Abstract
The aim of this study was to investigate the changes induced by high tidal volume ventilation (HVTV) in pulmonary expression of micro-RNAs (miRNAs) and identify potential target genes and corresponding miRNA-gene networks. Using a real-time RT-PCR-based array in RNA samples from lungs of mice subjected to HVTV for 1 or 4 h and control mice, we identified 65 miRNAs whose expression changed more than twofold upon HVTV. An inflammatory and a TGF-β-signaling miRNA-gene network were identified by in silico pathway analysis being at highest statistical significance (P = 10−43 and P = 10−28, respectively). In the inflammatory network, IL-6 and SOCS-1, regulated by miRNAs let-7 and miR-155, respectively, appeared as central nodes. In TGF-β-signaling network, SMAD-4, regulated by miR-146, appeared as a central node. The contribution of miRNAs to the development of lung injury was evaluated in mice subjected to HVTV treated with a precursor or antagonist of miR-21, a miRNA highly upregulated by HVTV. Lung compliance was preserved only in mice treated with anti-miR-21 but not in mice treated with pre-miR-21 or negative-control miRNA. Both alveolar-arterial oxygen difference and protein levels in bronchoalveolar lavage were lower in mice treated with anti-miR-21 than in mice treated with pre-miR-21 or negative-control miRNA (DA-a: 66 ± 27 vs. 131 ± 22, 144 ± 10 mmHg, respectively, P < 0.001; protein concentration: 1.1 ± 0.2 vs. 2.3 ± 1, 2.1 ± 0.4 mg/ml, respectively, P < 0.01). Our results show that HVTV induces changes in miRNA expression in mouse lungs. Modulation of miRNA expression can affect the development of HVTV-induced lung injury.
Keywords: inflammation, miR-21, TGF-β, miRNA-gene network
mechanical ventilation is a life-saving intervention for patients with lung injury and respiratory failure. During mechanical ventilation, lungs can be exposed to high distending forces, which induce inflammation and disruption of alveolar barrier function, resulting in lung injury, a process referred to as ventilator-induced lung injury (VILI) (11). Experimental and clinical studies have shown that VILI is associated with increased levels of proinflammatory cytokines, such as interleukin 6 (IL-6), tumor necrosis factor-α (TNF-α) (1, 24), as well as transforming growth factor-β (TGF-β) (5, 16). The mechanisms that regulate VILI have not been completely defined (24), and no pharmacological intervention has been identified that can decrease mortality associated with VILI.
Micro-RNAs (miRNAs) are small, noncoding RNA sequences that regulate gene expression (39). MiRNAs typically bind to sequences in the 3′ untranslated region of mRNAs and decrease their stability and/or translation. The roles of miRNAs in the regulation of all major cellular functions, including cell proliferation, differentiation, metabolism, and apoptosis, are increasingly being recognized (39). Recent studies have identified miRNAs as critical regulators of lung and systemic inflammation (2, 39, 45) and of cell responses to TGF-β signaling (17). Importantly, promising therapeutic interventions targeting miRNAs in inflammatory diseases have been recently reported (2, 20).
The aim of the present study was to characterize the changes in the lung miRNA expression profile induced by injurious mechanical ventilation and to identify miRNA target transcripts and miRNA-gene networks involved in VILI. Our hypothesis was that injurious ventilation with high tidal volume (HVTV) would induce changes in lung miRNAs, which would precede measurable physiological changes, such as increase in airway pressure. Moreover, we hypothesized that some of these changes could be involved in the pathogenesis of VILI. To examine the changes in lung miRNA expression profile, we performed an miRNA array using lung samples from mice included in our previous study (47). We evaluated lung miRNA expression at an early time point, 1 h after HVTV, when airway pressure has not increased, and after 4 h of HVTV, when VILI has developed. To examine whether changes in miRNA expression contribute to the development of VILI, we evaluated the effect of administration of a miRNA precursor and antagonist in mice subjected to HVTV. We used miR-21, which is highly induced by HVTV and has been previously used in vivo in another model of lung injury (25).
MATERIALS AND METHODS
MiRNA expression.
To characterize miRNA expression profile in VILI, RNA was extracted from whole lung homogenates of C57BL/6 mice subjected to HVTV, included in our previously published study (47). Briefly, 8-wk-old male C57BL/6 mice were allocated to one of three groups, n = 5 per group; group 1 was exposed to HVTV (VT = 40 ml/kg, peak inspiratory pressure = 35 cmH2O) for 1 h, group 2 was exposed to HVTV for 4 h, and group 3 were control mice. RNA was extracted using TRIZOL reagent (Invitrogen, Carlsbad, CA) and used for miRNA and mRNA expression analysis. The lung expression of 335 miRNAs was measured using TaqMan Low Density Arrays (TLDA rodent miRNA v1.0; Applied Biosystems, Carlsbad, CA) in the Dana-Farber Molecular Diagnostics Facility (Dana-Farber Cancer Institute, Boston, MA). MiRNA expression data were normalized to 18S expression levels. All differentially expressed microRNAs identified by the TLDA arrays were validated by real-time PCR analysis using the mirVana qRT-PCR miRNA Detection Kit and qRT-PCR Primer Sets (Ambion, Austin, TX).
Prediction of miRNA gene targets.
Potential miRNA gene targets were identified using the miRBase (http://microRNA.sanger.ac.uk), PicTar (http://dorina.mdc-berlin.de/rbp_browser/dorina.html), and TargetScan version 5.1 (http://www.targetscan.org/index.html) search engines. Each bioinformatic program uses different criteria to predict an interaction between the 3′UTR of a gene and the seed sequence (nucleotide positions 2–8) of the microRNA. Specifically, the miRBase program is based on the sequence complementarity between the 3′UTR of a gene and the seed sequence of a microRNA, considering the conservation of this interaction in different species and the free energy of the microRNA-3′UTR duplex. The miRBase program has information for 711 microRNAs, and the mapped microRNA-3′UTR interactions are 956,664. The PicTar program is based on the same parameters as the miRBase program and in addition it includes information about multiple binding sites for a specific microRNA in a specific 3′UTR. The PicTar program has information for 129 microRNAs, and the mapped microRNA-3′UTR interactions are 17,224. Finally, the TargetScan program is based on the sequence complementarity between the 3′UTR of a gene and the seed sequence of microRNA, considering the conservation of this interaction in different species, the local AU content, and examines the surrounding sequence. The TargetScan program has information for 675 microRNAs, and the mapped microRNA-3′UTR interactions are 189,075. To optimize the accuracy of prediction, a potential gene target was required to be predicted by a minimum of two out of three of the above programs, as previously described (14).
Gene network analysis.
Gene networks were constructed and analyzed using Ingenuity Gene Network Software Analysis as previously described (33). Interactions between highly interconnected miRNAs, and predicted target genes were identified by statistical likelihood using the following equation:
| (Eq. 1) |
where N is the number of genes in the network, of which G are central nodes genes, for a pathway of s genes of which f are central node genes. C (n, k) is the binomial coefficient. A central node is defined as the gene in a network that has the highest number of inputs (genes that regulate the central node gene) and outputs (genes that are regulated by the central node gene) (33). Statistically significant networks are considered those with a score greater than 5 (P < 10−5).
Measurement of mRNA and miR-21 levels.
RNA was extracted from frozen lung samples and bronchoalveolar lavage fluid (BALF) cells using TRIZOL reagent (Invitrogen). cDNA was synthesized for mRNA measurements using MMLV-RT (Invitrogen) and for miR-21 measurement using TaqMan micro-RNA-RT (Ambion). Levels of mRNAs encoding IL-6, SOCS1, SMAD4, BMPR2, and PTEN, as well as miR-21, were measured using a Realplex2 system (Eppendorf, Westbury, NY). Changes in relative gene expression were normalized to levels of 18S rRNA using the relative threshold cycle method.
In situ hybridization for miR-21.
Formalin-fixed paraffin-embedded lung sections from control mice and mice subjected for 4 h to HVTV and BALF collection, cut 3 μm thick, were used for in situ hybridization with LNA-enhanced miR-21 and control (U6 snRNA) detection probes, using Mercury LNA microRNA ISH optimization kit (Exiqon, Woburn, MA), according to manufacturer's instruction. Briefly, lung sections were subjected to deparaffination, incubation with proteinase-K (15 μg/ml for 15 min at 37°C), dehydration, and hybridization with either 50 nM double DIG LNA miR-21 probe or 1 nM U6 snRNA probe, at 51°C for 1 h. Subsequently sections were washed, blocked with 2% sheep serum in PBS-T, and incubated with anti-DIG-AP antibody 1:800 for 1 h and alkaline phosphatase solution containing nitro-blue tetrazolium and 5-bromo-4-chloro-3′-indolyphosphate for 2 h. Finally sections were dehydrated, mounted, and examined under microscope. Counterstaining was omitted for clarity. A pathologist blinded to the interventions analyzed the results.
