Abstract

Many small molecules, including bioactive molecules and approved drugs, spontaneously form colloidal aggregates in aqueous solution at micromolar concentrations. Though it is widely accepted that aggregation leads to artifacts in screens for ligands of soluble proteins, the effects of colloid formation in cell-based assays have not been studied. Here, seven anticancer drugs and one diagnostic reagent were found to form colloids in both biochemical buffer and in cell culture media. In cell-based assays, the antiproliferative activities of three of the drugs were substantially reduced when in colloidal form as compared to monomeric form; a new formulation method ensured the presence of drug colloids versus drug monomers in solution. We also found that Evans Blue, a dye classically used to measure vascular permeability and to demonstrate the “enhanced permeability and retention (EPR) effect” in solid tumors, forms colloids that adsorb albumin, as opposed to older literature that suggested the reverse.
Colloidal aggregates, which are formed by many small organic molecules in aqueous solution, have long plagued early drug discovery.1,2 Ranging from 50 to 500 nm in radius, these colloids form spontaneously and reversibly in buffer, governed by a characteristic critical aggregation concentration (CAC) similar to a critical micelle concentration (CMC).3 When a colloid has formed, soluble proteins adsorb to its surface causing partial denaturation and nonspecific inhibition.4,5 Colloid formation can be disrupted by the addition of nonionic detergents.6,7 It is now well-accepted that promiscuous inhibition caused by small molecule aggregation is a major source of false positive results in high-throughput and virtual screening.2,7,8
Colloid formation is not limited to screening molecules but is a common property of many organic molecules, reagents, and even approved drugs, which aggregate at micromolar and even submicromolar concentrations in solution.9−11 Further, many colloidal aggregates are stable in biological media, including simulated gastric and intestinal fluids,11,12 and in media with high concentrations of serum proteins.13 Apart from evidence that colloidal aggregates are active in yeast cell culture, inhibiting protein fibril formation,14 the effects of colloid formation of bioactive molecules in cell-based assays have been poorly studied.
The observation that many approved drugs aggregate, and hints of colloid stability in cell culture media, led us to question if several recent anticancer drugs and reagents form aggregates under common cell culture conditions and if this, in turn, affects their activities. We investigated seven anticancer drugs for colloidal aggregation, bexarotene, crizotinib, fulvestrant, lapatinib, nilotinib, sorafenib, and vemurafenib, and one diagnostic reagent, Evans Blue. We were drawn to these molecules based on their importance in physiology and medicine and because their physical properties resemble well characterized aggregators.9,11,15 The anticancer drugs include those approved within the past decade with a range of molecular targets and activity against several cancers (Supplementary Table 1). Evans Blue is a reagent widely used to measure vascular permeability.16
Here we not only study the behavior of these drugs and reagent in biochemical buffer but also develop techniques to do the same in cell culture, enabling us to investigate the effects of the colloids on cell growth. We find that the efficacy of the antineoplastics is profoundly diminished once their concentrations cross the critical aggregation threshold in cell-culture medium, a threshold that is well within the range at which these molecules are typically tested. Thus, these studies may provide tools and strategies to control the effects of drug colloid formation in cell culture and reveal the significance of colloids in cell-based assays. They also advance our understanding of the distribution and tissue penetration of Evans Blue, a reagent widely used to measure vascular permeability and to visibly mark tumor tissue. In contrast to many decades of work that has assumed that albumin binds and transports Evans Blue, we found the reverse: Evans Blue forms colloids that subsequently adsorb albumin and likely transport it. Some implications for our understanding of the penetration mapped by this reagent in vivo are considered.
