Abstract
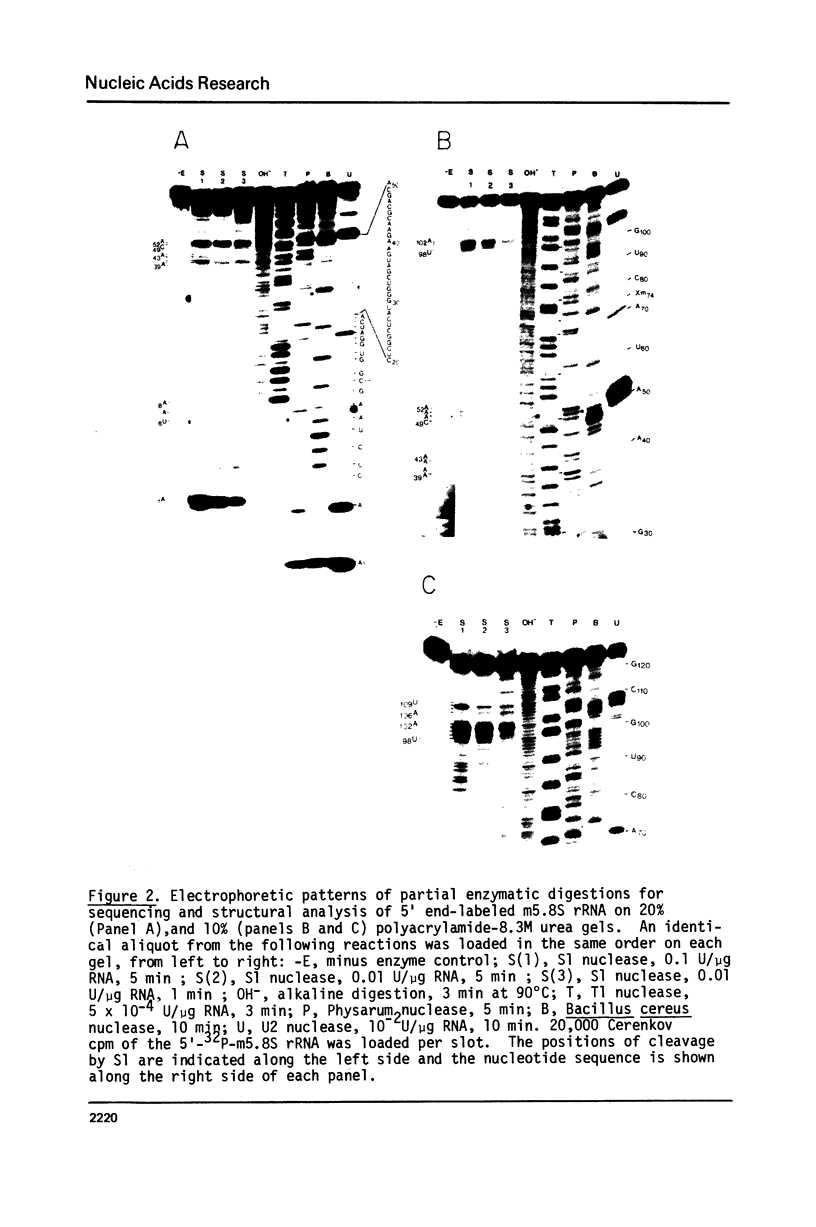
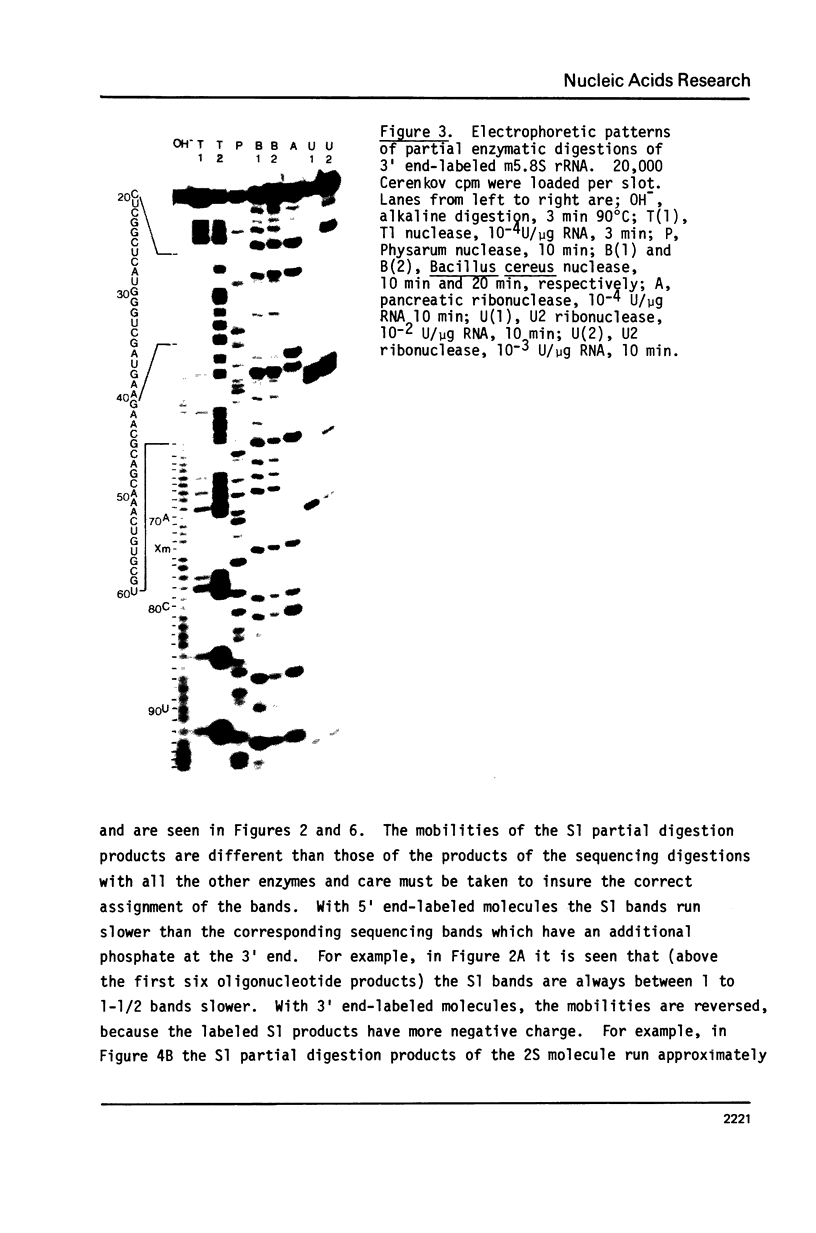
Drosophila melanogaster 5.8S and 2S rRNAs were end-labeled with 32p at either the 5' or 3' end and were sequenced. 5.8S rRNA is 123 nucleotides long and homologous to the 5' part of sequenced 5.8S molecules from other species. 2S rRNA is 30 nucleotides long and homologous to the 3' part of other 5.8S molecules. The 3' end of the 5.8S molecule is able to base-pair with the 5' end of the 2S rRNA to generate a helical region equivalent in position to the "GC-rich hairpin" found in all previously sequenced 5.8S molecules. Probing the structure of the labeled Drosophila 5.8S molecule with S1 nuclease in solution verifies its similarity to other 5.8S rRNAs. The 2S rRNA is shown to form a stable complex with both 5.8S and 26S rRNAs separately and together. 5.8S rRNA can also form either binary or ternary complexes with 2S and 26S rRNA. It is concluded that the 5.8S rRNA in Drosophila melanogaster is very similar both in sequence and structure to other 5.8 rRNAs but is split into two pieces, the 2S rRNA being the 3' part. 2S anchors the 5.8S and 26S rRNA. The order of the rRNA coding regions in the ribosomal DNA repeating unit is shown to be 18S - 5.8S - 2S - 26S. Direct sequencing of ribosomal DNA shows that the 5.8S and 2S regions are separated by a 28 nucleotide spacer which is A-T rich and is presumably removed by a specific processing event. A secondary structure model is proposed for the 26S-5.8S ternary complex and for the presumptive precursor molecule.