Animal experiments.
Male, 8-wk-old, C57BL/6 mice were anesthetized, tracheostomized, and treated with either pre-miR-21, anti-miR-21, or negative control miRNA (oligonucleotides specifically designed to serve as negative controls, all from Ambion; n = 5 per treatment group), at a dose of 4 μg/mouse in 2 μl/g normal saline injected intratracheally before mechanical ventilation. Mice in three treatment groups were subjected to HVTV for 4 h, whereas control mice were ventilated briefly until paralyzed and then subjected to sample collection including an inspiratory pressure-volume curve of the respiratory system, followed by BALF and tissue collection, as previously described (47). All animal experiments were approved by the Research Animal Care Committee of University of Crete Medical School and Heraklion Prefecture Veterinary Authority.
Evaluation of lung injury.
The development of lung injury was assessed by measuring lung mechanics, alveolar-arterial oxygen gradient (DA-a), BALF protein, and cytokine concentration. A pressure-volume curve of the respiratory system was obtained by slow lung inflation to peak airway pressure of 25 cmH2O (47). As an indicator of lung compliance we used the inspiratory capacity, defined as the volume inflated at airway pressure of 25 cmH2O, expressed as ml/kg. Alveolar-arterial oxygen gradient was calculated from arterial blood gas analysis using standard formula. Protein concentration in BALF was measured with the bicinchoninic acid assay (Pierce Chemical, Rockford, IL). IL-6 and macrophage inflammatory protein (MIP)-2 concentrations were measured using ELISA (R&D Systems, Minneapolis, MN).
Statistical analysis.
mRNA and miR-21 expression levels and indices of lung injury are expressed as means ± SD and compared by one-way ANOVA (nonparametric test), using SigmaStat statistical software. Significance was defined as P < 0.05.
RESULTS
Changes in lung miRNA expression profile induced by HVTV.
Out of the 335 miRNAs examined, the expression of 50 miRNAs increased more than twofold, and expression of 15 miRNAs decreased by more than half (Table 1). A progressive change in miRNA expression from 1 h to 4 h was observed in 74% of miRNAs, consistent with the progressive development of lung injury. The miRNAs with the greatest increase in expression after 4 h of HVT were miR-7b, miR-189, and miR-223, whereas the miRNAs with the greatest decrease in expression were miR-503 and miR-211.
Table 1.
Changes in lung miRNA expression profile induced by HVTV
| Control |
Fold Change in miRNA Expression |
||||||
|---|---|---|---|---|---|---|---|
| After 1 h of HVTV |
After 4 h of HVTV |
||||||
| miRNA | Array | Validation | Array | Validation | Array | Validation | miRNA Predicted Target Genes |
| miR-7b | 1.2 ± 0.2 | 1.1 ± 0.2 | 10.8 ± 0.4 | 10.4 ± 0.3 | 24.2 ± 5.6 | 22.1 ± 0.5 | IRS2, OXR1, GSK3B, NFAT5 |
| miR-189 | 1.0 ± 0.1 | 1.0 ± 0.1 | 2.6 ± 0.1 | 2.5 ± 0.2 | 18.6 ± 0.4 | 18.1 ± 0.2 | STK33, HIF3A, CDKN1B, E2F3 |
| miR-223 | 1.0 ± 0.1 | 1.0 ± 0.1 | 7.0 ± 0.8 | 6.8 ± 0.3 | 14.9 ± 2.0 | 15.4 ± 0.5 | RHOB, IGF1R, FOXO1, ECT2 |
| miR-135a | 1.0 ± 0.0 | 1.0 ± 0.0 | 9.5 ± 0.9 | 9.1 ± 0.3 | 14.3 ± 2.3 | 14.9 ± 0.3 | SMAD5, JAK2, GK5, ANGPT2 |
| miR-9 | 1.0 ± 0.0 | 1.0 ± 0.1 | 5.2 ± 0.1 | 5.0 ± 0.2 | 13.4 ± 4.2 | 12.8 ± 1.2 | CDH1, TNC, ITGA6, CTNNA1 |
| miR-132 | 1.0 ± 0.0 | 1.0 ± 0.0 | 3.2 ± 0.2 | 3.1 ± 0.3 | 11.0 ± 2.8 | 11.4 ± 2.2 | ACHE, APAF1, EP300, ETNK1 |
| miR-344 | 1.0 ± 0.1 | 1.0 ± 0.0 | 13.4 ± 0.5 | 12.8 ± 0.2 | 9.2 ± 0.3 | 9.9 ± 0.2 | ITGB8, ESRRG, YES1, SATB1 |
| miR-345 | 0.9 ± 0.1 | 1.0 ± 0.0 | 3.7 ± 0.3 | 3.2 ± 0.6 | 7.8 ± 0.9 | 8.5 ± 0.3 | SMAD1, GPR3, RUNX3, CDKN1A |
| miR-96 | 1.0 ± 0.1 | 1.0 ± 0.0 | 3.6 ± 0.9 | 3.3 ± 0.3 | 7.3 ± 0.9 | 7.8 ± 0.3 | SOX5, ADCY6, FUT9, CPEB1 |
| miR-21 | 1.0 ± 0.1 | 1.0 ± 0.0 | 4.1 ± 0.5 | 4.6 ± 0.2 | 7.1 ± 1.1 | 7.3 ± 0.6 | PTEN, PDCD4, SPRY1, BMPR2 |
| miR-1 | 1.0 ± 0.1 | 1.0 ± 0.0 | 7.9 ± 0.5 | 8.1 ± 0.2 | 6.1 ± 0.4 | 6.6 ± 0.3 | PAX3, ETS1, FOXP1, TRIM2 |
| miR-501 | 1.0 ± 0.0 | 1.0 ± 0.2 | 1.7 ± 0.1 | 1.9 ± 0.2 | 5.2 ± 2.5 | 4.8 ± 0.7 | ADAMTS3, SEMA3C, RBMS1, ESRRG |
