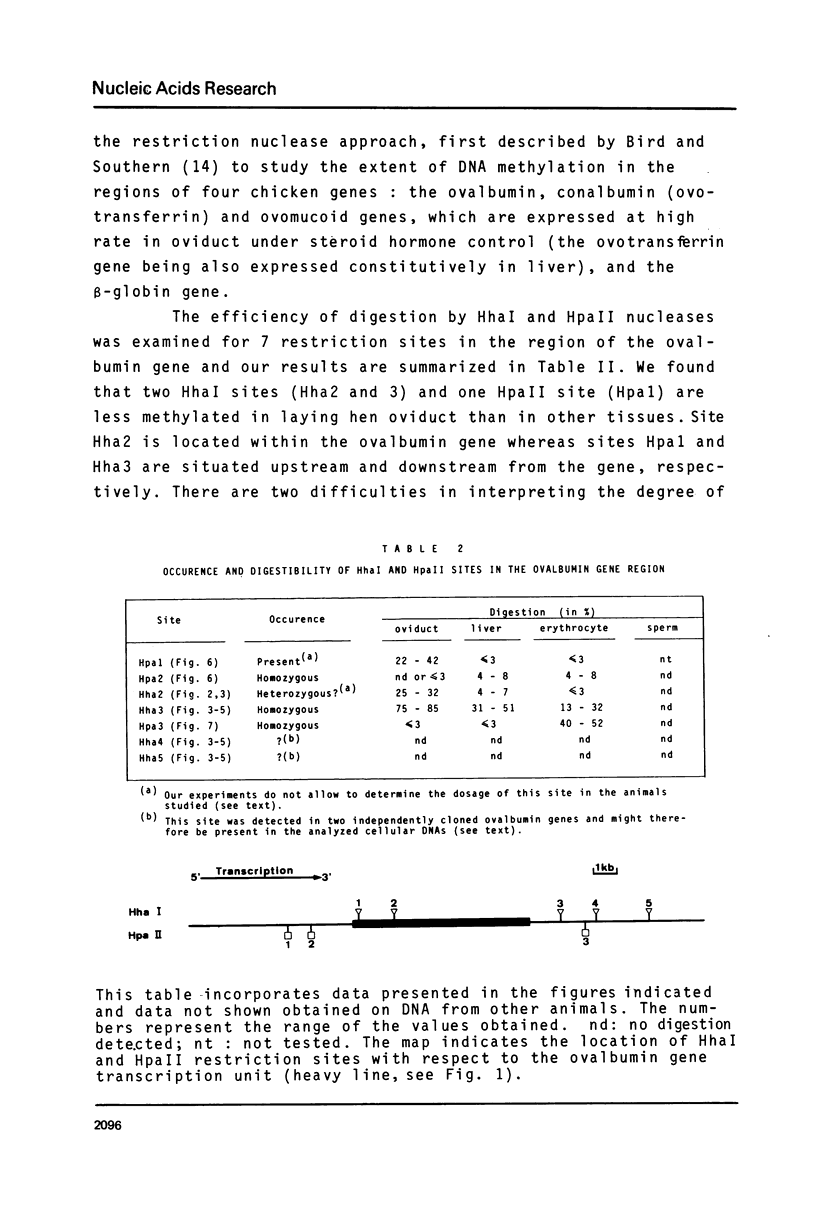
Full text
PDF


















Images in this article
Selected References