Full text
PDF












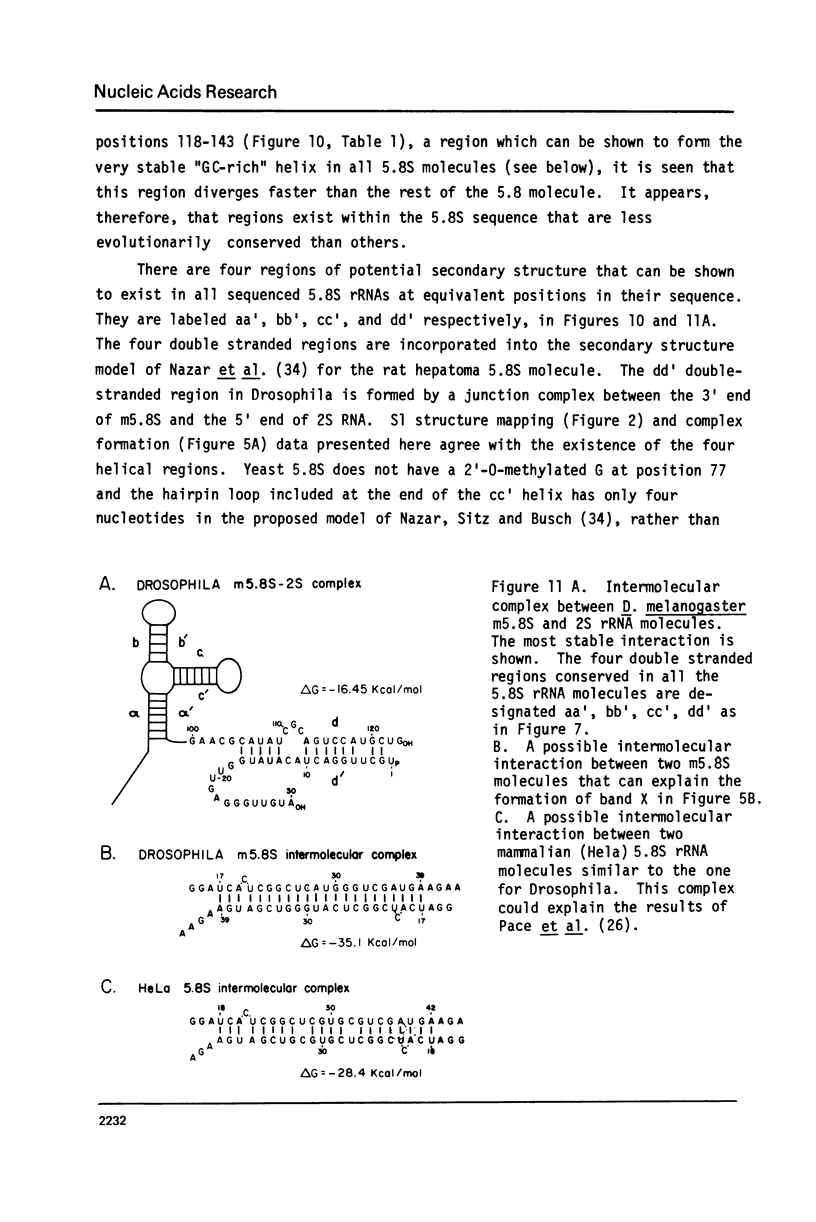


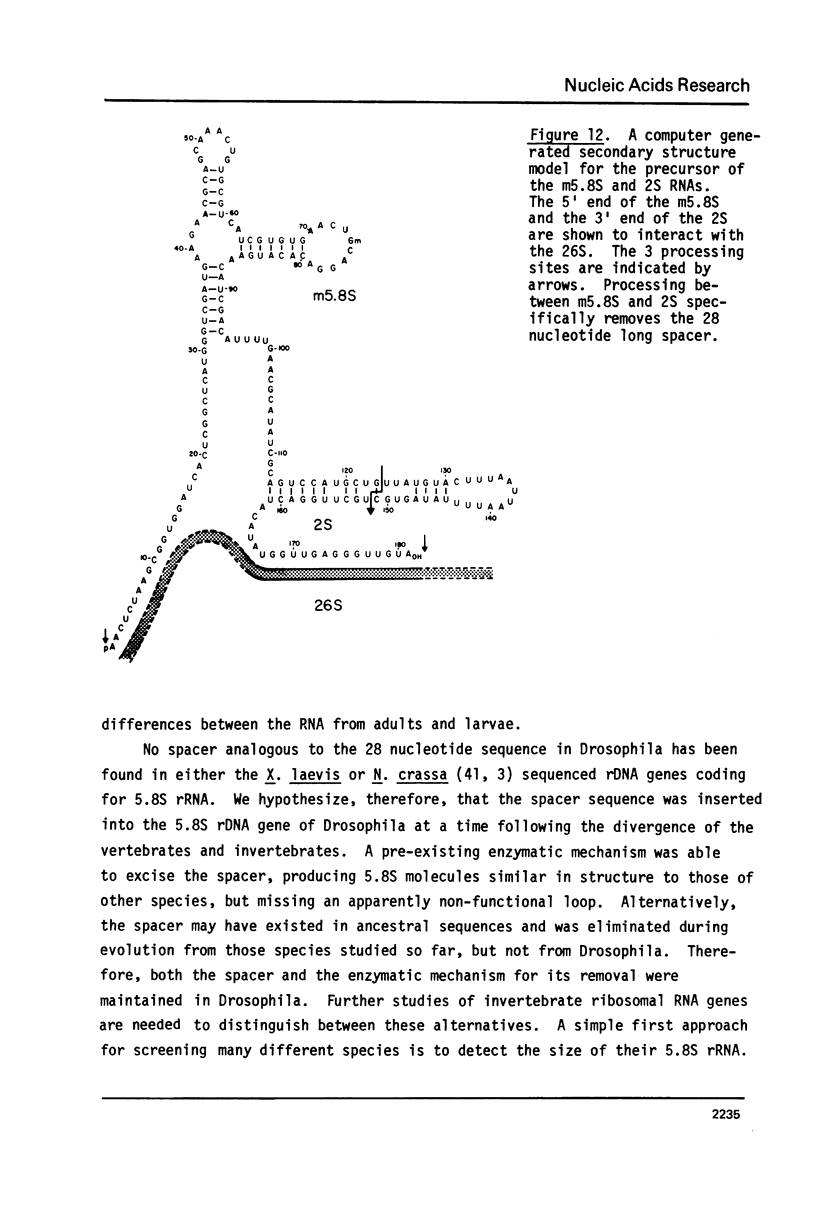



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bell G. I., DeGennaro L. J., Gelfand D. H., Bishop R. J., Valenzuela P., Rutter W. J. Ribosomal RNA genes of Saccharomyces cerevisiae. I. Physical map of the repeating unit and location of the regions coding for 5 S, 5.8 S, 18 S, and 25 S ribosomal RNAs. J Biol Chem. 1977 Nov 25;252(22):8118–8125. [PubMed] [Google Scholar]
- Cory S., Adams J. M. A very large repeating unit of mouse DNA containing the 18S, 28S and 5.8S rRNA genes. Cell. 1977 Aug;11(4):795–805. doi: 10.1016/0092-8674(77)90292-6. [DOI] [PubMed] [Google Scholar]
- Efstratiadis A., Vournakis J. N., Donis-Keller H., Chaconas G., Dougall D. K., Kafatos F. C. End labeling of enzymatically decapped mRNA. Nucleic Acids Res. 1977 Dec;4(12):4165–4174. doi: 10.1093/nar/4.12.4165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- England T. E., Uhlenbeck O. C. 3'-terminal labelling of RNA with T4 RNA ligase. Nature. 1978 Oct 12;275(5680):560–561. doi: 10.1038/275560a0. [DOI] [PubMed] [Google Scholar]
- Erdmann V. A. Collection of published 5S and 5.8S RNA sequences and their precursors. Nucleic Acids Res. 1979 Jan;6(1):r29–r44. doi: 10.1093/nar/6.1.419-c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fedoroff N. V. On spacers. Cell. 1979 Apr;16(4):697–710. doi: 10.1016/0092-8674(79)90086-2. [DOI] [PubMed] [Google Scholar]
- Frankel G., Cockburn A. F., Kindle K. L., Firtel R. A. Organization of the ribosomal RNA genes of Dictyostelium discoideum. Mapping of the transcribed region. J Mol Biol. 1977 Feb 5;109(4):539–558. doi: 10.1016/s0022-2836(77)80090-9. [DOI] [PubMed] [Google Scholar]