| miR-146b | 1.0 ± 0.1 | 1.0 ± 0.0 | 2.5 ± 1.0 | 2.7 ± 0.5 | 4.5 ± 1.2 | 4.1 ± 0.5 | IRAK1, TRAF6, IGSF1, SMAD4 |
| miR-106a | 1.0 ± 0.1 | 1.0 ± 0.0 | 2.2 ± 0.2 | 2.3 ± 0.1 | 4.5 ± 0.5 | 4.0 ± 0.3 | IL10, NFAT5, EPHA4, TGFBR2 |
| miR-485-3p | 0.9 ± 0.2 | 1.0 ± 0.0 | 1.3 ± 0.3 | 1.6 ± 0.2 | 4.4 ± 0.2 | 4.0 ± 0.1 | SMG1, SOX5, PLXNA4, STK10 |
| miR-685 | 1.0 ± 0.1 | 1.0 ± 0.0 | 2.1 ± 0.1 | 2.2 ± 0.2 | 4.3 ± 0.2 | 3.9 ± 0.5 | SIRT1, BMPER, WNT1, FOXO1 |
| miR-543 | 1.0 ± 0.0 | 1.0 ± 0.1 | 3.3 ± 0.2 | 3.1 ± 0.4 | 3.9 ± 0.3 | 3.8 ± 0.2 | IL1A, KLF6, ITGB8, FGF12 |
| miR-153 | 1.0 ± 0.1 | 1.0 ± 0.1 | 3.0 ± 0.1 | 2.9 ± 0.5 | 3.8 ± 0.1 | 3.8 ± 0.1 | ANGPT1, KLF5, KCNA1, GALNT7 |
| miR-210 | 1.0 ± 0.1 | 1.0 ± 0.0 | 1.6 ± 0.1 | 1.4 ± 0.2 | 3.8 ± 0.7 | 3.7 ± 0.3 | IGF2, EFNA3, GIT2, ISCU |
| miR-337 | 1.0 ± 0.1 | 1.0 ± 0.0 | 2.0 ± 0.1 | 2.2 ± 0.3 | 3.8 ± 0.2 | 3.9 ± 0.5 | IL13, RA1, KRAS, RAB22A, USP15 |
| miR-10a | 0.9 ± 0.2 | 1.0 ± 0.0 | 1.1 ± 0.1 | 1.2 ± 0.4 | 3.7 ± 0.8 | 3.1 ± 0.2 | TFAP2C, HOXA3, KLF11, GATA6 |
| miR-125b | 1.0 ± 0.0 | 1.0 ± 0.1 | 2.9 ± 0.3 | 3.3 ± 0.2 | 3.6 ± 0.4 | 3.5 ± 0.7 | TNFA, IRF4, BAK1, KLF13 |
| miR-7 | 1.0 ± 0.1 | 1.0 ± 0.1 | 1.5 ± 0.2 | 2.1 ± 0.3 | 3.5 ± 0.3 | 3.7 ± 0.2 | IRS2, OXR1, GSK3B, NFAT5 |
| miR-196a | 1.0 ± 0.1 | 0.9 ± 0.2 | −5.7 ± 1.9 | −5.7 ± 1.0 | 3.5 ± 0.0 | 3.3 ± 0.1 | HOXC8, HOXB8, NRAS, GATA6 |
| miR-146a | 1.0 ± 0.1 | 1.0 ± 0.1 | 1.7 ± 0.4 | 1.9 ± 0.2 | 3.4 ± 0.2 | 3.1 ± 0.2 | IRAK1, TRAF6, IGSF1, SMAD4 |
| miR-214 | 1.0 ± 0.1 | 1.0 ± 0.1 | 2.4 ± 0.1 | 2.5 ± 0.2 | 3.4 ± 0.4 | 3.0 ± 0.5 | FGFR1, SMYD5, KLK10, GPR6 |
| miR-155 | 1.0 ± 0.1 | 1.0 ± 0.2 | 1.8 ± 0.1 | 2.2 ± 0.2 | 3.3 ± 0.6 | 3.1 ± 0.2 | SOCS1, SHIP, TNFA, TRAF6 |
| miR-18a | 1.0 ± 0.1 | 1.0 ± 0.0 | 1.4 ± 0.1 | 1.8 ± 0.5 | 3.3 ± 0.3 | 3.1 ± 0.6 | ESR1, DICER1, CA13, CCND2 |
| miR-542-3p | 1.0 ± 0.0 | 1.0 ± 0.1 | 4.1 ± 0.1 | 4.4 ± 0.2 | 2.9 ± 0.1 | 2.7 ± 0.3 | TBR1, HSPG2, RC3H1 |
| miR-186 | 1.0 ± 0.1 | 1.0 ± 0.1 | 1.8 ± 0.1 | 2.2 ± 0.3 | 2.7 ± 0.4 | 2.4 ± 0.3 | MIB1, PDE10A, IRF8, XIAP |
| miR-151 | 1.0 ± 0.1 | 1.0 ± 0.2 | 1.4 ± 0.1 | 1.6 ± 0.2 | 2.6 ± 0.6 | 2.5 ± 0.3 | CLK1, AKT3, FXR1, DLC1 |
| miR-194 | 1.1 ± 0.2 | 1.0 ± 0.0 | 1.7 ± 0.2 | 1.4 ± 0.3 | 2.5 ± 0.4 | 2.4 ± 0.8 | ANGPT1, TMED5, TRAF6, SETD8 |
| miR-205 | 1.1 ± 0.2 | 1.0 ± 0.0 | 1.9 ± 0.1 | 2.1 ± 0.5 | 2.4 ± 0.4 | 2.3 ± 0.5 | YES1, MED1, COX11, LRP1 |
| miR-350 | 1.0 ± 0.0 | 1.0 ± 0.1 | −1.5 ± 0.1 | −1.2 ± 0.2 | 2.3 ± 0.6 | 2.2 ± 0.3 | EEA1, DNER, OTUD1, SUZ12 |
| miR-467b | 1.0 ± 0.1 | 1.0 ± 0.1 | 1.6 ± 0.4 | 1.8 ± 0.2 | 2.3 ± 0.1 | 2.1 ± 0.5 | SNX5, E2F2, E2F7, DOCK2 |
| miR-142-3p | 1.0 ± 0.0 | 1.0 ± 0.0 | 1.3 ± 0.0 | 1.5 ± 0.2 | 2.3 ± 0.5 | 2.1 ± 0.2 | BOD1, LCOR, RAB2A, ZEB2 |
| miR-137 | 1.0 ± 0.1 | 1.0 ± 0.1 | 3.6 ± 0.9 | 3.9 ± 0.3 | 2.3 ± 0.1 | 2.3 ± 0.1 | CDC42, PDLIM3, EPHA7, KLF12 |
| miR-330 | 0.9 ± 0.1 | 1.0 ± 0.1 | 1.3 ± 0.0 | 1.5 ± 0.2 | 2.3 ± 0.2 | 2.1 ± 0.4 | ITGA5, ELK1, EFNA3, ABCC1 |
| miR-27a | 1.0 ± 0.0 | 1.0 ± 0.0 | 1.4 ± 0.0 | 1.7 ± 0.2 | 2.2 ± 0.5 | 2.4 ± 0.7 | PLK2, FBXW7, LIN28B, CCNG1 |
| miR-455-3p | 1.0 ± 0.1 | 1.0 ± 0.2 | 1.6 ± 0.4 | 1.8 ± 0.2 | 2.2 ± 0.0 | 2.1 ± 0.2 | CUL3, HOXC4, TAZ, ITGA1 |
| miR-203 | 1.0 ± 0.1 | 1.0 ± 0.1 | 1.9 ± 0.2 | 2.2 ± 0.4 | 2.2 ± 0.4 | 2.2 ± 0.7 | LIFR, UBR1, ELL2, IL24 |
| miR-129-3p | 1.0 ± 0.0 | 1.0 ± 0.2 | 1.0 ± 0.2 | 1.0 ± 0.1 | 2.2 ± 0.2 | 2.1 ± 0.3 | ACACA, DOK4, MEMO1 |
| miR-199a | 1.0 ± 0.0 | 1.0 ± 0.0 | 1.2 ± 0.3 | 1.0 ± 0.1 | 2.1 ± 0.2 | 2.3 ± 0.5 | DDR1, AKAP1, ONECUT2, JAG1 |
| miR-700 | 1.0 ± 0.1 | 1.0 ± 0.1 | 1.0 ± 0.1 | 1.0 ± 0.1 | 2.1 ± 0.4 | 2.2 ± 0.7 | MARVELD1 |
| miR-34a | 1.0 ± 0.0 | 1.0 ± 0.2 | 1.6 ± 0.1 | 1.3 ± 0.3 | 2.1 ± 0.6 | 2.2 ± 0.2 | MYCN, SATB2, RRAS, E2F5 |
| miR-376a | 1.0 ± 0.1 | 1.0 ± 0.0 | 2.2 ± 0.1 | 2.5 ± 0.2 | 1.9 ± 0.1 | 2.0 ± 0.2 | MLL, CACHD1 |
| miR-369-5p | 1.0 ± 0.1 | 1.0 ± 0.1 | 2.4 ± 0.8 | 2.2 ± 0.4 | 1.8 ± 0.8 | 1.9 ± 0.3 | DXH15, TTI1, MAP3K13, RNF111 |
| miR-721 | 1.0 ± 0.0 | 1.0 ± 0.1 | 2.3 ± 0.2 | 2.7 ± 0.1 | 1.8 ± 0.1 | 1.7 ± 0.2 | TSC1, FMR1, DDX6, RAI2 |