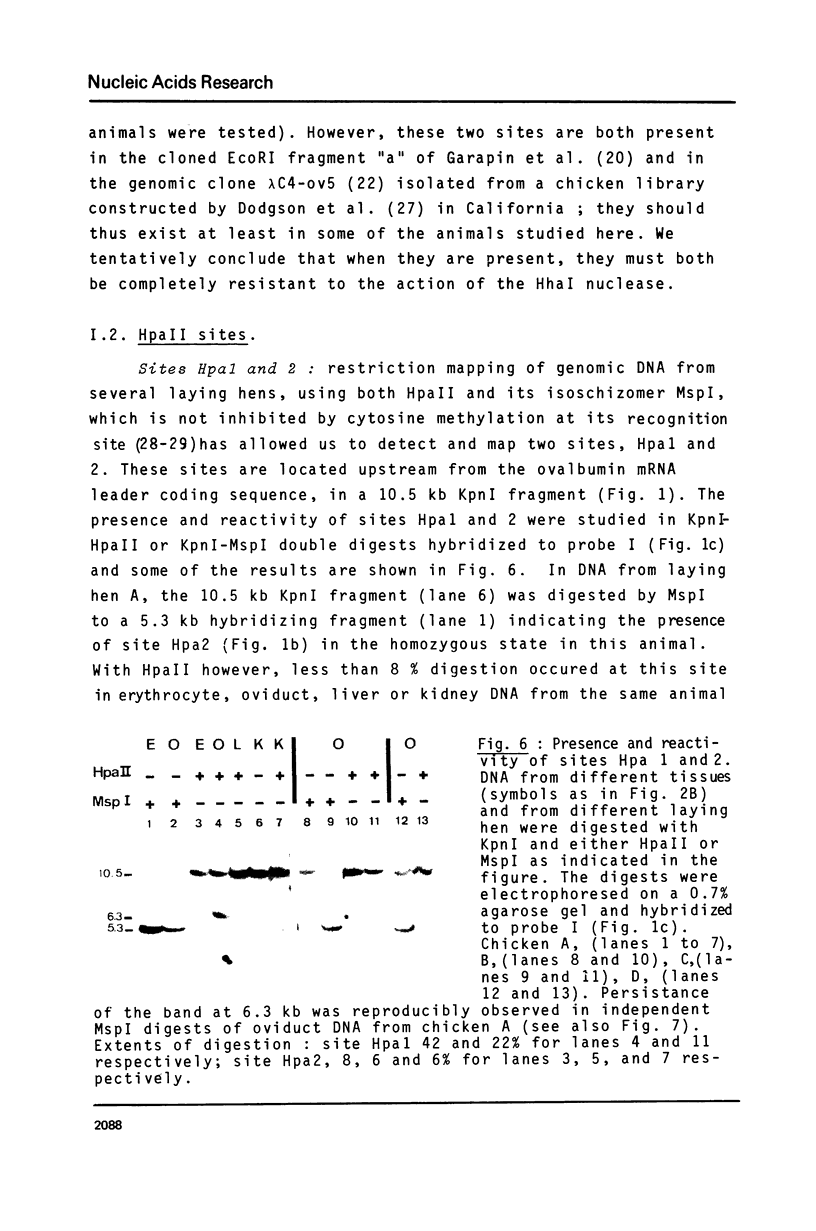
These references are in PubMed. This may not be the complete list of references from this article.
