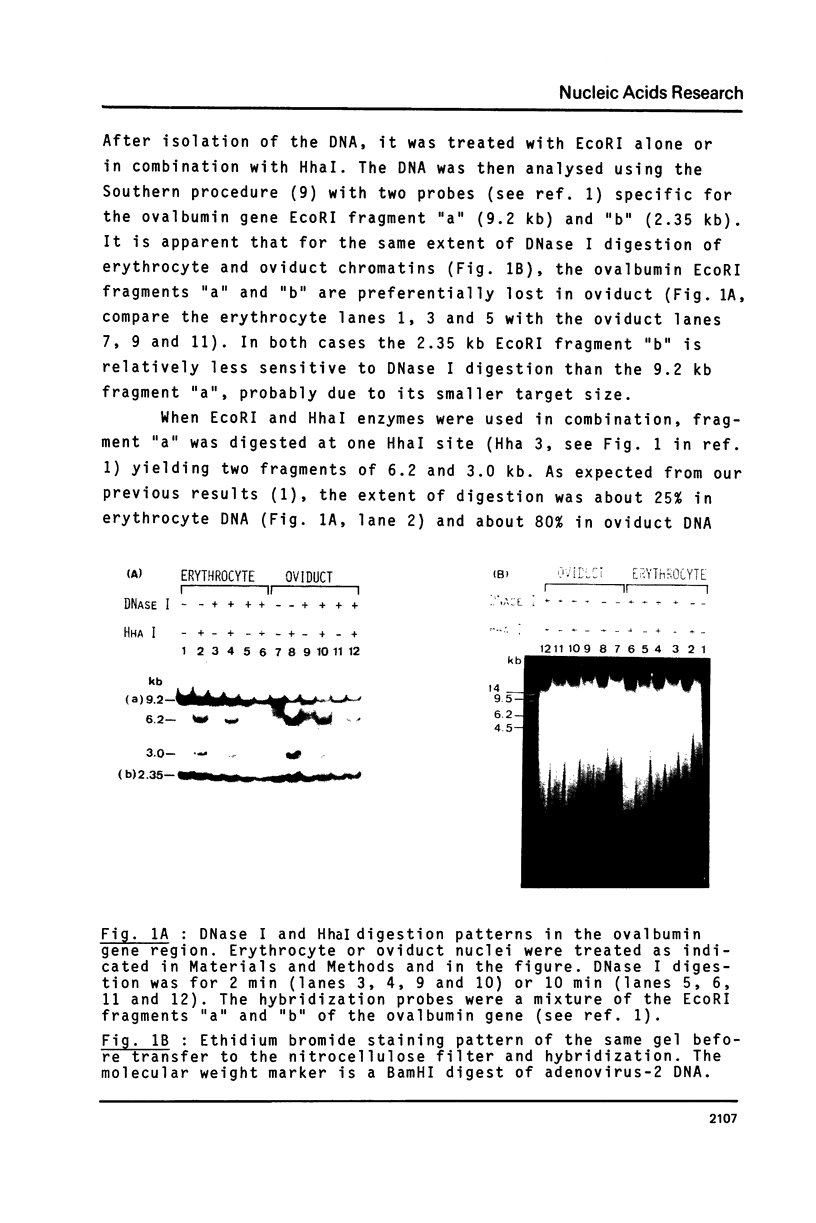
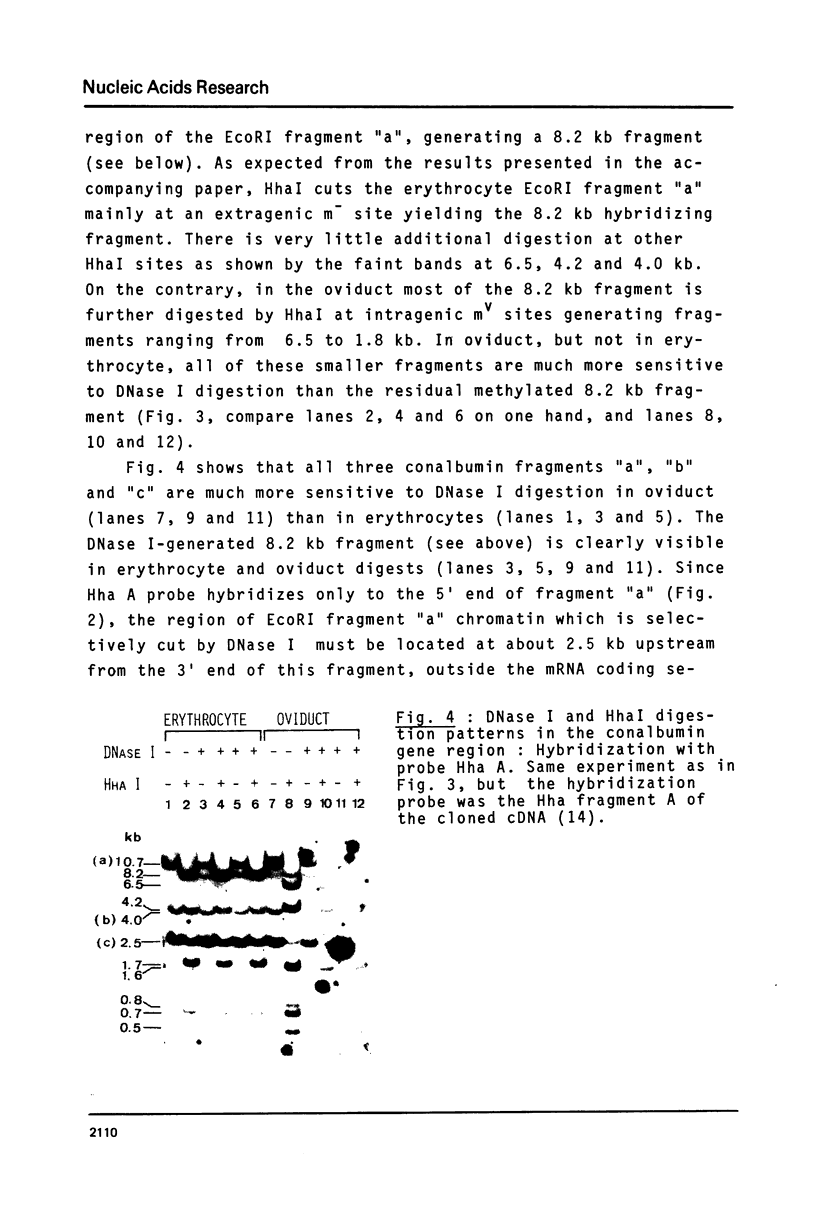
Abstract
To analyse the relationship between DNA undermethylation at some sites in the ovalbumin and conalbumin gene regions (1) and the expression of these genes in chick oviduct, digestions with HhaI, which differentiates between methylated and unmethylated HhaI restriction sites, was performed on DNA isolated from chicken erythrocyte or oviduct chromatin treated with DNase I which degrades preferentially "active" chromatin. This was followed by analysis with ovalbumin- and conalbumin-specific hybridization probes. We conclude that the residual DNA methylation found at some sites of the ovalbumin and conalbumin gene regions is derived from the fraction of cells in which the chromatin of these genes is not in an "active" form. On the other hand, the ovalbumin and conalbumin sites which are partially unmethylated in erythrocyte DNA correspond to chromatin regions which are not DNase I-senitive. We have also detected a site about 1 kb downstream from the 3' end of the conalbumin gene that is hypersensitive to DNase I in all tissues tested.
Full text
PDF






Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Adams R. L., McKay E. L., Douglas J. T., Burdon R. H. Methylation of nucleosomal and nuclease sensitive DNA. Nucleic Acids Res. 1977 Sep;4(9):3097–3108. doi: 10.1093/nar/4.9.3097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bellard M., Gannon F., Chambon P. Nucleosome structure III: the structure and transcriptional activity of the chromatin containing the ovalbumin and globin genes in chick oviduct nuclei. Cold Spring Harb Symp Quant Biol. 1978;42(Pt 2):779–791. doi: 10.1101/sqb.1978.042.01.078. [DOI] [PubMed] [Google Scholar]