Results and Discussion
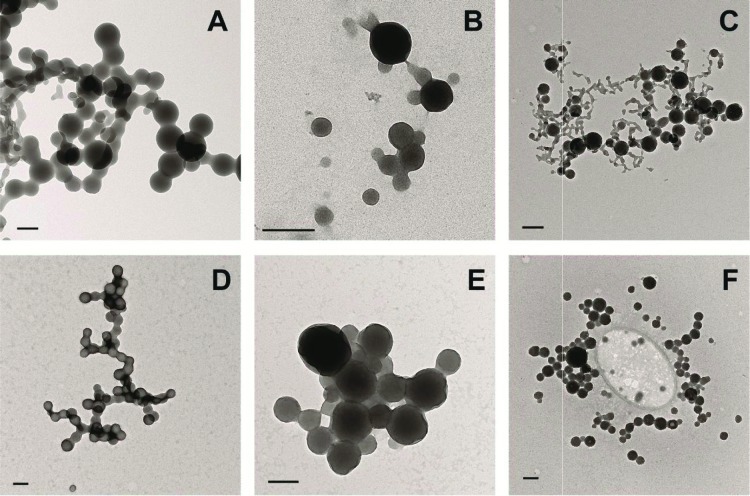
We first investigated the aggregation properties of seven anticancer drugs in aqueous buffer. Two defining characteristics for colloidal aggregation are the formation of particles on the submicrometer scale and the detergent-reversible inhibition of soluble enzymes by such particles. Consistent with their large molar mass (ranging from 349 g/mol for bexarotene to 607 g/mol for fulvestrant) and high hydrophobicity,9,17 each molecule formed clearly detectable particles, ranging from 67 to 164 nm in radius, by dynamic light scattering (DLS) (Table 1). Measurements of critical aggregation concentrations (CACs) suggest that, of the over 2000 observed aggregators to date, these chemotherapeutics are among the molecules most prone to aggregation, with three, fulvestrant, nilotinib, and sorafenib, having a CAC below 1 μM. Transmission electron microscopy (TEM) also revealed characteristic colloidal aggregation (Figure 1A, B, C). These drug colloids all exhibited characteristic enzyme sequestration and detergent-reversible inhibition of the model enzyme cruzain,18 with IC50 values in the low micromolar range (Table 1). Thus, the behavior of all seven drugs is consistent with colloidal aggregation.
Table 1. Colloid Formation by Anticancer Drugs.
Measured at twice the Critical Aggregation Concentration (CAC).
Figure 1.
Transmission electron micrographs of aggregating drugs in phosphate buffer (top row) and 10% FBS (bottom row): (A,D) fulvestrant, (B, E) lapatinib, (C, F) sorafenib. Bars represent 200 nm.
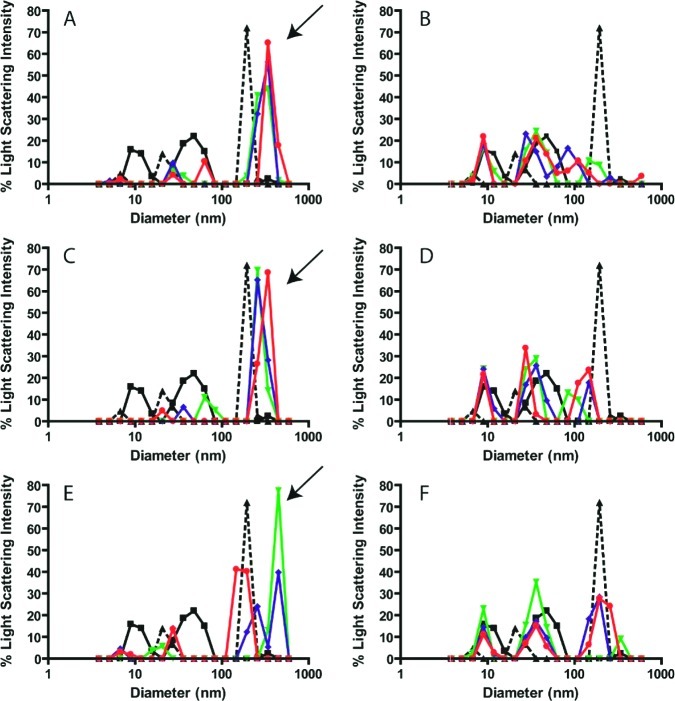
A more stringent and germane test was whether the drug colloids were stable in cell culture media. Previous studies have shown colloid stability in the presence of up to 1 mg mL–1 albumin.13 However, common cell culture media contains approximately 4 mg mL–1 albumin. Even under these stringent conditions, the colloids could be observed by TEM (Figure 1D, E, F). We also used DLS to measure particle formation in this media. Notably, colloids formed by three of the anticancer drugs, fulvestrant, lapatinib, and sorafenib, were stable for at least 24 h in cell culture media at 37 °C (Figure 2A, B, C) enabling us to investigate their effects on cancer cells.
Figure 2.
Colloid formation by anticancer drugs in cell culture media. Particle formation was measured by DLS for fulvestrant (A, B), lapatinib (C, D), and sorafenib (E, F) in the absence (A, C, E) and presence (B, D, F) of 0.025% Tween-80 at 0 h (▼), 12 h (◆), and 24 h (●). Media alone (■) and containing standard beads (▲) were measured for comparison. Arrows indicate colloids that are present in cell media. These peaks are disrupted by the addition of Tween-80, indicating the loss of colloidal aggregates.
