Abstract
The manifestation of RNA interference (RNAi)-based therapeutics lies in safe and successful delivery of small interfering RNAs (siRNAs), the molecular entity that triggers and guides sequence-specific degradation of target mRNAs. Optimizing the chemistry and structure of siRNAs to achieve maximum efficacy is an important parameter in the development of siRNA therapeutics. The RNAi protein machinery can tolerate a variety of non-canonical modifications made to siRNAs, each of which imparts advantageous properties. Here, we review these modifications to siRNAs in pre-clinical and clinical studies.
Introduction
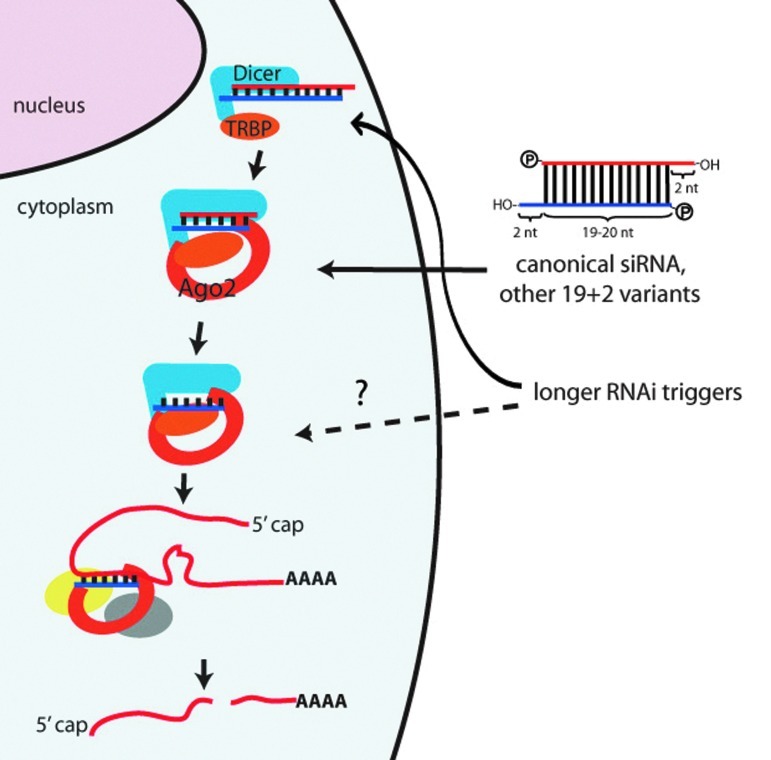
RNA interference (RNAi) is an endogenous protein suppression mechanism by which short double-stranded RNA (dsRNA) mediates sequence-specific degradation of mRNA, thereby preventing translation of the protein encoded by the target mRNA. The short dsRNA, designed such that one of the strands is complementary to the target mRNA (called the guide, or antisense, strand) and the other strand its complement (called the passenger, or sense, strand), is the trigger to initiate RNAi. Depending on the organism, there exist different, specific forms of RNAi, including the microRNA (miRNA) pathway, transcriptional gene silencing, and post-transcriptional gene silencing. Several key proteins facilitate the RNAi mechanism, including Dicer and the Argonaute (Ago) proteins. The RNAi protein machinery processes and orients a single strand of the dsRNA to execute gene silencing. For this review, we will be focusing on mammalian RNAi with perfectly complementary RNAi-trigger/mRNA pairing, leading to Ago2-mediated destruction of target mRNA (Fig. 1).
FIG. 1.
Schematic depicting perfectly complementary argonaute (Ago)2-mediated RNA interference (RNAi) in mammalian cells. RNAi triggers with the canonical siRNA structure do not require Dicer processing, whereas longer RNAi triggers will be processed by Dicer to enter the RNAi pathway. There is some evidence that longer RNAi triggers can be loaded into Ago2/RNA induced silencing complex (RISC) directly, but this model remains to be thoroughly tested. Red strands=passenger strands. Blue strands=guide strands. Figure is not drawn to scale.
RNAi has already risen as a gold standard for validating gene function in basic science studies, but also holds great promise as a new therapeutic paradigm. When looking at the broad concept of traditional drug design (i.e., small molecule inhibitors and therapeutic monoclonal antibodies), the point of therapeutic intervention occurs at the protein level (i.e., after a disease-causing protein has already been translated). These traditional drugs were “discovered” by high-throughput screening and trial-and-error chemical modifications to lead compounds. Structure-guided drug design claims to assist in “rational” drug design, but at some level, even these drug discovery strategies are iterative. Many important and groundbreaking traditional therapeutics have been developed to date and will continue to be important for improving human health. There remain, however, additional challenging diseases and therapeutic spaces in which traditional drug discovery may not succeed. Conceptually, an RNAi-based therapeutic would intervene at the post-transcriptional level of a disease-causing protein, and the sequence-specific nature when designing an efficacious and specific RNAi trigger cannot be matched by traditional drug design. Other potential advantages of RNAi-based therapeutics include relatively fast initial screening and the ability to target proteins that are deemed “un-druggable” by traditional drug design strategies. It is for these reasons that researchers and clinicians are exploring the possibility of making RNAi-based therapies into a new platform by which to make therapeutics.