- Free S. J., Rice P. W., Metzenberg R. L. Arrangement of the genes coding for ribosomal ribonucleic acids in Neurospora crassa. J Bacteriol. 1979 Mar;137(3):1219–1226. doi: 10.1128/jb.137.3.1219-1226.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glover D. M. Cloned segment of Drosophila melanogaster rDNA containing new types of sequence insertion. Proc Natl Acad Sci U S A. 1977 Nov;74(11):4932–4936. doi: 10.1073/pnas.74.11.4932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glover D. M., Hogness D. S. A novel arrangement of the 18S and 28S sequences in a repeating unit of Drosophila melanogaster rDNA. Cell. 1977 Feb;10(2):167–176. doi: 10.1016/0092-8674(77)90212-4. [DOI] [PubMed] [Google Scholar]
- Glover D. M., White R. L., Finnegan D. J., Hogness D. S. Characterization of six cloned DNAs from Drosophila melanogaster, including one that contains the genes for rRNA. Cell. 1975 Jun;5(2):149–157. doi: 10.1016/0092-8674(75)90023-9. [DOI] [PubMed] [Google Scholar]
- Holt G. R., Davis W. E., Ailor E. I., Warren A. H., Elyassi H. Massive airway hemorrhage after transtracheal aspiration. South Med J. 1978 Mar;71(3):325–327. doi: 10.1097/00007611-197803000-00031. [DOI] [PubMed] [Google Scholar]
- Jordan B. R. '2S' RNA, a new ribosomal RNA component in cultured Drosophila cells. FEBS Lett. 1974 Aug 15;44(1):39–42. doi: 10.1016/0014-5793(74)80301-7. [DOI] [PubMed] [Google Scholar]
- Jordan B. R., Glover D. M. 5.8 S and 2 S rDNA is located in the 'transcribed spacer' region between the 18 S and 26 S rRNA genes in Drosophila melanogaster. FEBS Lett. 1977 Jun 15;78(2):271–274. doi: 10.1016/0014-5793(77)80321-9. [DOI] [PubMed] [Google Scholar]
- Jordan B. R., Jourdan R., Jacq B. Late steps in the maturation of Drosophila 26 S ribosomal RNA: generation of 5-8 S and 2 S RNAs by cleavages occurring in the cytoplasm. J Mol Biol. 1976 Feb 15;101(1):85–105. doi: 10.1016/0022-2836(76)90067-x. [DOI] [PubMed] [Google Scholar]
- Khan M. S., Maden B. E. Nucleotide sequence relationships between vertebrate 5.8 S ribosomal RNAs. Nucleic Acids Res. 1977 Jul;4(7):2495–2505. doi: 10.1093/nar/4.7.2495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klessig D. F. Two adenovirus mRNAs have a common 5' terminal leader sequence encoded at least 10 kb upstream from their main coding regions. Cell. 1977 Sep;12(1):9–21. doi: 10.1016/0092-8674(77)90181-7. [DOI] [PubMed] [Google Scholar]