Anticancer drug candidates are often tested for antiproliferative effects through large-scale screens,19 and often at concentrations above the CACs determined here. Hence, we reasoned that colloids may be present in many cell culture experiments and may perturb efficacy. To evaluate the effects of drug colloids on cell proliferation, we aimed to treat cell lines with a colloidal formulation and a noncolloidal (monomer) formulation using equal drug concentrations. Since we had shown that colloids were stable in cell media, we needed a method to disrupt colloid formation under the same conditions. Colloid formation can be disrupted by the addition of surfactants to media; however, detergents are seldom used in cell culture due to their toxicity. We tested three detergents for cell toxicity: Tween-80, Tween-20, and Triton X-100 (Supplementary Figure 1). Only 0.025% Tween-80 had no significant effect on cell growth and was considered nontoxic. DLS experiments were repeated for fulvestrant, lapatanib, and sorafenib in the presence of 0.025% Tween-80 to verify that aggregation was indeed disrupted (Figure 2D, E, F). The large colloids (∼200 nm in diameter, indicated by arrows in Figure 2) were disrupted by the detergent. Although low levels (less than 10%) of some aggregates remained, we considered these to be insignificant compared to the detergent-free samples.
To measure the antiproliferative activities of colloidal versus monomeric forms of fulvestrant, lapatinib, and sorafenib, we used the above formulations, excluding or including 0.025% Tween-80, in cell-based assays. Media with vehicle additives alone were tested for cell toxicity and showed no significant impact on proliferation (Figure 3). It is well-known that these chemotherapeutics inhibit cancer cell growth when in solution, and to assess the antiproliferative effects, relevant cell lines were chosen for each drug based on its mechanism of action (Supplementary Table 1). The cells were treated for 72 h, with fresh drug formulations administered every 24 h to ensure consistent presence/absence of colloids throughout the treatment course. The free, soluble drug formulations (containing Tween-80) displayed typical cell growth inhibition profiles, whereas inhibition was essentially eliminated in the colloidal formulations (without detergent) (Figure 4). For example, when fulvestrant (15 μM) was used to treat MCF-7 cells, the free (monomer) drug formulation inhibited cell proliferation by 69.4 ± 6.0%, but inhibition by the colloidal formulation, at 8.2 ± 5.2%, was barely detectable. Similar results were observed for lapatinib (100 μM) with MDA-MB-231/H2N cells: the free drug inhibited cell proliferation by 69.5 ± 8.0%, while the colloidal form was within error of the no-drug control. Sorafenib (100 μM) followed the same trend in MDA-MB-231 cells: the free drug showed substantial cytotoxicity (55.4 ± 5.0% inhibition of proliferation) while inhibition by the colloidal form could not be detected. The differences in cell proliferation between noncolloidal and colloidal formulations were significant (p < 0.001) for all three drugs. Overall, the monomeric drug formulations inhibited cancer cell growth as expected and were substantially more toxic than the colloidal forms, which consistently showed no significant antiproliferative effects.
Figure 3.
Cell toxicity was tested for each vehicle formulation. Colloidal formulations contained 0.1% DMSO (□) while monomeric, free drug formulations contained 1% DMSO with 0.025% Tween-80 (■) in media (Columns, mean relative cell proliferation; Bars, standard deviation, n = 6).
Figure 4.
Colloidal (□) versus noncolloidal (■) formulations of three anticancer agents, fulvestrant, lapatinib, and sorafenib, were used to measure antiproliferative effects in relevant cell lines (Columns, mean relative cell proliferation; Bars, standard deviation; n = 6, *** denotes p < 0.001).