- Adams R. L., McKay E. L., Douglas J. T., Burdon R. H. Methylation of nucleosomal and nuclease sensitive DNA. Nucleic Acids Res. 1977 Sep;4(9):3097–3108. doi: 10.1093/nar/4.9.3097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bird A. P., Southern E. M. Use of restriction enzymes to study eukaryotic DNA methylation: I. The methylation pattern in ribosomal DNA from Xenopus laevis. J Mol Biol. 1978 Jan 5;118(1):27–47. doi: 10.1016/0022-2836(78)90242-5. [DOI] [PubMed] [Google Scholar]
- Bird A. P., Taggart M. H., Smith B. A. Methylated and unmethylated DNA compartments in the sea urchin genome. Cell. 1979 Aug;17(4):889–901. doi: 10.1016/0092-8674(79)90329-5. [DOI] [PubMed] [Google Scholar]
- Breathnach R., Benoist C., O'Hare K., Gannon F., Chambon P. Ovalbumin gene: evidence for a leader sequence in mRNA and DNA sequences at the exon-intron boundaries. Proc Natl Acad Sci U S A. 1978 Oct;75(10):4853–4857. doi: 10.1073/pnas.75.10.4853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breathnach R., Mandel J. L., Chambon P. Ovalbumin gene is split in chicken DNA. Nature. 1977 Nov 24;270(5635):314–319. doi: 10.1038/270314a0. [DOI] [PubMed] [Google Scholar]
- Browne M. J., Cato A. C., Burdon R. H. The distribution of modified and non-modified C-G doublets in BHK-21 cell DNA. FEBS Lett. 1978 Jul 1;91(1):69–73. doi: 10.1016/0014-5793(78)80019-2. [DOI] [PubMed] [Google Scholar]
- Burdon R. H., Adams R. L. The in vivo methylation of DNA in mouse fibroblasts. Biochim Biophys Acta. 1969 Jan 21;174(1):322–329. doi: 10.1016/0005-2787(69)90257-3. [DOI] [PubMed] [Google Scholar]
- Cochet M., Perrin F., Gannon F., Krust A., Chambon P., McKnight G. S., Lee D. C., Mayo K. E., Palmiter R. Cloning of an almost full-length chicken conalbumin double-stranded cDNA. Nucleic Acids Res. 1979 Jun 11;6(7):2435–2452. doi: 10.1093/nar/6.7.2435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coulondre C., Miller J. H., Farabaugh P. J., Gilbert W. Molecular basis of base substitution hotspots in Escherichia coli. Nature. 1978 Aug 24;274(5673):775–780. doi: 10.1038/274775a0. [DOI] [PubMed] [Google Scholar]
- DOSKOCIL J., SORM F. Distribution of 5-methylcytosine in pyrimidine sequences of deoxyribonucleic acids. Biochim Biophys Acta. 1962 Jun 11;55:953–959. doi: 10.1016/0006-3002(62)90909-5. [DOI] [PubMed] [Google Scholar]
- Dawid I. B., Brown D. D., Reeder R. H. Composition and structure of chromosomal and amplified ribosomal DNA's of Xenopus laevis. J Mol Biol. 1970 Jul 28;51(2):341–360. doi: 10.1016/0022-2836(70)90147-6. [DOI] [PubMed] [Google Scholar]
- Dodgson J. B., Strommer J., Engel J. D. Isolation of the chicken beta-globin gene and a linked embryonic beta-like globin gene from a chicken DNA recombinant library. Cell. 1979 Aug;17(4):879–887. doi: 10.1016/0092-8674(79)90328-3. [DOI] [PubMed] [Google Scholar]
- Engel J. D., Dodgson J. B. Analysis of the adult and embryonic chicken globin genes in chromosomal DNA. J Biol Chem. 1978 Nov 25;253(22):8239–8246. [PubMed] [Google Scholar]
- Fisher E. F., Caruthers M. H. Studies on gene control regions XII. The functional significance of a lac operator constitutive mutation. Nucleic Acids Res. 1979 Sep 25;7(2):401–416. doi: 10.1093/nar/7.2.401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gannon F., O'Hare K., Perrin F., LePennec J. P., Benoist C., Cochet M., Breathnach R., Royal A., Garapin A., Cami B. Organisation and sequences at the 5' end of a cloned complete ovalbumin gene. Nature. 1979 Mar 29;278(5703):428–434. doi: 10.1038/278428a0. [DOI] [PubMed] [Google Scholar]