To bring RNAi-based therapeutics to the clinic, the research and development effort can be divided into 2 broad areas: small interfering RNA (siRNA)–target combination optimization and delivery. Delivery, meaning the development of carrier materials to protect the therapeutic RNA payload, as well as safely and efficaciously delivery relevant quantities to the cells of interest, is arguably the largest challenge for realizing RNAi-based therapeutics. Many chemists, material scientists, nanotechnologists, and virologists are actively working on solving the delivery challenge. We will not focus on the delivery challenge in this review, but refer instead to review articles on this subject (de Fougerolles et al., 2007; Kim et al., 2007; Lu et al., 2008; Ford et al., 2010).
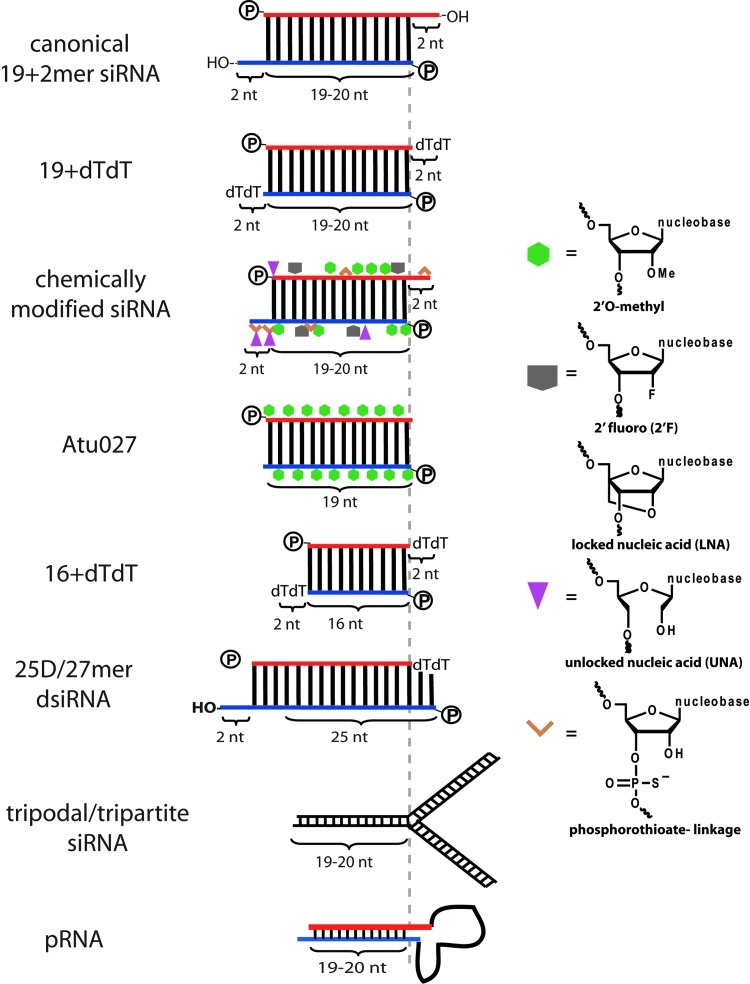
This review will focus on the current strategies used for RNAi trigger optimization. The basis for optimizing RNAi triggers include: (1) facilitating chemical synthesis; (2) increasing the stability against biological fluids; (3) influencing strand selection towards the desired (guide) strand; (4) avoiding the activation of the innate immune response; and (5) reducing off-target effects, which is further subcategorized into guide strand- or passenger strand-mediated off-target effects. Our focus will be on exogenously delivered RNAi triggers (Figure 2), not DNA-encoded (or DNA-directed, ddRNAi) RNAi triggers.
FIG. 2.
Schematic of a canonical siRNA and several chemical and structural variations. The chemically modified small interfering RNA (siRNA) is not any specific siRNA reported in the literature, but rather a generic illustration of where some of the indicated chemical modifications can be placed. Red strands=passenger strands. Blue strands=guide strands. Dashed vertical grey line denotes the 5′ end of the guide strand of a canonical siRNA and the predicted 5′ end of the guide strand after Dicer processing for longer RNAi triggers. Figures not drawn to scale.