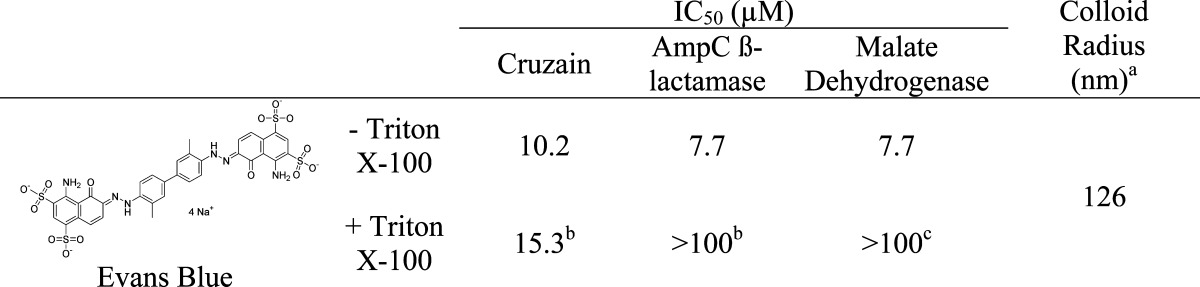
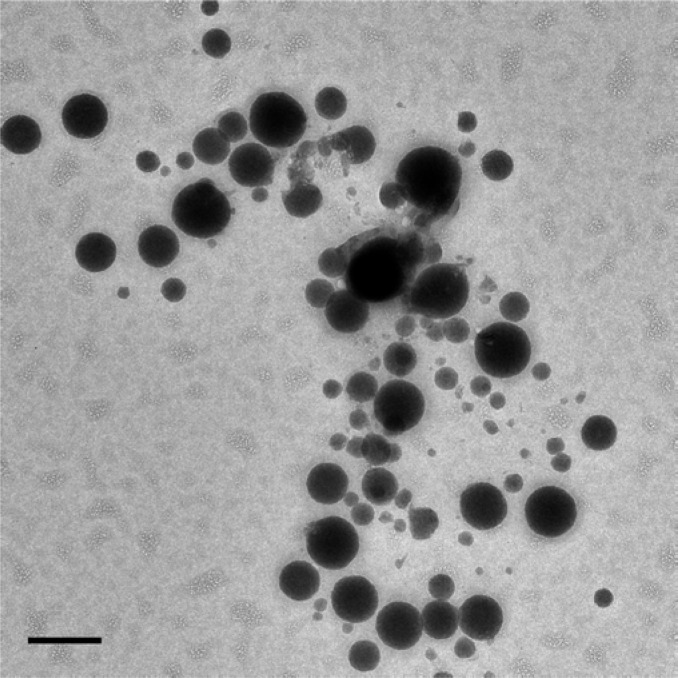
Just as the physical properties of the seven antineoplastics tested resembled those of known aggregators, Evans Blue resembles canonical dye aggregators like Congo Red, Disperse Yellow, and Methylene Blue. Consistent with this, Evans Blue aggregated at low micromolar concentrations in biochemical buffers, forming colloids with radii of 126 nm by DLS (Table 2). The dye colloids inhibited three unrelated enzymes, cruzain, AmpC β-lactamase, and malate dehydrogenase, at micromolar concentrations; as is true for other colloids, this inhibition was reversible by the addition of Triton X-100 (Table 2). Evans Blue also formed stable colloids in 10% FBS, which were imaged by TEM (Figure 5).
Table 2. Evans Blue Dye Forms Colloids in Biochemical Buffer.
100 μM.
0.1% Triton X-100.
0.01% Triton X-100.
Figure 5.
Transmission electron micrograph of Evans Blue aggregates in phosphate buffer containing 10% FBS. Bar represents 100 nm.
The enzyme inhibition supports the idea that Evans Blue colloids adsorb proteins. Thus, rather than albumin acting as a carrier for Evans Blue in serum, it may be that Evans Blue colloids adsorb and carry serum albumin instead. To test this hypothesis directly, we used microscale thermophoresis20 to measure the binding of bovine serum albumin (BSA) to Evans Blue colloids (Figure 6). BSA was labeled with Alexa Fluor 647 using N-Hydroxysuccinimide (NHS)-ester chemistry, and the thermophoresis of the protein was monitored. Thermophoresis measures the change in fluorescence signal over time while the sample is heated by an infrared laser. The thermophoretic profile of BSA was altered upon adsorption to Evans Blue colloids, and the change in fluorescence could be directly correlated to binding. Addition of Evans Blue to 10 nM of labeled BSA had no effect on the protein until the CAC of the dye was approached, after which the protein thermophoresis profile began to dramatically change, reaching a plateau at 100 μM Evans Blue. This concentration of dye corresponds to complete stoichiometric adsorption of BSA to the colloids. Addition of 0.01% Triton X-100 disrupted the colloidal interaction between Evans Blue and albumin and raised the apparent Kd by at least 500-fold, essentially eliminating binding. Similarly, repeating the binding isotherm with BSA at 100 nM instead of 10 nM increased the apparent Kd by almost 50-fold (Figure 6). There is no classical mechanism that explains perturbation of affinity by raising protein concentration in this range; rather, this effect is characteristic of colloid-based sequestration.3,4,21 Taken together, these results support a model where Evans Blue colloids sequester albumin, in contrast to current models that suggest albumin binds and transports Evans Blue in vivo.
Figure 6.