| miR-34b | 1.0 ± 0.1 | 1.0 ± 0.1 | 2.1 ± 0.1 | 2.6 ± 0.2 | 1.7 ± 0.2 | 1.6 ± 0.7 | MYCN, SATB2, RRAS, E2F5 |
| miR-541 | 1.0 ± 0.0 | 1.0 ± 0.2 | 2.4 ± 0.2 | 2.8 ± 0.3 | 1.2 ± 0.2 | 1.5 ± 0.4 | STK3, CPEB4, RUNX1, GK5 |
| miR-486 | 1.0 ± 0.1 | 1.0 ± 0.0 | −2.7 ± 0.3 | −2.3 ± 0.2 | 1.6 ± 0.2 | 1.3 ± 0.3 | PTEN, AFF3, DOCK3, FGF9 |
| miR-9 | 1.0 ± 0.1 | 1.0 ± 0.1 | −3.0 ± 1.2 | −2.8 ± 1.3 | −1.2 ± 0.0 | −1.1 ± 0.5 | CDH1, TNC, ITGA6, CTNNA1 |
| miR-682 | 1.0 ± 0.2 | 1.0 ± 0.2 | −6.1 ± 0.2 | −6.5 ± 0.5 | −1.3 ± 0.1 | −1.6 ± 0.2 | STK39, ATRN, UBR1, AAK1 |
| miR-381 | 1.0 ± 0.0 | 1.0 ± 0.0 | −15.6 ± 0.4 | −12.5 ± 0.2 | −2.1 ± 0.0 | −1.9 ± 0.6 | STX12, PNN, TET3, LRP6 |
| miR-701 | 1.0 ± 0.1 | 1.0 ± 0.1 | −1.3 ± 0.1 | −1.5 ± 0.1 | −2.3 ± 0.5 | −2.7 ± 0.2 | OSR1, ZNF827 |
| miR-706 | 1.0 ± 0.0 | 1.0 ± 0.2 | −2.5 ± 0.2 | −2.7 ± 0.4 | −2.3 ± 0.5 | −2.2 ± 0.3 | ENAH, MTX3, CYC1, PARG |
| let-7b | 1.0 ± 0.1 | 1.0 ± 0.0 | −1.5 ± 0.1 | −1.8 ± 0.3 | −2.3 ± 0.5 | −2.6 ± 0.4 | IL6, TLR4, RAS, MYC |
| miR-192 | 1.0 ± 0.0 | 1.0 ± 0.0 | −2.4 ± 0.2 | −2.6 ± 0.5 | −4.1 ± 1.6 | −3.6 ± 0.5 | EREG, DICER1, ZEB2, ATF1 |
| miR-487b | 1.0 ± 0.0 | 1.0 ± 0.2 | −1.0 ± 0.2 | −1.1 ± 0.2 | −4.3 ± 0.5 | −4.9 ± 0.1 | NELF, IRS1, ASTN1, EPHA3 |
| let-7d | 1.0 ± 0.1 | 1.0 ± 0.0 | −1.6 ± 0.3 | −1.3 ± 0.5 | −4.3 ± 0.2 | −4.3 ± 0.2 | LIN28B, HMGA2, GDF6, HIC2 |
| miR-200c | 1.0 ± 0.0 | 1.0 ± 0.1 | −2.4 ± 0.4 | −2.1 ± 0.3 | −7.1 ± 2.5 | −7.2 ± 0.8 | ZEB2, ZEB1, RECK, TIMP2 |
| let-7a | 1.0 ± 0.1 | 1.0 ± 0.0 | −3.8 ± 0.5 | −3.2 ± 0.2 | −7.0 ± 1.9 | −7.1 ± 0.3 | IL6, TLR4, RAS, MYC |
| miR-676 | 1.0 ± 0.1 | 1.0 ± 0.2 | −2.8 ± 0.2 | −2.4 ± 0.4 | −8.9 ± 3.8 | −9.5 ± 0.6 | PDE4D, BAGE5, IGFBP5, AUP1 |
| miR-211 | 1.0 ± 0.1 | 1.0 ± 0.0 | −16.7 ± 4.0 | −15.4 ± 0.6 | −9.6 ± 0.2 | −9.2 ± 0.2 | EPHA5, SOX4, TCF12, SOS1 |
| miR-503 | 1.0 ± 0.1 | 1.0 ± 0.0 | 1.2 ± 0.1 | 0.8 ± 0.5 | −12.5 ± 1.9 | −12.7 ± 0.7 | MYB, RICTOR, SYT3, CCNE1 |
Values are means ± SD. HVTV, high tidal volume ventilation; miR, microRNA.
Identification of miRNA downstream targets.
To identify potential targets for differentially expressed miRNAs, we screened their sequences against the mouse genome database, using the miRNA target identification programs miRBase, PicTar, and TargetScan v.5.1, as described in materials and methods. Table 1 presents potential target genes of miRNAs whose expression was altered by HVTV.
Gene network analysis.
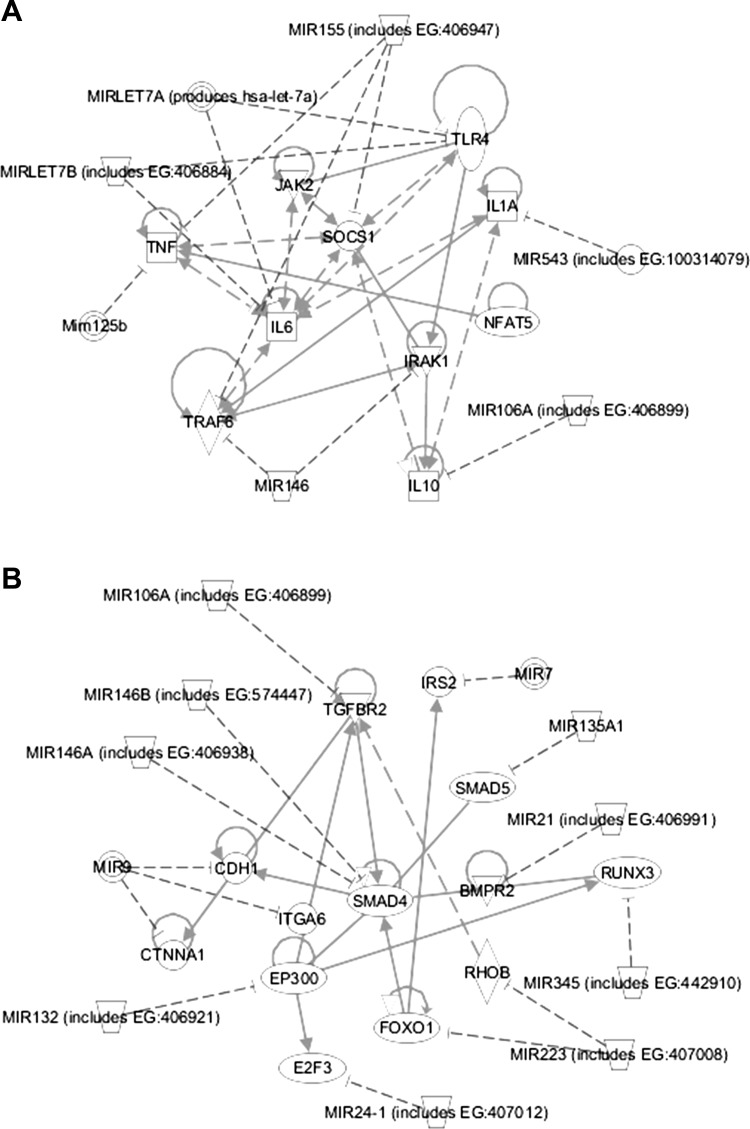
Gene network analysis was performed by integrating the miRNA expression data and their predicted gene targets to identify enrichment of pathways related to VILI. The miRNA-gene network with the highest statistical significance score (P = 10−43) was a network of inflammatory mediators, as shown in Fig. 1A. This network includes seven miRNAs whose expression was modulated during HVTV (miR-155, let-7a, let-7b, miR-125b, miR-146, miR-106a, and miR-543) and 10 genes that are targets of the miRNAs (TNF-α, JAK2, SOCS1, IRAK1, TLR4, IL1A, NFAT5, IL6, TRAF6, and IL10). According to the gene network analysis, IL6 and SOCS1 are the central nodes of this inflammatory network.
Fig. 1.
MicroRNA/gene networks induced by mechanical ventilation with high tidal volume (HVTV) in mice. A: inflammatory signaling microRNA-gene network. B: TGF-β signaling microRNA-gene network. Lines indicate the predicted interactions between microRNAs and their downstream targets. TLR4, Toll-like-receptor 4; TNF, tumor-necrosis-factor; JAK2, Janus kinase 2; SOCS1, suppressor of cytokine signaling 1; IL1A, interleukin 1A; IL6, interleukin 6; TRAF6, TNF receptor-associated factor 6; IRAK1, interleukin-1 receptor-associated kinase 1; NFAT5, nuclear factor of activated T-cells 5; IL10, interleukin 10; TGFBR2, transforming growth factor β receptor type II; IRS2, insulin receptor substrate 2; CDH1, cadherin 1; ITGA6, integrin α6; BMPR2, bone morphogenetic protein receptor, type II; EP300, E1A binding protein p300; RHOB, ras homolog gene family, member B; FOXO1, forkhead box O1; E2F3, E2F transcription factor 3; CTNNA1, catenin (cadherin-associated protein) α1; RUNX3, runt-related transcription factor 3.