RNAi triggers used in basic science and pre-clinical reports
Unmodified siRNAs
RNAi was first described in Caenorhabditis elegans and subsequently in plants and flies. The triggers used in these early discoveries were dsRNAs of ∼30–800 bp (Fire et al., 1998). Initially, researchers were skeptical about RNAi's utility in mammals, as mammalian cells and organs contain more developed adaptive and innate immune systems. It was known that dsRNA longer than ∼30 bp would trigger interferon production via foreign RNA-sensing receptors such as the Toll-like receptors (TLRs), the retinoic acid inducible gene 1 (RIG-1), and PKR, leading to non-RNAi-related gene expression effects (Jackson et al., 2010) and possibly cell death.
After scrutinizing the small duplex RNAs from flies, studies from Tuschl and colleagues revealed that RNAi could be achieved in mammalian cells with the use of synthetic 19+2 small RNA duplexes (Elbashir et al., 2001). These small interfering RNAs (siRNAs), composed of 2 strands that basepair perfectly from nucleotides 1–19 counting from the 5′ end and posses 2 nucleotide 3′ overhangs, were efficiently loaded into the RNAi machinery resulting in specific silencing of the expression of the target protein of interest without triggering an innate immune response. For completeness, these RNAi triggers possess a 5′ hydroxyl (5′OH) group, and it is known that, while not rate limiting (Kenski et al., 2012), the 5′OH is monophosphorylated by Clp-1 kinase (Weitzer et al., 2007). These siRNAs, composed entirely of RNA nucleotides, helped jumpstart the RNAi revolution in basic biological research. Several basic science and pre-clinical studies began exploring the use of canonical and partially backbone modified siRNAs.
Chemical modifications
Nucleotide substitutions and backbone chemical modifications
In the seminal publication by Tuschl and colleagues, RNA duplexes with the same 19+2 structure but with DNA nucleotides in the 2-nt 3′ overhangs (Fig. 2) were also tested and shown to be equally efficacious. The use of DNA nucleotides in the 3′ overhang facilitate the solid phase synthesis manufacturing of these oligos as well as provide protection against RNases. This relatively simple nucleotide chemical modification is still widely used in siRNAs today.
The DNA overhangs on siRNAs led to other nucleotide substitutions. Nearly half the length of an siRNA's guide or passenger strand can be replaced with DNA nucleotides and remain functional (Ui-Tei et al., 2008). Several bulky imaging groups on nucleobases in select positions along the siRNA strand are tolerated by the RNAi machinery (Hoerter et al., 2011).
Other chemical modifications to siRNAs, inspired by the research done with chemically modified antisense oligonucleotides (ASOs), also emerged. Specifically, instead of substituting nucleotides, modifications to the backbone (i.e., either to the phosphodiester bond or to functional groups of the ribose ring) of siRNAs can confer stability against nucleases as well as enable other functional characteristics. The strategy for screening compatible chemical modifications for some groups' RNAi trigger variants appear iterative (Allerson et al., 2005; Kubo et al., 2007; Kubo et al., 2008; Bramsen et al., 2009), perhaps mimicking the mindset used for traditional small molecule inhibitor drug lead compound chemical optimization, whereas other groups' strategies to integrate chemical modifications are more focused.
A wide range of backbone chemical modifications have been reported in the literature, the most popular modifications, especially in RNAi-based therapeutics in clinical trials, being phosphorothioate, 2′fluoro, 2′O-methyl, locked nucleic acid (LNA), and unlocked nucleic acid (UNA) (Fig. 2). All of the above-mentioned backbone modifications confer enhanced nuclease stability (Layzer et al., 2004; Dande et al., 2006; Bramsen et al., 2009). Phosphorothioate modifications should not, however, be placed between nucleotides 9–10 of the designed passenger strand, as a modification in this position would block Ago2's endonuclease function, a step required in the maturation of the RNA induced silencing complex (RISC) (Matranga et al., 2005). The 2′fluoro (Layzer et al., 2004) and 2′O-methyl modifications are well tolerated in most positions, although they can affect efficacy when placed in certain positions (Allerson et al., 2005). Additionally, selection of the passenger strand can be abolished by a 5′O-methyl group (Chen et al., 2008), and off-target effects can be mitigated with a 2′O-methyl at position 2 of the guide strand (Jackson et al., 2006). Moderate, judicious placement of LNA nucleotides, especially in the overhangs or in selected positions on the passenger strand, is tolerated by the RNAi machinery, whereas heavily LNA-modified siRNAs lose silencing ability (Elmen et al., 2005). The LNA modification has also been used as a molecular tool to understand the biological underpinnings of RNAi, such as siRNAs with a pre-nicked passenger strand (small internally segmented siRNAs, sisiRNAs) (Bramsen et al., 2007; Chorn et al., 2010) and the strand selection and off-targeting capability of LNA-stabilized siRNAs (Petri et al., 2011). The UNA modification is tolerated in most positions except as the first nucleotide of the guide strand (Kenski et al., 2010). Placing a UNA modification on the first nucleotide of the passenger strand, however, confers guide strand selectivity by blocking passenger strand incorporation into RISC (Vaish et al., 2011). A UNA modification at the seventh position of the guide strand was shown to improve on-target silencing while discouraging off-target silencing (Bramsen et al., 2010). These different chemical modifications can be combined to generate heterogeneous backbone chemically modified siRNAs (Allerson et al., 2005; Bramsen et al., 2009). Other less-commonly used modifications include 3′ propanediol (Ocampo et al., 2012), 2′O-methoxyethyl, and 2′-deoxy-2′-fluoro-beta-D-arabinonucleic acid (Dowler et al., 2006; Bramsen et al., 2009).