Aggregating dye, Evans Blue, binds albumin via a colloidal mechanism. Thermophoresis was measured at increasing concentrations of Evans Blue in (■) 10 nM BSA, 0.001% Triton X-100, (▲) 10 nM BSA, 0.01% Triton X-100, and (▼) 100 nM BSA, 0.001% Triton X-100. Data represent the mean and range for repeat experiments.
Mechanistic and Biological Implications
Two key observations emerge from this work. First, several cancer drugs form stable colloids not only in biochemical buffers but also in cell culture media. In cell culture, these colloids appear to act as reservoirs that reduce the free, effective concentration of the drugs, diminishing their true efficacy. Second, Evans Blue may be acting as a diagnostic marker for the critical EPR effect not because it is transported by albumin through the tumor vasculature, but rather because the dye colloids themselves permeate the tumor tissue, with albumin, bound to the colloidal surface, in tow. These observations, and the development of approaches to control, or arguably even exploit, colloidal aggregation, will influence how we execute and interpret assays with other drugs and reagents.
The observation that colloidal aggregation reduces—often essentially eliminates—the antiproliferative activity of these drugs might lead to false negative results in cell culture assays. Likely, this reflects limited drug exposure to the cells: while the free drug can diffuse through the cell membrane to reach the target of action, drug sequestered in colloidal aggregates cannot. Colloids, like micelles, are in equilibrium with their soluble form and can be converted back to monomeric form by diluting below the CAC.22 Therefore, colloids may act as drug reservoirs; the drug will remain inactive as long as colloids remain intact, but as the colloids destabilize due to decreasing local concentration or other environmental changes, the free drug will be released. There is a certain irony in this: small molecule aggregation has thus far been solely associated with false positive hits from HTS; here it leads to false negative results in cell-based assays. As with techniques to detect the false positive hits from screening, the development here of a simple technique to detect and eliminate aggregators—i.e., low concentrations of Tween-80—in cell culture may be broadly useful to the field.
The observation that Evans Blue aggregates and then sequesters albumin may be the most peculiar observation to emerge from these studies, but perhaps also the one most pregnant with implications. This dye is routinely used to assess vascular permeability and, in cancer physiology, to display the leaky vasculature of solid tumors. Tumor blood vessel architecture is thought to be hyperpermeable, and the tumors are, at the same time, thought to lack effective lymphatic drainage. Consequently, macromolecules larger than 40 kDa preferentially accumulate in tumor tissue, a phenomenon known as the “enhanced permeability and retention (EPR) effect.”16 Evans Blue was, in fact, the dye first used to characterize the EPR effect, based on observations dating to the 1930s that it selectively disseminates into tumors.23 Meanwhile, studies from the 1930s through 1940s suggested that serum albumin acts as a carrier of Evans Blue.24,25 Taken together, the hypothesis has developed that albumin binds Evans Blue, and it is the albumin-bound form of the dye that selectively leaks into and accumulates in solid tumors.16,26
Whereas we do not dispute Evans Blue’s preferential localization in tumor tissues, our results throw doubt on the accepted mechanism. Rather than binding to albumin and being transported through the tumor vasculature, it is rather more likely that albumin is adsorbed by the colloidal particles of the dye, and it is the colloid–protein conjugate that finds itself, by the EPR effect, in the tumor. Indeed, close reading of the original literature suggests that the apparent stoichiometry of binding of the dye to the protein was not only high, ranging from 8 to 70 mols of dye to 1 mol of albumin,24,27 but also essentially unbounded by the techniques of the time. In retrospect, this atypical behavior fits well with a colloidal, rather than a classical, mechanism of albumin binding. In fact, once one realizes that Evans Blue is an aggregator, the role of protein localization becomes secondary. Evans Blue colloids are 120 nm in radius and will likely, themselves, permeate tumor tissue via the EPR effect, absent any protein. Indeed, such behavior might, under the right circumstances, be true of other molecules, including colloid forming drugs. This would have a profound effect on their distribution and efficacy in vivo and may merit further study.
Methods
Materials
Fulvestrant, lapatinib, nilotinib, and sorafenib were purchased from AK Scientific; crizotinib and vemurafenib were purchased from Selleck Chemicals; bexarotene was purchased from Toronto Research Chemicals. Dulbecco’s phosphate buffered saline (DPBS) and McCoy’s 5A cell culture media were purchased from Multicell Technologies. Fetal bovine serum (FBS) was purchased from the UCSF Cell Culture Facility. Cell lines MDA-MB-231 (HTB-26), MCF-7 (HTB-22), SK-BR-3 (HTB-30), and HT-1080 (CCL-121) were purchased from ATCC. The MDA-MB-231, HER2-transfected subclone, 231-H2N, was kindly provided by Dr. Robert S. Kerbel (Sunnybrook Health Sciences Centre).28 The MTS cell proliferation assay was purchased from Promega. Duke Standards NIST Traceable Polymer Microspheres were purchased from Thermo Scientific. All other chemicals and reagents were purchased from Sigma-Aldrich or TCI America.