The miRNA-gene network with the second highest statistical significance score was a TGF-β signaling network (P = 10−28; Fig. 1B). This network includes 11 miRNAs whose expression was modulated during HVTV (miR-106a, miR-7, miR-135, miR-21, miR-345, miR-223, miR-24, miR-132, miR-9, miR-146a, miR-146b) and 13 genes that are targets of the miRNAs (TGFBR2, IRS2, SMAD5, CDH1, ITGA6, SMAD4, BMPR2, EP300, RHOB, FOXO1, E2F3, CTNNA1, and RUNX3). According to the gene network analysis, SMAD4 is the central node of the network.
Expression levels of miRNA target genes.
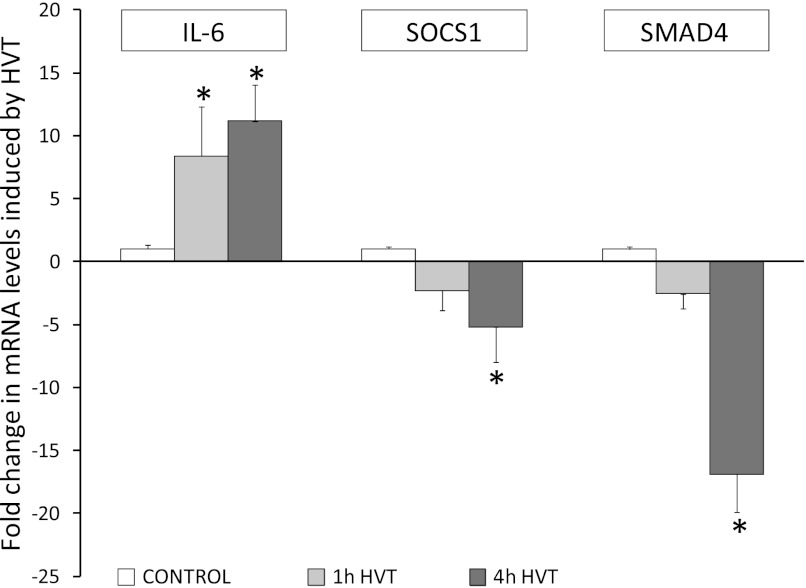
We measured pulmonary levels of mRNAs targeted by miRNAs represented in the inflammatory gene and TGFβ-regulated networks (Fig. 2). The expression of let-7, which targets IL-6, decreased with HVTV. Pulmonary IL-6 mRNA levels increased after HVTV (11-fold increase from control after 4 h of HVTV, P < 0.05), consistent with the decrease in let-7 expression. The expression of miR-155 and miR-146, which target SOCS1 and SMAD4, respectively, increased with HVTV. Pulmonary SOCS1 and SMAD4 mRNA levels decreased after HVTV (5-fold and 25-fold decrease from control after 4 h of HVTV, respectively, P < 0.05), consistent with the increase in miR-155 and miR-146 expression.
Fig. 2.
Changes in the levels of mRNAs encoding IL-6, SOCS1, and SMAD4 in lungs of mice exposed to mechanical HVTV for 1 or 4 h compared with control mice: *P < 0.05 vs. control.
In vivo modulation of lung miRNA in HVTV.
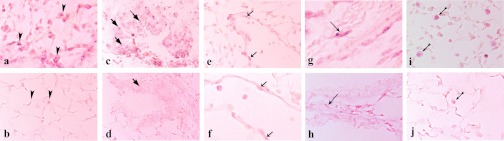
We tested the hypothesis that changes in miRNA expression contribute to the pathogenesis of VILI, focusing on miR-21, a miRNA highly upregulated by HVTV. To identify the cells that contribute to the observed increase in miR-21 expression, we measured miR-21 expression levels in BAL cells and lung tissues from mice subjected to HVTV for 4 h. MiR-21 expression after HVTV increased 21 ± 5-fold in BAL cells (consisting of macrophages at 90%) and 6 ± 1-fold in lung homogenates. Additionally, we performed in situ hybridization in paraffin-embedded sections from lungs of control mice and mice subjected to 4 h of HVTV after BAL cell collection. Increased miR-21 expression was detected in epithelial cells (type II alveolar pneumocytes and bronchiole lining cells), endothelial cells, stromal myofibroblasts, as well as in alveolar macrophages (Fig. 3).
Fig. 3.
In situ hybridization with double DIG-LNA miR-21 probe, anti-DIG-AP antibody, and AP solution containing NBT-BCIP (purple staining) of formalin-fixed, paraffin-embedded lung sections, from mice subjected for 4 h to HVTV (a, c, e, g, i), and control mice (b, d, f, h, j). MiR-21 staining is observed in lungs from mice subjected to HVTV in alveolar type II pneumocytes (a and b), bronchiole lining cells (c and d), endothelial cells (e and f), myofibroblasts (g and h), as well as in alveolar macrophages (i and j; arrows indicating specific cell type; a, b, c, d, e, h, i, j: magnification ×400, f and g: magnification ×600).
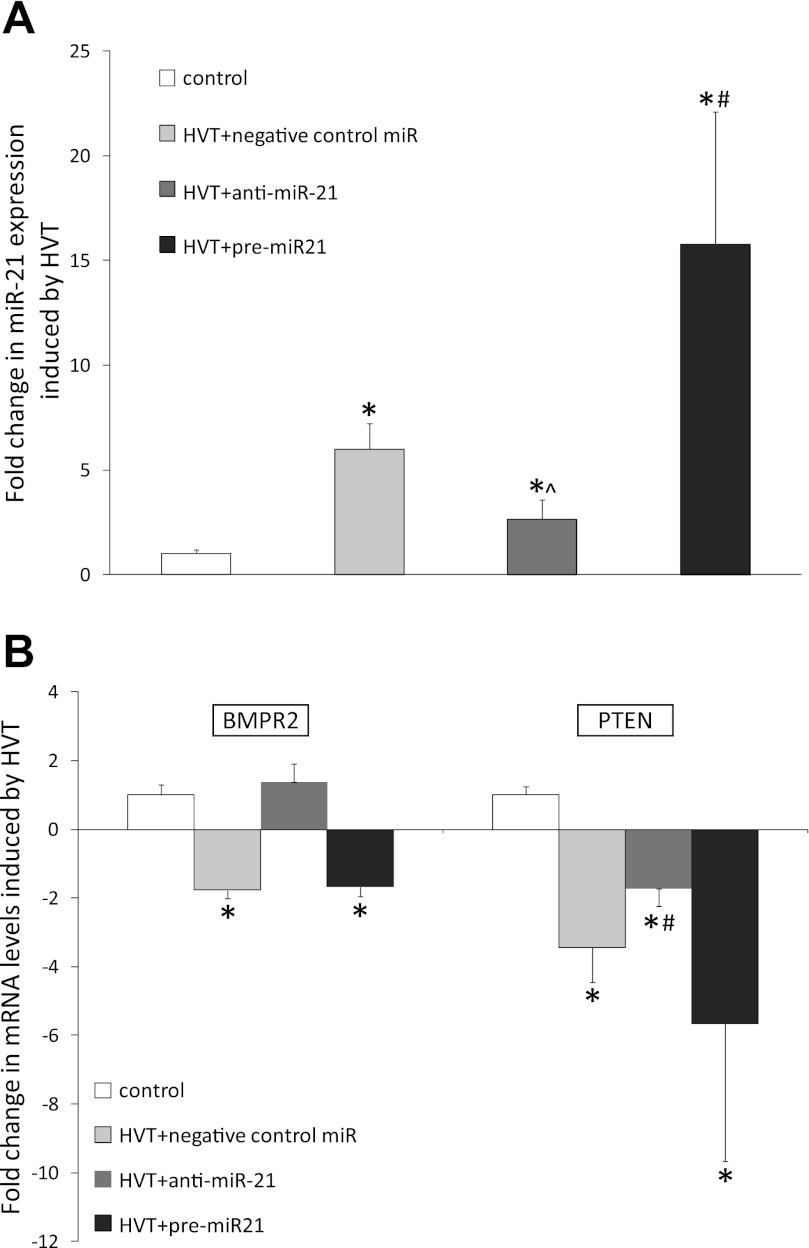
Subsequently we modulated the expression of miR-21 by intratracheal administration of a precursor (pre-miR-21) or antagonist (anti-miR-21) of miR-21 before mechanical HVTV. The increase in pulmonary miR-21 levels after HVTV was greater in mice treated with pre-miR-21 than in mice treated with negative control miRNA (16 ± 6- vs. 6 ± 1-fold increase from control, P < 0.05; Fig. 4A). Treatment with the anti-miR-21 attenuated the increase in miR-21 induced by HVTV (3 ± 1- vs. 6 ± 1-fold increase, P < 0.005). Pulmonary levels of miR-21 target mRNAs, BMPR2 and PTEN (15, 37), were greater in mice treated with anti-miR-21 than in mice treated with pre-miR-21 or negative control-miR (Fig. 4B).