Ligand-conjugated modifications
In order to provide additional functionality to siRNAs, several ligands have been conjugated to siRNAs without compromising gene-silencing activity. Imaging agents, such as Cy3, Cy5, and TAMRA, are commonly conjugated to allow fluorescent imaging of siRNAs. Small molecule ligands, such as folate, have been conjugated to siRNAs to provide cell-specific targeting against folate receptor-overexpressing cancer cells (Guo et al., 2006; Dohmen et al., 2012). Peptide-conjugated siRNAs can also allow specific cell targeting (e.g., arginine-glycine-aspartate [RGD]-conjugated siRNA) (Alam et al., 2011) or cell permeability (e.g., HIV transactivator of transcription [TAT]-conjugated siRNA) (Moschos et al., 2007). Conjugating cholesterol to siRNAs targeting apoB allowed enhanced cellular uptake in gastrointestinal organs and successfully reduced total cholesterol in mice (Soutschek et al., 2004).
Structural modifications with canonical siRNA chemistry
The 19+2 structure of an siRNA is symmetric, thereby allowing the RNAi machinery to select either the desired (guide) or undesired (passenger) for gene silencing. To date, though not perfect, the most robust criteria influencing strand selection of a symmetric canonical siRNA is the thermodynamic end structure, such that the strand of siRNA with the less stable 5′ end is selected for RNA silencing (Khvorova et al., 2003; Schwarz et al., 2003; Reynolds et al., 2004).
Similar to the different motivations for chemical modifications, the strategy for siRNA structure modifications for some groups is arguably iterative and extensive (Kubo et al., 2007; Kubo et al., 2008), whereas the modification strategy for other groups is more hypothesis driven. Several variations on the canonical structure of an siRNA have been reported, each with different biological properties or advantages compared to canonical siRNAs. For example, it was demonstrated that the 3′ overhangs of RNAi triggers are an important design criteria, and that, regardless of the siRNA sequence, strands containing 3′ overhangs are more likely to be selected by the RNAi machinery than 3′ blunt-ended strands (Sano et al., 2008). Thus, one can shorten the passenger strand to generate a unilateral 3′ overhang exclusively on the guide strand to promote guide strand selection. In a different example of shortening variations to canonical siRNAs designed to reduce passenger strand-mediated off-target silencing capability, RNAi triggers with a shorter passenger strand have been designed, the rationale being that selection of the undesired shorter passenger strand does not meet the length requirements to be used for RNAi. From a screen of short passenger strand RNAi trigger variants targeting green fluorescent protein (GFP), a 16+dTdT (containing 2 deoxythymidine, dT, overhangs) was shown to be as good or better, depending on the target choice, than its sequence-matched canonical 19+dTdT (Chu et al., 2008). This report, however, was not fully reproducible (Sierant et al., 2010). In a different context, the use of a 14+dTdT RNAi trigger was explored for use in the treatment of a mouse model of age-related macular degeneration. The rationale here was that a 14+dTdT was below the length threshold for detection by the innate immune system's TLR3 (Kleinman et al., 2012). It remains to be seen, however, whether this short RNAi trigger can function in RNAi. Asymmetric siRNAs with short passenger strands have been reported, where a 19-21nt guide strand overlaps a short passenger strand of at least 12 nucleotides on either or both the 5′ and 3′ ends (Sun et al., 2008; Chang et al., 2009). These asymmetric RNAi triggers were still functional in gene silencing and showed decreased passenger strand-mediated off-target effects (Chang et al., 2009).
Instead of shortening canonical siRNAs, several groups have demonstrated that longer dsRNAs, yet still below the innate immune system-sensing threshold, were more efficacious in gene silencing. Indeed, siRNA hairpin precursors, called short hairpin RNAs (shRNAs), are more efficacious in silencing target gene expression compared to their sequence-matched synthetic canonical siRNAs (Siolas et al., 2005). Due to their longer length, these triggers are substrates for the key upstream RNAi processing enzyme Dicer, as opposed to canonical siRNAs, which are the size of Dicer products. While not completely understood, the hypothesis is that the Dicer processing step facilitates a Dicer substrate's introduction into the RNAi pathway more effectively than a Dicer product. As a more calculated design approach using the knowledge about longer dsRNAs yielding more gene knockdown, as well as the knowledge of the importance of 3′ overhangs on the desired strand, another variant, called Dicer substrate interfering RNA (dsiRNA), was designed. While one study synthesized and screened 70 different dsiRNAs and their sequence-matched canonical siRNAs and saw little, if any, difference in on-target potency (Foster et al., 2012), the original papers describing dsiRNAs (Kim et al., 2005; Rose et al., 2005; Collingwood et al., 2008) showed improved silencing compared to sequence matched canonical siRNAs, and multiple independent reports (Dore-Savard et al., 2008; Howard et al., 2009; Bohle et al., 2011; Lundberg et al., 2011; Darniot et al., 2012; Kubo et al., 2012) have used dsiRNAs in their studies.