Flow Cytometry
Critical aggregation concentrations (CACs) were determined using a BD Gentest Solubility Scanner, as previously described.22 CACs were measured for all drugs in 50 mM potassium phosphate, pH 7.0, and in MCoy’s 5A plus 10% FBS for fulvestrant, lapatinib, and sorafenib. All measurements were taken with a final concentration of 1% DMSO; no serial dilutions were made. Values reported were obtained by running duplicate samples in three independent experiments.
Enzyme Inhibition Assays
Inhibition of AmpC β-lactamase, cruzain, and malate dehydrogenase was measured as previously described.9,11,22 The final concentration of DMSO was 1% for all samples. Values reported are the average of duplicate samples run in two independent experiments.
Dynamic Light Scattering
Particle formation was measured using a DynaPro MS/X (Wyatt Technology) as previously described.22 Colloid sizes were measured in 50 mM potassium phosphate, pH 7.0, at twice the CAC concentration. For stability studies in cell media, light scattering was measured in McCoy’s 5A with 10% FBS with the optical resolution set to 4. Standard beads of 200 nm diameter were used for reference at approximately 250 aM. Samples were mixed at RT and then incubated at 37 °C for 24 h. Fulvestrant was measured at 15 μM, and lapatinib and sorafenib were measured at 100 μM, in the absence and presence of 0.025% Tween-80. Experiments were repeated thrice. Each histogram shows a single representative sample.
Transmission Electron Microscopy
Solutions were prepared by diluting concentrated DMSO stocks with 50 mM potassium phosphate, pH 7.0, with or without 10% FBS. Drug colloids were imaged at the following concentrations: 75 μM fulvestrant, 100 μM lapatinib, and 100 μM sorafenib; Evans Blue was prepared at 1 mM. TEM was performed on a Tecnai T12 microscope (FEI) at 120 kV, and images were taken using an UltraScan 4000 CCD camera (Gatan, Inc.). Samples were negatively stained with ammonium molybdate on 200 or 400 mesh carbon-coated copper grids (Ted Pella, Inc.)
Drug Formulations
RPMI 1640 cell growth media with 10% FBS was used for all experiments. Stock solutions of each drug were prepared in DMSO at 100 mM for lapatinib and sorafenib and 15 mM for fulvestrant. For colloidal formulations, stock solutions (1 μL) were added into RPMI media (999 μL) to give 1000-fold dilutions (100 μM for lapatinib and sorafenib and 15 μM for fulvestrant). For noncolloidal (free drug) formulations, stock drug solutions were first diluted 10-fold in DMSO (40 μL); RPMI media (3959 μL) containing Tween 80 (1 μL) were added onto the DMSO stock solutions to give final 1000-fold drug dilutions with 1% DMSO (v/v) and 0.025% Tween 80 (v/v). Vehicle controls were prepared in the same manner.
Cell Culture
Cell lines were maintained (<8 passages) in a tissue culture incubator (37 °C, 5% CO2, 95% humidified) in plastic culture flasks in RPMI 1640 growth medium with 10% FBS, 10 UI mL–1 penicillin, and 10 μg mL–1 streptomycin. Cells were seeded at 10,000 cells (cm2)−1 and allowed to adhere overnight. Drug formulations (described above) and the control medium were made fresh and exchanged every 24 h for a total incubation of 72 h. Cells were then washed with fresh RPMI media, and proliferation was determined using an MTS assay according to the manufacturer’s instructions. Relative cell proliferation is defined as (absorbance of treated cells)/(absorbance of untreated cells) × 100.
Thermophoresis
Binding of bovine serum albumin (BSA) to Evans Blue colloids was measured using a Monolith NT.115 (NanoTemper Technologies) as described.20 BSA was labeled with Alexa Fluor 647 using N-hydroxysuccinimide (NHS)-ester chemistry. Labeled BSA was used for all dilutions and in all subsequent experiments. BSA was kept at a constant concentration (10 or 100 nM), and Evans Blue was added at increasing concentrations. The fluorescence was monitored using an excitation wavelength of 650 nm and an emission wavelength of 680 nm. Samples were prepared, and measurements were acquired at RT in small glass capillaries. Thermophoresis measures the change in fluorescence signal while the sample is heated by an infrared laser. The thermophoretic shift of BSA is altered upon binding an Evans Blue colloid; therefore, the change in fluorescent shift can be directly correlated to binding. Results shown were obtained from two independent experiments.