- Levis R., Penman S. Processing steps and methylation in the formation of the ribosomal RNA of cultured Drosophila cells. J Mol Biol. 1978 May 15;121(2):219–238. doi: 10.1016/s0022-2836(78)80006-0. [DOI] [PubMed] [Google Scholar]
- Lightfoot D. Thermodynamics of a stable yeast 5.8S rRNA hairpin helix. Nucleic Acids Res. 1978 Oct;5(10):3565–3577. doi: 10.1093/nar/5.10.3565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lockard R. E., Alzner-Deweerd B., Heckman J. E., MacGee J., Tabor M. W., RajBhandary U. L. Sequence analysis of 5'[32P] labeled mRNA and tRNA using polyacrylamide gel electrophoresis. Nucleic Acids Res. 1978 Jan;5(1):37–56. doi: 10.1093/nar/5.1.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luoma G. A., Marshall A. G. Laser Raman evidence for new cloverleaf secondary structures for eukaryotic 5.8S RNA and prokaryotic 5S RNA. Proc Natl Acad Sci U S A. 1978 Oct;75(10):4901–4905. doi: 10.1073/pnas.75.10.4901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maxam A. M., Gilbert W. A new method for sequencing DNA. Proc Natl Acad Sci U S A. 1977 Feb;74(2):560–564. doi: 10.1073/pnas.74.2.560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nazar R. N., Roy K. L. Nucleotide sequence of rainbow trout (Salmo gairdneri) ribosomal 5.8 S ribonucleic acid. J Biol Chem. 1978 Jan 25;253(2):395–399. [PubMed] [Google Scholar]
- Nazar R. N., Sitz T. O., Busch H. Structural analyses of mammalian ribosomal ribonucleic acid and its precursors. Nucleotide sequence of ribosomal 5.8 S ribonucleic acid. J Biol Chem. 1975 Nov 25;250(22):8591–8597. [PubMed] [Google Scholar]
- Pace N. R., Walker T. A., Schroeder E. Structure of the 5.8S RNA component of the 5.8S-28S ribosomal RNA junction complex. Biochemistry. 1977 Nov 29;16(24):5321–5328. doi: 10.1021/bi00643a025. [DOI] [PubMed] [Google Scholar]