Other longer dsRNA structures with distinctive shapes and unique features have also been described. The phage phi29 packaging RNA (pRNA), which adopts a handshake-like structure, was adapted to accommodate an siRNA sequence to participate in RNAi (Guo et al., 2006). The additional utility of these siRNA-modified pRNAs lies in their ability to form multimers, thereby forming multivalent RNA molecules (GUO, 2010). Tripodal (Chang et al., 2012a) and branched tripartite RNAi triggers (Chang et al., 2012b), which adopt a 3-pointed star structure, can deliver multiple siRNAs per RNA unit. These long dsRNA variant structures will need to be assessed further, particularly for the claim that despite being >30 bp, they do not induce an innate immune system response nor do they necessarily require Dicer processing for RNAi function.
Combined structural variations and chemical modifications
In attempts to maximize the nuclease stability and functionality conferred by chemically modified nucleotides as well as the favorable properties of structurally modified RNAi trigger variants, some researchers have combined structural and chemical modifications. For example, the structural dsiRNA variants mentioned earlier contain deoxyribonucleotides at the 3′ blunt end of the passenger strand to introduce functional polarity (Rose et al., 2005), and can be further selectively modified with DNA, 2′F, or 2′O-methyl ribonucleotides without compromising the gene silencing ability (Collingwood et al., 2008; Foster et al., 2012). Proprietary lipophile-conjugated (Accell, Thermo Fisher) and single strand tailed chemically modified siRNA (sd-rxRNA, RXi Pharmaceuticals) have been designed for use as self-delivering siRNAs. In contrast to what is known about the advantages of the 3′ overhang's role in effective RNAi triggers, Atu027 is a blunt-ended, alternating 2′O-methyl 23mer dsRNA targeting the oncogene PKN3 and has demonstrated therapeutic silencing in mouse and primate models of various primary and metastatic tumors (Aleku et al., 2008; Santel et al., 2010). Lastly, while the goal of some therapeutic siRNAs may be to gently modulate the gene expression of a diseased cell, the therapeutic goal of most anti-cancer treatments is to completely eradicate (i.e., kill) cancer cells. Cancer cells, and the tumor niche, are sensitive to local inflammation (Bui et al., 2007). Hence, while many RNAi triggers are designed to avoid triggering inflammation-inducing innate immune responses, it may be beneficial to design anti-cancer RNAi triggers that simultaneously intentionally trigger local inflammation for tumor therapy. This proof of principle was demonstrated in a metastatic melanoma mouse model with a blunt-ended 19mer dsRNA containing a 5′ triphosphate group, a ligand for the type 1 interferon-inducing receptor RIG-1, on the 5′ ends (Poeck et al., 2008).
RNAi in clinical trials
At the time of preparation of this manuscript, ClinicalTrials.gov reports that ∼30 siRNA- or shRNA-based therapeutics are being evaluated in clinical trials. These clinical trials range from viral vector- or plasmid DNA-delivered shRNAs to exogenously delivered synthetic siRNA, and from applications encompassing ex vivo treatment of human immunodeficiency virus, age-related macular degeneration (AMD), cancer, and others. Only one of these trials had progressed to phase 3 but has been withdrawn. Here, we focus on the trials employing synthetic siRNAs.