- Garapin A. C., Cami B., Roskam W., Kourilsky P., Le Pennec J. P., Perrin F., Gerlinger P., Cochet M., Chambon P. Electron microscopy and restriction enzyme mapping reveal additional intervening sequences in the chicken ovalbumin split gene. Cell. 1978 Jul;14(3):629–639. doi: 10.1016/0092-8674(78)90247-7. [DOI] [PubMed] [Google Scholar]
- Garapin A. C., Lepennec J. P., Roskam W., Perrin F., Cami B., Krust A., Breathnach R., Chambon P., Kourilsky P. Isolation by molecular cloning of a fragment in the split ovalbumin gene. Nature. 1978 Jun 1;273(5661):349–354. doi: 10.1038/273349a0. [DOI] [PubMed] [Google Scholar]
- Grippo P., Iaccarino M., Parisi E., Scarano E. Methylation of DNA in developing sea urchin embryos. J Mol Biol. 1968 Sep 14;36(2):195–208. doi: 10.1016/0022-2836(68)90375-6. [DOI] [PubMed] [Google Scholar]
- Gross-Bellard M., Oudet P., Chambon P. Isolation of high-molecular-weight DNA from mammalian cells. Eur J Biochem. 1973 Jul 2;36(1):32–38. doi: 10.1111/j.1432-1033.1973.tb02881.x. [DOI] [PubMed] [Google Scholar]
- Holliday R., Pugh J. E. DNA modification mechanisms and gene activity during development. Science. 1975 Jan 24;187(4173):226–232. [PubMed] [Google Scholar]
- Humphries P., Cochet M., Krust A., Gerlinger P., Kourilsky P., Chambon P. Molecular cloning of extensive sequences of the in vitro synthesized chicken ovalbumin structural gene. Nucleic Acids Res. 1977 Jul;4(7):2389–2406. doi: 10.1093/nar/4.7.2389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hynes N. E., Groner B., Sippel A. E., Nguyen-Huu M. C., Schütz G. mRNA complexity and egg white protein mRNA content in mature and hormone-withdrawn oviduct. Cell. 1977 Aug;11(4):923–932. doi: 10.1016/0092-8674(77)90303-8. [DOI] [PubMed] [Google Scholar]
- Jeffreys A. J. DNA sequence variants in the G gamma-, A gamma-, delta- and beta-globin genes of man. Cell. 1979 Sep;18(1):1–10. doi: 10.1016/0092-8674(79)90348-9. [DOI] [PubMed] [Google Scholar]
- Lai E. C., Woo S. L., Dugaiczyk A., O'Malley B. W. The ovalbumin gene: alleles created by mutations in the intervening sequences of the natural gene. Cell. 1979 Jan;16(1):201–211. doi: 10.1016/0092-8674(79)90201-0. [DOI] [PubMed] [Google Scholar]
- Lee D. C., McKnight G. S., Palmiter R. D. The action of estrogen and progesterone on the expression of the transferrin gene. A comparison of the response in chick liver and oviduct. J Biol Chem. 1978 May 25;253(10):3494–3503. [PubMed] [Google Scholar]
- Mandel J. L., Breathnach R., Gerlinger P., Le Meur M., Gannon F., Chambon P. Organization of coding and intervening sequences in the chicken ovalbumin split gene. Cell. 1978 Jul;14(3):641–653. doi: 10.1016/0092-8674(78)90248-9. [DOI] [PubMed] [Google Scholar]
- Mann M. B., Smith H. O. Specificity of Hpa II and Hae III DNA methylases. Nucleic Acids Res. 1977 Dec;4(12):4211–4221. doi: 10.1093/nar/4.12.4211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGhee J. D., Ginder G. D. Specific DNA methylation sites in the vicinity of the chicken beta-globin genes. Nature. 1979 Aug 2;280(5721):419–420. doi: 10.1038/280419a0. [DOI] [PubMed] [Google Scholar]
- McReynolds L., O'Malley B. W., Nisbet A. D., Fothergill J. E., Givol D., Fields S., Robertson M., Brownlee G. G. Sequence of chicken ovalbumin mRNA. Nature. 1978 Jun 29;273(5665):723–728. doi: 10.1038/273723a0. [DOI] [PubMed] [Google Scholar]