Fig. 4.
A: changes in levels of miR-21 in lung homogenates from mice treated with negative control miR, anti-miR-21, or pre-miR-21 followed by mechanical HVTV for 4 h, compared with control mice: *P < 0.05 for all ventilated mice compared with control mice, #P < 0.05 for pre-miR-21 treatment vs. negative control miR, ^P < 0.05 for anti-miR-21 treatment vs. negative control miR. B: changes in levels of BMPR2 mRNA in lung homogenates from mice treated with negative control miR, anti-miR-21, or pre-miR-21 followed by HVTV for 4 h, compared with control mice: *P < 0.001 for negative-control-miR and pre-miR-21 treatment vs. control, and PTEN mRNA; *P < 0.001 for all ventilated mice vs. control, #P < 0.05 for anti-miR-21 treatment vs. negative-control-miR and pre-miR-21.
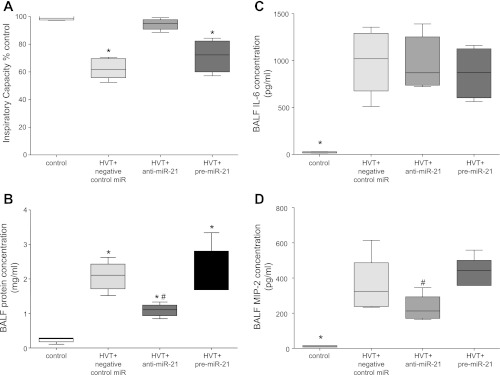
Mice treated with anti-miR-21 before HVTV had improved oxygenation, as indicated by the DA-a, than did mice treated with pre-miR-21 or the negative control miRNA (66 ± 27 vs. 131 ± 22 and 144 ± 10 mmHg, respectively, P < 0.001) and preserved lung compliance, as indicated by the inspiratory capacity (% of control: 94 ± 4% vs. 72 ± 12%, 62 ± 7%, P < 0.001, Fig. 5A). BALF protein concentration increased from control in all ventilated mice, but it was lower in mice treated with anti-miR-21 than in mice treated with pre-miR-21 or negative control miR (1.1 ± 0.2 vs. 2.3 ± 1, 2.1 ± 0.4 mg/ml, respectively, P < 0.01, Fig. 5B). BALF concentrations of IL-6 and MIP-2 were greater in all ventilated mice than control mice. BALF IL-6 was not different between groups of ventilated mice (Fig. 5C), whereas MIP-2 was lower in mice treated with anti-miR-21 than mice treated with pre-miR-21 (Fig. 5D).
Fig. 5.
Evaluation of lung injury in mice treated with negative control miR, anti-miR-21, or pre-miR-21 followed by mechanical HVTV for 4 h, compared with control mice. All data are presented in whisker blots (min-max, line at mean). A: inspiratory capacity as % control: *P < 0.001 for negative-control-miR and pre-miR-21 treatment vs. control. B: bronchoalveolar lavage fluid (BALF) protein concentration: *P < 0.001 for control vs. all ventilated mice, #P < 0.05 for anti-miR-21 treatment vs. negative-control-miR and pre-miR-21. C: BALF concentrations of IL-6: *P < 0.001 for control vs. all ventilated mice. D: BALF concentrations of macrophage inflammatory protein (MIP)-2: *P < 0.001 for control vs. all ventilated mice, #P < 0.05 for anti-miR-21 vs. pre-miR-21 treatment.
Taken together, our results indicate that in vivo modulation of pulmonary miR-21 levels, using anti-miR-21, ameliorates indices of high-permeability pulmonary edema associated with VILI.
DISCUSSION
This study reports the impact of mechanical HVTV on the miRNA expression profile in lungs of mice. We found that injurious mechanical ventilation induced rapid and progressive changes in lung miRNA expression, suggesting that these miRNAs might play a role in the development of lung injury. The miRNA-gene networks identified using miRNA target prediction databases at highest statistical significance were an inflammatory and a TGF-β signaling network.
Whereas the role of inflammation in VILI is well established, the role of miRNAs in the regulation of inflammation has only recently been recognized (45). Innate immune responses have been associated with increases in levels of miR-146, miR-155, miR-125, and miR-9, as well as a decrease in let-7 levels (2, 4, 7, 29, 42, 43). We observed that most miRNAs, previously reported to be associated with inflammation (45), were similarly modulated by injurious mechanical ventilation. The predicted targets of differentially expressed miRNAs include cytokines, such as IL-6 and TNF-α, known to play an important role in VILI (11). IL-6 has been recognized as a central modulator of acute lung injury (ALI) and VILI in several experimental and clinical studies (9, 34, 41). Additionally, genes such as SOCS1, which have been associated with inflammation (19, 32) but not previously described in VILI, were identified in silico as important components of the inflammatory miRNA-gene network in VILI.
Patients in the intensive care unit suffering from ALI are at highest risk for developing VILI. A previous study, using a murine model of lung injury induced by inhaled LPS, examined the changes in lung miRNA expression profile associated with ALI (29). Although the miRNA primer/probe sets and mouse strain were different in our study, 15 miRNAs were similarly changed in lung injury induced by LPS and HVTV, including let-7, miR-21, miR-106, miR-146, miR-132, miR-210, miR-214, and miR-223. These findings suggest that these miRNAs are critical modulators of lung inflammation and may play an important role in human ALI, where both infectious and mechanical stimuli are present.
Interestingly, we observed that many of the miRNAs induced by HVTV (and their target mRNAs) participated in a TGF-β-signaling miRNA-gene network. TGF-β signals are involved in lung barrier function and inflammation (10, 26, 28). The role of TGF-β signaling in the late phase of ALI is well established (10), but this study adds to the increasing evidence suggesting that TGF-β signaling may also play a role in the early phase of lung injury (18, 38). MiRNAs play an important role in the regulation of TGF-β signaling (17, 27). SMAD proteins, downstream molecules of TGF-β signaling pathway, as well as TGFβR2 and BMPR2, TGF-β, and TGF-β ligands receptors, have all been identified as direct targets of miRNAs including miR-146, miR-106, and miR-21 (27).
Among the differentially expressed miRNAs, those exhibiting the greater increase (more than 10-fold) after 4 h of HVTV were miR-7b, miR-189, miR-223, miR-135a, miR-9, and miR-132. MiR-7b targets the early response gene fos, which has been shown to increase upon HVTV (21, 44). MiR-189 and miR-135a target the TGF-β signaling molecules furin, Net1A, and SMAD5 (23, 35, 48), emphasizing the important role of miRNA regulation of TGF-β signaling in VILI. MiRNAs miR-223 and miR-132 are highly expressed in myeloid cells (8, 42), and therefore the observed increase in their expression could be attributed to inflammatory cell infiltration in lungs induced by HVTV. Additionally, activation of TLR signaling, which occurs in VILI, increases the expression of both miR-132 and miR-9 (4, 42). Both miRNAs may have an anti-inflammatory role, miR-9 by targeting NFkB1 (4), and miR-132 by targeting acetylcholinesterase (40). The miRNAs exhibiting the greatest decrease (more than 7-fold) were miR-503, miR-211, miR-676, let-7a, and miR-200c. All these miRNAs are considered tumor suppressors, suggesting that the observed decrease in their expression could facilitate cell proliferation as a response to injury. Let-7a and 200c were the only highly expressed miRNAs in lungs of control mice. Both may control barrier function because miR-200c targets e-cadherin transcriptional repressor ZEB1, and let-7a targets integrin b3 (13, 30). Let-7a also targets IL-6 (14); thus decrease of let-7a could contribute to the increase of IL-6 observed in VILI.
To investigate whether the observed changes in lung miRNAs play a role in the development of VILI, we examined the effect of modulating pulmonary miRNA expression in vivo on the development of VILI. We chose miR-21 because it is significantly induced by HVTV in both resident alveolar cells and infiltrating macrophages. Moreover, pulmonary miR-21 antagonism in vivo was shown to ameliorate pulmonary fibrosis in mice treated with bleomycin (25). We were able to increase and decrease pulmonary miR-21 levels by intratracheal administration of a precursor and antagonist of miR-21, respectively. We observed that treatment with anti-miR-21 ameliorated indices of VILI. The preserved lung compliance and oxygenation, together with the reduction in BALF protein levels, suggest that treatment with anti-miR-21 can prevent HVTV-induced barrier dysfunction. Target genes of miR-21 that could play a role in VILI include BMPR2 and PTEN, both of which have a protective role in barrier function (3, 6, 22). We found that the observed increase of miR-21 in lungs of mice subjected to HVTV was associated with decrease in both BMPR2 and PTEN mRNA levels. Moreover, treatment with anti-miR-21 was associated with preservation of BMPR2 and PTEN mRNA levels.