Clinical trials using unmodified exogenously delivered synthetic siRNAs
The first generation of siRNA-based therapeutics, starting as early as July 2005, delivered unmodified siRNA duplexes. These trials were designed prior to the finding that in the case of the eye, unmodified siRNAs can trigger TLR3 (Kleinman et al., 2008) and lead to inflammation-based, not RNAi-based, gene silencing. The target tissues were often in superficial organs and did not involve any lipid- or polymer-based delivery agent. For example, a group of three companies—Opko, Allergan, and Quark—initiated phase 1 and 2 trials for treatment of eye disease (AMD, macular edema, etc.) using siRNAs targeting vascular endothelial growth factor (VEGF) in order to reduce deleterious neovascularization, using direct injection into the eye (Dejneka et al., 2008). For the treatment of pachyonychia congenita, TransDerm uses direct needle injection of an siRNA, targeting point mutations of the keratin gene, composed of unmodified nucleotides [albeit with proprietary ligand conjugation to promote delivery reagent-independent cell uptake (Hickerson et al., 2011)] into the diseased skin tissue (Hickerson et al., 2008a). Completing a phase 1-b clinical trial (Leachman et al., 2010), the authors suggest that an advantage of using unmodified siRNAs in their application is that any siRNA that does not quickly enter the target tissue (i.e., is delivered to the bloodstream) will be rapidly degraded and decrease non-target cell delivery and side effects (Hickerson et al., 2008b). Transderm is investing in microneedle technology to assist in a less painful delivery mechanism (Lara et al., 2012). As a last example, employing a tumor-cell-targeted polymer-based delivery vehicle (cyclodextran polycation), Calando Pharmaceuticals' CALAA-01 unmodified siRNA nanoparticle (RONDEL) was used in a phase 1 clinical trial targeting metastatic melanoma (Davis et al., 2010). Target gene knockdown, verified by 5′ rapid amplification of cDNA ends (RACE) assay, was observed for one patient, but due to the small patient size of the trial at the time of publication, final conclusions will have to wait for the completion of the trial. In this case, the use of a delivery agent may shield unmodified siRNAs from triggering extracellular innate immune system receptors. Nonetheless, the TLR3 finding has undoubtedly made these and other companies re-think their approach of using unmodified siRNAs and instead employ chemically modified nucleotides, both to resist RNase degradation and to mitigate innate immune system activation.
Next generation siRNA therapeutics in clinical trials
The next generation of exogenously delivered siRNA therapeutics in clinical trials not only incorporate delivery vehicles but also employ chemically and structurally modified siRNA variants. Though only in phase 1 clinical trials, these next-generation siRNA-based therapeutics have taken a lesson from the basic science and earlier clinical trials in designing their therapeutic materials and overall strategy. Combined with their own lipid delivery formulation (Santel et al., 2006a; Santel et al., 2006b), Silence Therapeutics' aforementioned Atu027 was safely tolerated in the highest tested dose in a phase 1 clinical trial (Strumberg et al., 2012). Despite recent legal disputes, the 2 companies with the most mature clinical trial profiles are Alnylam Pharmaceuticals and Tekmira Pharmaceuticals, tackling diseases as diverse as hypercholesterolemia, Ebola, respiratory syncytial virus, transthyretin-mediated amyloidosis, and various cancers. Their RNAi triggers maintain a 19+2 structure with specifically placed DNA, 2′F, 2′O-methyl, and phosphorothioate modifications (Akinc et al., 2008; Akinc et al., 2009; Semple et al., 2010).
Results from these clinical trials demonstrate progress as well as future challenges. Depending on the study, the most encouraging results include safety as well as some positive therapeutic response despite single sub-therapeutic doses, while consistent biomarker results are still being investigated. Tekmira and Alnylam have generated arguably the most insightful clinical data using polyethylene glycol modified (PEGylated) lipid nanoparticle technology. A 2009 phase 1 clinical trial employing the PEGylated lipid nanoparticle delivery system developed by Tekmira, delivering siRNA against ApoB to curb hypercholesterolemia, was terminated after only a year. Two subjects had adverse effects during treatment, possibly but not certainly due to the treatment. Tekmira had, however, already continued studying and optimizing its formulation for safety and efficacy, including the use of chemically modified immune-dampening siRNAs, in pre-clinical studies and animal models. Subsequently, the phase 1 clinical trial using PEGylated lipid nanoparticles for patients with refractory solid tumors (ALN-VSP02) reported that the majority of the patients did not experience serious or emergent adverse events (www.alnylam.com/capella/category/presentations), although 1 patient with extensive pre-existing liver metastasis died during the trial. From 15 patients' biopsied samples, 3 exhibited evidence of RNAi via the 5′ RACE assay for 1 of the mRNA targets (VEGF). The 5′ RACE assay, however, is not easy, and performing the assay on the small amount of biopsied samples may pose additional technical challenges. Alnylam's phase 1 placebo-controlled ALN-PCSK9 clinical trial, using Tekmira's advanced-generation PEGylated lipid nanoparticle delivery system, aiming to curb hypercholesterolemia, demonstrated encouraging safety and some efficacy even at single administrations of low doses (www.alnylam.com/capella/category/presentations). Lastly, based on preclinical studies combining the strategies of lipophile conjugation (Wolfrum et al., 2007) and lipid-carbohydrate PEGylated nanoparticle formulation (Akinc et al., 2010), another phase 1 clinical trial employing a hepatocyte-targeted N-acetyl galactose-conjugated siRNA to combat liver transthyretin-mediated amyloidosis (www.alnylam.com/capella/category/presentations) is scheduled to begin in the second half of 2012.