- Nordstrom J. L., Roop D. R., Tsai M. J., O'Malley B. W. Identification of potential ovomucoid mRNA precursors in chick oviduct nuclei. Nature. 1979 Mar 22;278(5702):328–331. doi: 10.1038/278328a0. [DOI] [PubMed] [Google Scholar]
- O'Malley B. W., McGuire W. L., Kohler P. O., Korenman S. G. Studies on the mechanism of steroid hormone regulation of synthesis of specific proteins. Recent Prog Horm Res. 1969;25:105–160. doi: 10.1016/b978-0-12-571125-8.50006-5. [DOI] [PubMed] [Google Scholar]
- Palmiter R. D. Regulation of protein synthesis in chick oviduct. I. Independent regulation of ovalbumin, conalbumin, ovomucoid, and lysozyme induction. J Biol Chem. 1972 Oct 25;247(20):6450–6461. [PubMed] [Google Scholar]
- Perrin F., Cochet M., Gerlinger P., Cami B., LePennec J. P., Chambon P. The chicken conalbumin gene: studies of the organization of cloned DNAs. Nucleic Acids Res. 1979 Jun 25;6(8):2731–2748. doi: 10.1093/nar/6.8.2731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pollock J. M., Jr, Swihart M., Taylor J. H. Methylation of DNA in early development: 5-methyl cytosine content of DNA in sea urchin sperm and embryos. Nucleic Acids Res. 1978 Dec;5(12):4855–4861. doi: 10.1093/nar/5.12.4855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riggs A. D. X inactivation, differentiation, and DNA methylation. Cytogenet Cell Genet. 1975;14(1):9–25. doi: 10.1159/000130315. [DOI] [PubMed] [Google Scholar]
- Riggs A. D. X inactivation, differentiation, and DNA methylation. Cytogenet Cell Genet. 1975;14(1):9–25. doi: 10.1159/000130315. [DOI] [PubMed] [Google Scholar]
- Salser W. Globin mRNA sequences: analysis of base pairing and evolutionary implications. Cold Spring Harb Symp Quant Biol. 1978;42(Pt 2):985–1002. doi: 10.1101/sqb.1978.042.01.099. [DOI] [PubMed] [Google Scholar]
- Scarano E. The control of gene function in cell differentiation and in embryogenesis. Adv Cytopharmacol. 1971 May;1:13–24. [PubMed] [Google Scholar]
- Simon D., Grunert F., von Acken U., Döring H. P., Kröger H. DNA-methylase from regenerating rat liver: purification and characterisation. Nucleic Acids Res. 1978 Jun;5(6):2153–2167. doi: 10.1093/nar/5.6.2153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singer J., Roberts-Ems J., Riggs A. D. Methylation of mouse liver DNA studied by means of the restriction enzymes msp I and hpa II. Science. 1979 Mar 9;203(4384):1019–1021. doi: 10.1126/science.424726. [DOI] [PubMed] [Google Scholar]
- Sümegi J., Breedveld D., Hossenlopp P., Chambon P. A rapid procedure for purification of EcoRI endonuclease. Biochem Biophys Res Commun. 1977 May 9;76(1):78–85. doi: 10.1016/0006-291x(77)91670-9. [DOI] [PubMed] [Google Scholar]
- Turnbull J. F., Adams R. L. DNA methylase: purification from ascites cells and the effect of various DNA substrates on its activity. Nucleic Acids Res. 1976 Mar;3(3):677–695. doi: 10.1093/nar/3.3.677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanyushin B. F., Mazin A. L., Vasilyev V. K., Belozersky A. N. The content of 5-methylcytosine in animal DNA: the species and tissue specificity. Biochim Biophys Acta. 1973 Mar 28;299(3):397–403. doi: 10.1016/0005-2787(73)90264-5. [DOI] [PubMed] [Google Scholar]
- Weintraub H., Groudine M. Chromosomal subunits in active genes have an altered conformation. Science. 1976 Sep 3;193(4256):848–856. doi: 10.1126/science.948749. [DOI] [PubMed] [Google Scholar]