Our study has several limitations. The mouse model of VILI used in this study applies a tidal volume that is much higher than that used in clinical practice. However, it has been shown that in animals with healthy lungs a strain greater than 2 is required to induce injury (36), and the tidal volume used in this study resulted in a strain of 2.2. Moreover, we used a model of aseptic VILI, which may not recapitulate clinical practice but facilitates our understanding of the mechanisms of VILI. We did not compare the effect of different tidal volumes on lung miRNA expression profile because it has been previously reported that mechanical ventilation even at low tidal volume is injurious in mice (12, 46). In preliminary experiments, we observed that mechanical ventilation with normal tidal volume (8 ml/kg) moderately increased miR-21 expression.
The identification of the role and cellular source of each differentially expressed miRNA was beyond the scope of this study. It is probable that not all differentially expressed miRNAs contribute to the pathogenesis of VILI. Most likely several miRNAs whose expression changed in response to HVTV are part of protective mechanisms that aim in limiting lung injury. For example, the observed increase in miR-146 could contribute to inhibition of TGF-β signaling by targeting SMAD-4 and/or promote macrophage tolerance to TLR signals by targeting TRAF-6 and IRAK-1 (31), suggesting a protective role for miR-146.
In summary, the present study demonstrates that several miRNAs are differentially regulated during the development of VILI. The major networks these miRNAs participate in include an inflammatory and a TGF-β signaling miRNA-gene network. Moreover, this study provides evidence that local administration of precursors or anti-miRs can modulate pulmonary miRNA expression and affect the development of VILI, providing novel potential therapeutic approaches.
GRANTS
This work has been supported by US Public Health Service grant HL-42397 to W. M. Zapol, by the LeDucq Foundation and US Public Health Service grant HL74352 to K.D. Bloch, by European Society of Intensive Care Medicine research grant to K. Vaporidi, and by University of Crete Research Committee grants to D. Georgopoulos and K. Vaporidi (KA 2869, 3500).
DISCLOSURES
No conflicts of interest, financial or otherwise, are declared by the authors.
AUTHOR CONTRIBUTIONS
Author contributions: K.V., W.M.Z., K.D.B., and D.I. conception and design of research; K.V., E.V., E.K., and M.H. performed experiments; K.V., E.V., E.K., E.L., and D.I. analyzed data; K.V., E.L., D.G., and D.I. interpreted results of experiments; K.V., E.L., and D.I. prepared figures; K.V. drafted manuscript; K.V., D.G., K.D.B., and D.I. edited and revised manuscript; D.G., W.M.Z., K.D.B., and D.I. approved final version of manuscript.
ACKNOWLEDGMENTS
The authors thank the Dana-Farber microarray core services for their help with the high-throughput microRNA analysis. The authors also thank Dr. Eumorfia Kondili and Dr. George Prinianakis for help and support at the experimental Intensive Care Medicine laboratory at the University of Crete, and Eleutheria Tsendelierou for assistance in ISH assay.
Preliminary results of this study have been presented as an abstract at the ATS International Conference (May 14–19, 2010, New Orleans, Louisiana; A2031).
REFERENCES
- 1. Acute Respiratory Distress Syndrome Network Ventilation with lower tidal volumes compared with traditional tidal volumes for acute lung injury, and the acute respiratory distress syndrome. N Engl J Med 342: 1301–1308, 2000 [DOI] [PubMed] [Google Scholar]
- 2. Androulidaki A, Iliopoulos D, Arranz A, Doxaki C, Schworer S, Zacharioudaki V, Margioris AN, Tsichlis PN, Tsatsanis C. The kinase Akt1 controls macrophage response to lipopolysaccharide by regulating microRNAs. Immunity 31: 220–231, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Barbieri SS, Ruggiero L, Tremoli E, Weksler BB. Suppressing PTEN activity by tobacco smoke plus interleukin-1beta modulates dissociation of VE-cadherin/beta-catenin complexes in endothelium. Arterioscler Thromb Vasc Biol 28: 732–738, 2008 [DOI] [PubMed] [Google Scholar]
- 4. Bazzoni F, Rossato M, Fabbri M, Gaudiosi D, Mirolo M, Mori L, Tamassia N, Mantovani A, Cassatella MA, Locati M. Induction and regulatory function of miR-9 in human monocytes and neutrophils exposed to proinflammatory signals. Proc Natl Acad Sci USA 106: 5282–5287, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Budinger GR, Chandel NS, Donnelly HK, Eisenbart J, Oberoi M, Jain M. Active transforming growth factor-beta1 activates the procollagen I promoter in patients with acute lung injury. Intensive Care Med 31: 121–128, 2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Burton VJ, Ciuclan LI, Holmes AM, Rodman DM, Walker C, Budd DC. Bone morphogenetic protein receptor II regulates pulmonary artery endothelial cell barrier function. Blood 117: 333–341, 2011 [DOI] [PubMed] [Google Scholar]
- 7. Ceppi M, Pereira PM, Dunand-Sauthier I, Barras E, Reith W, Santos MA, Pierre P. MicroRNA-155 modulates the interleukin-1 signaling pathway in activated human monocyte-derived dendritic cells. Proc Natl Acad Sci USA 106: 2735–2740, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Chen CZ, Li L, Lodish HF, Bartel DP. MicroRNAs modulate hematopoietic lineage differentiation. Science 303: 83–86, 2004 [DOI] [PubMed] [Google Scholar]
- 9. Cross LJM, Matthay MA. Biomarkers in acute lung injury: insights into the pathogenesis of acute lung injury. Crit Care Clin 27: 355–377, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Dhainaut JF, Charpentier J, Chiche JD. Transforming growth factor-beta: a mediator of cell regulation in acute respiratory distress syndrome. Crit Care Med 31: S258–S264, 2003 [DOI] [PubMed] [Google Scholar]
- 11. Frank JA, Matthay MA. Science review: mechanisms of ventilator-induced injury. Crit Care 7: 233–241, 2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Hauber HP, Karp D, Goldmann T, Vollmer E, Zabel P. Effect of low tidal volume ventilation on lung function and inflammation in mice. BMC Pulm Med 10: 21, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Hurteau GJ, Carlson JA, Spivack SD, Brock GJ. Overexpression of the microRNA hsa-miR-200c leads to reduced expression of transcription factor 8 and increased expression of E-cadherin. Cancer Res 67: 7972–7976, 2007 [DOI] [PubMed] [Google Scholar]
- 14. Iliopoulos D, Hirsch HA, Struhl K. An epigenetic switch involving NF-kappaB, Lin28, Let-7 MicroRNA, and IL6 links inflammation to cell transformation. Cell 139: 693–706, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Iliopoulos D, Jaeger SA, Hirsch HA, Bulyk ML, Struhl K. STAT3 activation of miR-21 and miR-181b-1 via PTEN and CYLD are part of the epigenetic switch linking inflammation to cancer. Mol Cell 39: 493–506, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Imanaka H, Shimaoka M, Matsuura N, Nishimura M, Ohta N, Kiyono H. Ventilator-induced lung injury is associated with neutrophil infiltration, macrophage activation, and TGF-beta 1 mRNA upregulation in rat lungs. Anesth Analg 92: 428–436, 2001 [DOI] [PubMed] [Google Scholar]
- 17. Inui M, Martello G, Piccolo S. MicroRNA control of signal transduction. Nat Rev Mol Cell Biol 11: 252–263, 2010 [DOI] [PubMed] [Google Scholar]
- 18. Jain M, Budinger GRS, Lo A, Urich D, Rivera SE, Ghosh AK, Gonzalez A, Chiarella SE, Marks K, Donnelly HK, Soberanes S, Varga J, Radigan KA, Chandel NS, Mutlu GM. Leptin promotes fibroproliferative acute respiratory distress syndrome by inhibiting peroxisome proliferator-activated receptor-gamma. Am J Respir Crit Care Med 183: 1490–1498, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Kubo M, Hanada T, Yoshimura A. Suppressors of cytokine signaling and immunity. Nat Immunol 4: 1169–1176, 2003 [DOI] [PubMed] [Google Scholar]