Acknowledgments
Supported by NIH Grant GM71630 (to B.K.S. and M.S.S.) and by Grant AG02132 (to B.K.S. and S. B. Prusiner). We thank J. Karpiak and R. Coleman for reading this manuscript, M. Lee for preliminary work on this project, K. Ho for the Table of Contents graphic, NanoTemper Technologies for use of the Monolith NT.115, and M. Braunfield and the UCSF Macromolecular Structure Group for use and support of the Tecnai T12 microscope.
Supporting Information Available
A Supplementary figure, showing detergent toxicity, and a table, giving the mechanism of action and origins of the kinase drugs, are provided. This material is available free of charge via the Internet at http://pubs.acs.org.
The authors declare no competing financial interest.
Author Contributions
§ These authors contributed equally to this work.
Funding Statement
National Institutes of Health, United States
Supplementary Material
References
- Roche O.; Schneider P.; Zuegge J.; Guba W.; Kansy M.; Alanine A.; Bleicher K.; Danel F.; Gutknecht E. M.; Rogers-Evans M.; Neidhart W.; Stalder H.; Dillon M.; Sjogren E.; Fotouhi N.; Gillespie P.; Goodnow R.; Harris W.; Jones P.; Taniguchi M.; Tsujii S.; von der Saal W.; Zimmermann G.; Schneider G. (2002) Development of a virtual screening method for identification of “frequent hitters” in compound libraries. J. Med. Chem. 45, 137–142. [DOI] [PubMed] [Google Scholar]
- Feng B. Y.; Simeonov A.; Jadhav A.; Babaoglu K.; Inglese J.; Shoichet B. K.; Austin C. P. (2007) A high-throughput screen for aggregation-based inhibition in a large compound library. J. Med. Chem. 50, 2385–2390. [DOI] [PubMed] [Google Scholar]
- McGovern S. L.; Helfand B. T.; Feng B.; Shoichet B. K. (2003) A specific mechanism of nonspecific inhibition. J. Med. Chem. 46, 4265–4272. [DOI] [PubMed] [Google Scholar]
- McGovern S. L.; Caselli E.; Grigorieff N.; Shoichet B. K. (2002) A common mechanism underlying promiscuous inhibitors from virtual and high-throughput screening. J. Med. Chem. 45, 1712–1722. [DOI] [PubMed] [Google Scholar]
- Coan K. E.; Maltby D. A.; Burlingame A. L.; Shoichet B. K. (2009) Promiscuous aggregate-based inhibitors promote enzyme unfolding. J. Med. Chem. 52, 2067–2075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryan A. J.; Gray N. M.; Lowe P. N.; Chung C. W. (2003) Effect of detergent on “promiscuous” inhibitors. J. Med. Chem. 46, 3448–3451. [DOI] [PubMed] [Google Scholar]
- Feng B. Y.; Shoichet B. K. (2006) A detergent-based assay for the detection of promiscuous inhibitors. Nat. Prot. 1, 550–553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thorne N., Auld D. S., Inglese J.. Apparent activity in high-throughput screening: origins of compound-dependent assay interference. Curr. Opin. Chem. Biol. 2010, 14, 315–324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seidler J.; McGovern S. L.; Doman T. N.; Shoichet B. K. (2003) Identification and prediction of promiscuous aggregating inhibitors among known drugs. J. Med. Chem. 46, 4477–4486. [DOI] [PubMed] [Google Scholar]
- McGovern S. L.; Shoichet B. K. (2003) Kinase inhibitors: not just for kinases anymore. J. Med. Chem. 46, 1478–1483. [DOI] [PubMed] [Google Scholar]
- Doak A. K.; Wille H.; Prusiner S. B.; Shoichet B. K. (2010) Colloid formation by drugs in simulated intestinal fluid. J. Med. Chem. 53, 4259–4265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frenkel Y. V.; Clark A. D. Jr.; Das K.; Wang Y. H.; Lewi P. J.; Janssen P. A.; Arnold E. (2005) Concentration and pH dependent aggregation of hydrophobic drug molecules and relevance to oral bioavailability. J. Med. Chem. 48, 1974–1983. [DOI] [PubMed] [Google Scholar]
- Coan K. E.; Shoichet B. K. (2007) Stability and equilibria of promiscuous aggregates in high protein milieus. Molecular bioSystems 3, 208–213. [DOI] [PubMed] [Google Scholar]