Conclusions
After a rapid start to RNAi-based therapeutics, game-changing basic science and pre-clinical reports, as well as waning support from big pharma, has slowed RNAi therapeutics' progress. RNAi therapeutics researchers and companies, originally excited by the power of early RNAi reports, are now reevaluating their initial ambitious dream to target nearly any and every disease-causing gene. Nonetheless, several clinical trials are in progress employing state-of-the-art delivery and nucleic acid modifications to achieve safe and efficacious therapeutic gene silencing. Arguably, the target cells and organs of choice will play a large role in the feasibility of successfully delivering therapeutic RNA; organs such as skin, liver, or lung may be easier targets than solid tumors in internal organs.
Acknowledgments
JJR is supported by NIH grants R01A1042552 and R01HL074704.
References
- AKINC A. GOLDBERG M. QIN J. DORKIN J.R. GAMBA-VITALO C. MAIER M., et al. Development of lipidoid-siRNA formulations for systemic delivery to the liver. Mol. Ther. 2009;17:872–879. doi: 10.1038/mt.2009.36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- AKINC A. QUERBES W. DE S. QIN J. FRANK-KAMENETSKY M. JAYAPRAKASH K.N., et al. Targeted delivery of RNAi therapeutics with endogenous and exogenous ligand-based mechanisms. Mol. Ther. 2010;18:1357–1364. doi: 10.1038/mt.2010.85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- AKINC A. ZUMBUEHL A. GOLDBERG M. LESHCHINER E.S. BUSINI V. HOSSAIN N., et al. A combinatorial library of lipid-like materials for delivery of RNAi therapeutics. Nat. Biotechnol. 2008;26:561–569. doi: 10.1038/nbt1402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ALAM M.R. MING X. FISHER M. LACKEY J.G. RAJEEV K.G. MANOHARAN M., et al. Multivalent cyclic RGD conjugates for targeted delivery of small interfering RNA. Bioconjug. Chem. 2011;22:1673–1681. doi: 10.1021/bc200235q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ALEKU M. SCHULZ P. KEIL O. SANTEL A. SCHAEPER U. DIECKHOFF B., et al. Atu027, a liposomal small interfering RNA formulation targeting protein kinase N3, inhibits cancer progression. Cancer Res. 2008;68:9788–9798. doi: 10.1158/0008-5472.CAN-08-2428. [DOI] [PubMed] [Google Scholar]
- ALLERSON C.R. SIOUFI N. JARRES R. PRAKASH T.P. NAIK N. BERDEJA A., et al. Fully 2′-modified oligonucleotide duplexes with improved in vitro potency and stability compared to unmodified small interfering RNA. J. Med. Chem. 2005;48:901–904. doi: 10.1021/jm049167j. [DOI] [PubMed] [Google Scholar]
- BOHLE H. LORENZEN N. SCHYTH B.D. Species specific inhibition of viral replication using dicer substrate siRNAs (DsiRNAs) targeting the viral nucleoprotein of the fish pathogenic rhabdovirus viral hemorrhagic septicemia virus (VHSV) Antiviral Res. 2011;90:187–194. doi: 10.1016/j.antiviral.2011.03.174. [DOI] [PubMed] [Google Scholar]
- BRAMSEN J.B. LAURSEN M.B. DAMGAARD C.K. LENA S.W. BABU B.R. WENGEL J., et al. Improved silencing properties using small internally segmented interfering RNAs. Nucleic Acids Res. 2007;35:5886–5897. doi: 10.1093/nar/gkm548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- BRAMSEN J.B. LAURSEN M.B. NIELSEN A.F. HANSEN T.B. BUS C. LANGKJAER N., et al. A large-scale chemical modification screen identifies design rules to generate siRNAs with high activity, high stability and low toxicity. Nucleic Acids Res. 2009;37:2867–2881. doi: 10.1093/nar/gkp106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- BRAMSEN J.B. PAKULA M.M. HANSEN T.B. BUS C. LANGKJAER N. ODADZIC D., et al. A screen of chemical modifications identifies position-specific modification by UNA to most potently reduce siRNA off-target effects. Nucleic Acids Res. 2010;38:5761–573. doi: 10.1093/nar/gkq341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- BUI J.D. SCHREIBER R.D. Cancer immunosurveillance, immunoediting and inflammation: independent or interdependent processes? Curr. Opin. Immunol. 2007;19:203–208. doi: 10.1016/j.coi.2007.02.001. [DOI] [PubMed] [Google Scholar]
- CHANG C.I. LEE T.Y. KIM S. SUN X. HONG S.W. YOO J.W., et al. Enhanced intracellular delivery and multi-target gene silencing triggered by tripodal RNA structures. J. Gene Med. 2012a;14:138–146. doi: 10.1002/jgm.1653. [DOI] [PubMed] [Google Scholar]