- 20. Lanford RE, Hildebrandt-Eriksen ES, Petri A, Persson R, Lindow M, Munk ME, Kauppinen S, Orum H. Therapeutic Silencing of MicroRNA-122 in Primates with Chronic Hepatitis C Virus Infection. Science 327: 198–201, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Lee HJ, Palkovits M, Young WS., 3rd miR-7b, a microRNA up-regulated in the hypothalamus after chronic hyperosmolar stimulation, inhibits Fos translation. Proc Natl Acad Sci USA 103: 15669–15674, 2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Lee KS, Kim SR, Park SJ, Lee HK, Park HS, Min KH, Jin SM, Lee YC. Phosphatase and tensin homolog deleted on chromosome 10 (PTEN) reduces vascular endothelial growth factor expression in allergen-induced airway inflammation. Mol Pharmacol 69: 1829–1839, 2006 [DOI] [PubMed] [Google Scholar]
- 23. Li Z, Hassan MQ, Volinia S, van Wijnen AJ, Stein JL, Croce CM, Lian JB, Stein GS. A microRNA signature for a BMP2-induced osteoblast lineage commitment program. Proc Natl Acad Sci USA 105: 13906–13911, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Lionetti V, Recchia FA, Ranieri VM. Overview of ventilator-induced lung injury mechanisms. Curr Opin Crit Care 11: 82–86, 2005 [DOI] [PubMed] [Google Scholar]
- 25. Liu G, Friggeri A, Yang YP, Milosevic J, Ding QA, Thannickal VJ, Kaminski N, Abraham E. miR-21 mediates fibrogenic activation of pulmonary fibroblasts and lung fibrosis. J Exp Med 207: 1589–1597, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Lu Q, Harrington EO, Jackson H, Morin N, Shannon C, Rounds S. Transforming growth factor-β1-induced endothelial barrier dysfunction involves Smad2-dependent p38 activation and subsequent RhoA activation. J Appl Physiol 101: 375–384, 2006 [DOI] [PubMed] [Google Scholar]
- 27. McCoy CE. The role of miRNAs in cytokine signaling. Front Biosci 16: 2161–2171, 2011 [DOI] [PubMed] [Google Scholar]
- 28. Mokres LM, Parai K, Hilgendorff A, Ertsey R, Alvira CM, Rabinovitch M, Bland RD. Prolonged mechanical ventilation with air induces apoptosis and causes failure of alveolar septation and angiogenesis in lungs of newborn mice. Am J Physiol Lung Cell Mol Physiol 298: L23–L35, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Moschos SA, Williams AE, Perry MM, Birrell MA, Belvisi MG, Lindsay MA. Expression profiling in vivo demonstrates rapid changes in lung microRNA levels following lipopolysaccharide-induced inflammation but not in the anti-inflammatory action of glucocorticoids. BMC Genomics 8: 240, 2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Muller DW, Bosserhoff AK. Integrin beta 3 expression is regulated by let-7a miRNA in malignant melanoma. Oncogene 27: 6698–6706, 2008 [DOI] [PubMed] [Google Scholar]
- 31. Nahid MA, Satoh M, Chan EKL. MicroRNA in TLR signaling and endotoxin tolerance. Cell Mol Immunol 8: 388–403, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Nakashima T, Yokoyama A, Onari Y, Shoda H, Haruta Y, Hattori N, Naka T, Kohno N. Suppressor of cytokine signaling 1 inhibits pulmonary inflammation and fibrosis. J Allergy Clin Immunol 121: 1269–1276, 2008 [DOI] [PubMed] [Google Scholar]
- 33. Nakou M, Bertsias G, Stagakis I, Centola M, Tassiulas I, Hatziapostolou M, Kritikos I, Goulielmos G, Boumpas DT, Iliopoulos D. Gene network analysis of bone marrow mononuclear cells reveals activation of multiple kinase pathways in human systemic lupus erythematosus. PLoS One 5: 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Nonas SA, Finigan JH, Gao L, Garcia JG. Functional genomic insights into acute lung injury: role of ventilators and mechanical stress. Proc Am Thorac Soc 2: 188–194, 2005 [DOI] [PubMed] [Google Scholar]
- 35. Papadimitriou E, Vasilaki E, Vorvis C, Iliopoulos D, Moustakas A, Kardassis D, Stournaras C. Differential regulation of the two RhoA-specific GEF isoforms Net1/Net1A by TGF-beta and miR-24: role in epithelial-to-mesenchymal transition. Oncogene 31: 2862–2875, 2011 [DOI] [PubMed] [Google Scholar]
- 36. Protti A, Cressoni M, Santini A, Langer T, Mietto C, Febres D, Chierichetti M, Coppola S, Conte G, Gatti S, Leopardi O, Masson S, Lombardi L, Lazzerini M, Rampoldi E, Cadringher P, Gattinoni L. Lung stress and strain during mechanical ventilation: any safe threshold? Am J Respir Crit Care Med 183: 1354–1362, 2011 [DOI] [PubMed] [Google Scholar]
- 37. Qin W, Zhao B, Shi Y, Yao C, Jin L, Jin Y. BMPRII is a direct target of miR-21. Acta Biochim Biophys Sin (Shanghai) 41: 618–623, 2009 [DOI] [PubMed] [Google Scholar]
- 38. Roux J, Carles M, Koh H, Goolaerts A, Ganter MT, Chesebro BB, Howard M, Houseman BT, Finkbeiner W, Shokat KM, Paquet AC, Matthay MA, Pittet JF. Transforming growth factor beta1 inhibits cystic fibrosis transmembrane conductance regulator-dependent cAMP-stimulated alveolar epithelial fluid transport via a phosphatidylinositol 3-kinase-dependent mechanism. J Biol Chem 285: 4278–4290, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Schickel R, Boyerinas B, Park SM, Peter ME. MicroRNAs: key players in the immune system, differentiation, tumorigenesis and cell death. Oncogene 27: 5959–5974, 2008 [DOI] [PubMed] [Google Scholar]
- 40. Shaked I, Meerson A, Wolf Y, Avni R, Greenberg D, Gilboa-Geffen A, Soreq H. MicroRNA-132 potentiates cholinergic anti-inflammatory signaling by targeting acetylcholinesterase. Immunity 31: 965–973, 2009 [DOI] [PubMed] [Google Scholar]
- 41. Stuber F, Wrigge H, Schroeder S, Wetegrove S, Zinserling J, Hoeft A, Putensen C. Kinetic and reversibility of mechanical ventilation-associated pulmonary and systemic inflammatory response in patients with acute lung injury. Intensive Care Med 28: 834–841, 2002 [DOI] [PubMed] [Google Scholar]
- 42. Taganov KD, Boldin MP, Chang KJ, Baltimore D. NF-kappaB-dependent induction of microRNA miR-146, an inhibitor targeted to signaling proteins of innate immune responses. Proc Natl Acad Sci USA 103: 12481–12486, 2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Tili E, Michaille JJ, Cimino A, Costinean S, Dumitru CD, Adair B, Fabbri M, Alder H, Liu CG, Calin GA, Croce CM. Modulation of miR-155 and miR-125b levels following lipopolysaccharide/TNF-alpha stimulation and their possible roles in regulating the response to endotoxin shock. J Immunol 179: 5082–5089, 2007 [DOI] [PubMed] [Google Scholar]
- 44. Tremblay L, Valenza F, Ribeiro SP, Li J, Slutsky AS. Injurious ventilatory strategies increase cytokines and c-fos m-RNA expression in an isolated rat lung model. J Clin Invest 99: 944–952, 1997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Tsitsiou E, Lindsay MA. microRNAs and the immune response. Curr Opin Pharmacol 9: 514–520, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Vaneker M, Joosten LA, Heunks LM, Snijdelaar DG, Halbertsma FJ, van Egmond J, Netea MG, van der Hoeven JG, Scheffer GJ. Low-tidal-volume mechanical ventilation induces a toll-like receptor 4-dependent inflammatory response in healthy mice. Anesthesiology 109: 465–472, 2008 [DOI] [PubMed] [Google Scholar]
- 47. Vaporidi K, Francis RC, Bloch KD, Zapol WM. Nitric oxide synthase 3 contributes to ventilator-induced lung injury. Am J Physiol Lung Cell Mol Physiol 299: L150–L159, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Wang J, Huang W, Xu R, Nie Y, Cao X, Meng J, Xu X, Hu S, Zheng Z. MicroRNA-24 regulates cardiac fibrosis after myocardial infarction. J Cell Mol Med. In press [DOI] [PMC free article] [PubMed] [Google Scholar]