- Pellegrini M., Manning J., Davidson N. Sequence arrangement of the rDNA of Drosophila melanogaster. Cell. 1977 Feb;10(2):213–214. doi: 10.1016/0092-8674(77)90215-x. [DOI] [PubMed] [Google Scholar]
- Perry R. P. Processing of RNA. Annu Rev Biochem. 1976;45:605–629. doi: 10.1146/annurev.bi.45.070176.003133. [DOI] [PubMed] [Google Scholar]
- Pilly D., Niemeyer A., Schmidt M., Bargetzi J. P. Enzymes for RNA sequence analysis. Preparation and specificity of exoplasmodial ribonucleases I and II from Physarum polycephalum. J Biol Chem. 1978 Jan 25;253(2):437–445. [PubMed] [Google Scholar]
- Pipas J. M., McMahon J. E. Method for predicting RNA secondary structure. Proc Natl Acad Sci U S A. 1975 Jun;72(6):2017–2021. doi: 10.1073/pnas.72.6.2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Renkawitz R., Gerbi S. A., Glätzer K. H. Ribosomal DNA of fly Sciara coprophila has a very small and homogeneous repeat unit. Mol Gen Genet. 1979 May 23;173(1):1–13. doi: 10.1007/BF00267685. [DOI] [PubMed] [Google Scholar]
- Rushizky G. W., Shaternikov V. A., Mozejko J. H., Sober H. A. S1 nuclease hydrolysis of single-stranded nucleic acids with partial double-stranded configuration. Biochemistry. 1975 Sep 23;14(19):4221–4226. doi: 10.1021/bi00690a011. [DOI] [PubMed] [Google Scholar]
- Selker E., Yanofsky C. Nucleotide sequence and conserved features of the 5.8 S rRNA coding region of Neurospora crassa. Nucleic Acids Res. 1979 Jun 11;6(7):2561–2567. doi: 10.1093/nar/6.7.2561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shine J., Dalgarno L. Occurrence of heat-dissociable ribosomal RNA in insects: the presence of three polynucleotide chains in 26 S RNA from cultured Aedes aegypti cells. J Mol Biol. 1973 Mar 25;75(1):57–72. doi: 10.1016/0022-2836(73)90528-7. [DOI] [PubMed] [Google Scholar]