- Garel A., Zolan M., Axel R. Genes transcribed at diverse rates have a similar conformation in chromatin. Proc Natl Acad Sci U S A. 1977 Nov;74(11):4867–4871. doi: 10.1073/pnas.74.11.4867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gross-Bellard M., Oudet P., Chambon P. Isolation of high-molecular-weight DNA from mammalian cells. Eur J Biochem. 1973 Jul 2;36(1):32–38. doi: 10.1111/j.1432-1033.1973.tb02881.x. [DOI] [PubMed] [Google Scholar]
- Hewish D. R., Burgoyne L. A. Chromatin sub-structure. The digestion of chromatin DNA at regularly spaced sites by a nuclear deoxyribonuclease. Biochem Biophys Res Commun. 1973 May 15;52(2):504–510. doi: 10.1016/0006-291x(73)90740-7. [DOI] [PubMed] [Google Scholar]
- Palmiter R. D., Mulvihill E. R., McKnight G. S., Senear A. W. Regulation of gene expression in the chick oviduct by steroid hormones. Cold Spring Harb Symp Quant Biol. 1978;42(Pt 2):639–647. doi: 10.1101/sqb.1978.042.01.066. [DOI] [PubMed] [Google Scholar]
- Palmiter R. D. Regulation of protein synthesis in chick oviduct. I. Independent regulation of ovalbumin, conalbumin, ovomucoid, and lysozyme induction. J Biol Chem. 1972 Oct 25;247(20):6450–6461. [PubMed] [Google Scholar]
- Perrin F., Cochet M., Gerlinger P., Cami B., LePennec J. P., Chambon P. The chicken conalbumin gene: studies of the organization of cloned DNAs. Nucleic Acids Res. 1979 Jun 25;6(8):2731–2748. doi: 10.1093/nar/6.8.2731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weintraub H., Groudine M. Chromosomal subunits in active genes have an altered conformation. Science. 1976 Sep 3;193(4256):848–856. doi: 10.1126/science.948749. [DOI] [PubMed] [Google Scholar]
- Wu C., Bingham P. M., Livak K. J., Holmgren R., Elgin S. C. The chromatin structure of specific genes: I. Evidence for higher order domains of defined DNA sequence. Cell. 1979 Apr;16(4):797–806. doi: 10.1016/0092-8674(79)90095-3. [DOI] [PubMed] [Google Scholar]
- Wu C., Wong Y. C., Elgin S. C. The chromatin structure of specific genes: II. Disruption of chromatin structure during gene activity. Cell. 1979 Apr;16(4):807–814. doi: 10.1016/0092-8674(79)90096-5. [DOI] [PubMed] [Google Scholar]