- Feng B. Y.; Toyama B. H.; Wille H.; Colby D. W.; Collins S. R.; May B. C.; Prusiner S. B.; Weissman J.; Shoichet B. K. (2008) Small-molecule aggregates inhibit amyloid polymerization. Nat. Chem. Biol. 4, 197–199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng B. Y.; Shelat A.; Doman T. N.; Guy R. K.; Shoichet B. K. (2005) High-throughput assays for promiscuous inhibitors. Nat. Chem. Biol. 1, 146–148. [DOI] [PubMed] [Google Scholar]
- Fang J.; Nakamura H.; Maeda H. (2010) The EPR effect: Unique features of tumor blood vessels for drug delivery, factors involved, and limitations and augmentation of the effect. Adv. Drug Delivery Rev. 63, 136–151. [DOI] [PubMed] [Google Scholar]
- Lipinski C. A.; Lombardo F.; Dominy B. W.; Feeney P. J. (2001) Experimental and computational approaches to estimate solubility and permeability in drug discovery and development settings. Adv. Drug Delivery Rev. 46, 3–26. [DOI] [PubMed] [Google Scholar]
- Jadhav A.; Ferreira R. S.; Klumpp C.; Mott B. T.; Austin C. P.; Inglese J.; Thomas C. J.; Maloney D. J.; Shoichet B. K.; Simeonov A. (2009) Quantitative analyses of aggregation, autofluorescence, and reactivity artifacts in a screen for inhibitors of a thiol protease. J. Med. Chem. 53, 37–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melnick J. S.; Janes J.; Kim S.; Chang J. Y.; Sipes D. G.; Gunderson D.; Jarnes L.; Matzen J. T.; Garcia M. E.; Hood T. L.; Beigi R.; Xia G.; Harig R. A.; Asatryan H.; Yan S. F.; Zhou Y.; Gu X. J.; Saadat A.; Zhou V.; King F. J.; Shaw C. M.; Su A. I.; Downs R.; Gray N. S.; Schultz P. G.; Warmuth M.; Caldwell J. S. (2006) An efficient rapid system for profiling the cellular activities of molecular libraries. Proc. Natl. Acad. Sci. U.S.A. 103, 3153–3158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wienken C. J.; Baaske P.; Rothbauer U.; Braun D.; Duhr S. (2010) Protein-binding assays in biological liquids using microscale thermophoresis. Nat. Commun. 1, 100. [DOI] [PubMed] [Google Scholar]
- Shoichet B. K. (2006) Interpreting steep dose-response curves in early inhibitor discovery. J. Med. Chem. 49, 7274–7277. [DOI] [PubMed] [Google Scholar]
- Coan K. E.; Shoichet B. K. (2008) Stoichiometry and physical chemistry of promiscuous aggregate-based inhibitors. J. Am. Chem. Soc. 130, 9606–9612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore F. D.; Tobin L. H. (1942) Studies with Radioactive Di-Azo Dyes. 1. the Localization of Radioactive Di-Brom Trypan Blue in Inflammatory Lesions. J. Clin. Invest. 21, 471–481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rawson R. A. (1942) The binding of T-1824 and structurally related diazo dyes by the plasma proteins. Am. J. Physiol. 138, 10. [Google Scholar]
- von Porat B. (1951) Blood volume determinations with the Evans blue dye method. Acta Med. Scand. 139, 13–18. [PubMed] [Google Scholar]
- Matsumura Y.; Maeda H. (1986) A new concept for macromolecular therapeutics in cancer chemotherapy: mechanism of tumoritropic accumulation of proteins and the antitumor agent smancs. Cancer Res. 46, 6387–6392. [PubMed] [Google Scholar]
- Leveen H. H.; Fishman W. H. (1947) Combination of Evans blue with plasma protein: its significance in capillary permeability studies, blood dye disappearance curves, and its use as a protein tag. Am. J. Physiol. 151, 8. [DOI] [PubMed] [Google Scholar]
- du Manoir J. M.; Francia G.; Man S.; Mossoba M.; Medin J. A.; Viloria-Petit A.; Hicklin D. J.; Emmenegger U.; Kerbel R. S. (2006) Strategies for delaying or treating in vivo acquired resistance to trastuzumab in human breast cancer xenografts. Clin. Cancer Res. 12, 904–916. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.