- Simoncsits A., Brownlee G. G., Brown R. S., Rubin J. R., Guilley H. New rapid gel sequencing method for RNA. Nature. 1977 Oct 27;269(5631):833–836. doi: 10.1038/269833a0. [DOI] [PubMed] [Google Scholar]
- Southern E. M. Detection of specific sequences among DNA fragments separated by gel electrophoresis. J Mol Biol. 1975 Nov 5;98(3):503–517. doi: 10.1016/s0022-2836(75)80083-0. [DOI] [PubMed] [Google Scholar]
- Speirs J., Birnstiel M. Arrangement of the 5-8 S RNA cistrons in the genome of Xenopus laevis. J Mol Biol. 1974 Aug 5;87(2):237–256. doi: 10.1016/0022-2836(74)90146-6. [DOI] [PubMed] [Google Scholar]
- Tinoco I., Jr, Borer P. N., Dengler B., Levin M. D., Uhlenbeck O. C., Crothers D. M., Bralla J. Improved estimation of secondary structure in ribonucleic acids. Nat New Biol. 1973 Nov 14;246(150):40–41. doi: 10.1038/newbio246040a0. [DOI] [PubMed] [Google Scholar]
- Tonegawa S., Maxam A. M., Tizard R., Bernard O., Gilbert W. Sequence of a mouse germ-line gene for a variable region of an immunoglobulin light chain. Proc Natl Acad Sci U S A. 1978 Mar;75(3):1485–1489. doi: 10.1073/pnas.75.3.1485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker T. A., Pace N. R. Transcriptional organization of the 5.8S ribosomal RNA cistron in Xenopus laevis ribosomal DNA. Nucleic Acids Res. 1977 Mar;4(3):595–601. doi: 10.1093/nar/4.3.595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wellauer P. K., Dawid I. B., Tartof K. D. X and Y chromosomal ribosomal DNA of Drosophila: comparison of spacers and insertions. Cell. 1978 Jun;14(2):269–278. doi: 10.1016/0092-8674(78)90113-7. [DOI] [PubMed] [Google Scholar]
- Wellauer P. K., Dawid I. B. The structural organization of ribosomal DNA in Drosophila melanogaster. Cell. 1977 Feb;10(2):193–212. doi: 10.1016/0092-8674(77)90214-8. [DOI] [PubMed] [Google Scholar]
- Wurst R. M., Vournakis J. N., Maxam A. M. Structure mapping of 5'-32P-labeled RNA with S1 nuclease. Biochemistry. 1978 Oct 17;17(21):4493–4499. doi: 10.1021/bi00614a021. [DOI] [PubMed] [Google Scholar]