- CHANG C.I. LEE T.Y. YOO J.W. SHIN D. KIM M. KIM S., et al. Branched, tripartite-interfering RNAs silence multiple target genes with long guide strands. Nucleic Acid Ther. 2012b;22:30–39. doi: 10.1089/nat.2011.0315. [DOI] [PubMed] [Google Scholar]
- CHANG C.I. YOO J.W. HONG S.W. LEE S.E. KANG H.S. SUN X., et al. Asymmetric shorter-duplex siRNA structures trigger efficient gene silencing with reduced nonspecific effects. Mol. Ther. 2009;17:725–32. doi: 10.1038/mt.2008.298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- CHEN P. Y. WEINMANN L. GAIDATZIS D. PEI Y. ZAVOLAN M. TUSCHL T., et al. Strand-specific 5′-O-methylation of siRNA duplexes controls guide strand selection and targeting specificity. RNA. 2008;14:263–274. doi: 10.1261/rna.789808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- CHORN G. ZHAO L. SACHS A.B. FLANAGAN W.M. LIM L.P. Persistence of seed-based activity following segmentation of a microRNA guide strand. RNA. 2010;16:2336–2340. doi: 10.1261/rna.2296210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- CHU C.Y. RANA T.M. Potent RNAi by short RNA triggers. RNA. 2008;14:1714–1719. doi: 10.1261/rna.1161908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- COLLINGWOOD M.A. ROSE S.D. HUANG L. HILLIER C. AMARZGUIOUI M. WIIGER M.T., et al. Chemical modification patterns compatible with high potency dicer-substrate small interfering RNAs. Oligonucleotides. 2008;18:187–200. doi: 10.1089/oli.2008.0123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DANDE P. PRAKASH T.P. SIOUFI N. GAUS H. JARRES R. BERDEJA A., et al. Improving RNA interference in mammalian cells by 4′-thio-modified small interfering RNA (siRNA): effect on siRNA activity and nuclease stability when used in combination with 2′-O-alkyl modifications. J. Med. Chem. 2006;49:1624–1634. doi: 10.1021/jm050822c. [DOI] [PubMed] [Google Scholar]
- DARNIOT M. SCHILDGEN V. SCHILDGEN O. SPROAT B. KLEINES M. DITT V., et al. RNA interference in vitro and in vivo using DsiRNA targeting the nucleocapsid N mRNA of human metapneumovirus. Antiviral Res. 2012;93:364–373. doi: 10.1016/j.antiviral.2012.01.004. [DOI] [PubMed] [Google Scholar]
- DAVIS M.E. ZUCKERMAN J.E. CHOI C.H. SELIGSON D. TOLCHER A. ALABI C.A., et al. Evidence of RNAi in humans from systemically administered siRNA via targeted nanoparticles. Nature. 2010;464:1067–1070. doi: 10.1038/nature08956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DE FOUGEROLLES A. VORNLOCHER H. P. MARAGANORE J. LIEBERMAN J. Interfering with disease: a progress report on siRNA-based therapeutics. Nat Rev. Drug Discov. 2007;6:443–453. doi: 10.1038/nrd2310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DEJNEKA N.S. WAN S. BOND O.S. KORNBRUST D.J. REICH S.J. Ocular biodistribution of bevasiranib following a single intravitreal injection to rabbit eyes. Mol. Vis. 2008;14:997–1005. [PMC free article] [PubMed] [Google Scholar]
- DOHMEN C. FROHLICH T. LACHELT U. ROHL I. VORNLOCHER H.-P. HADWIGER P., et al. Defined Folate-PEG-siRNA Conjugates for Receptor-specific Gene Silencing. Mol. Ther. Nucleic Acids. 2012;1:e7. doi: 10.1038/mtna.2011.10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DORE-SAVARD L. ROUSSY G. DANSEREAU M.A. COLLINGWOOD M.A. LENNOX K.A. ROSE S.D., et al. Central delivery of Dicer-substrate siRNA: a direct application for pain research. Mol. Ther. 2008;16:1331–1339. doi: 10.1038/mt.2008.98. [DOI] [PubMed] [Google Scholar]
- DOWLER T. BERGERON D. TEDESCHI A.L. PAQUET L. FERRARI N. DAMHA M.J. Improvements in siRNA properties mediated by 2′-deoxy-2′-fluoro-beta-D-arabinonucleic acid (FANA) Nucleic Acids Res. 2006;34:1669–1675. doi: 10.1093/nar/gkl033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ELBASHIR S.M. HARBORTH J. LENDECKEL W. YALCIN A. WEBER K. TUSCHL T. Duplexes of 21-nucleotide RNAs mediate RNA interference in cultured mammalian cells. Nature. 2001;411:494–498. doi: 10.1038/35078107. [DOI] [PubMed] [Google Scholar]
- ELMEN J. THONBERG H. LJUNGBERG K. FRIEDEN M. WESTERGAARD M. XU Y., et al. Locked nucleic acid (LNA) mediated improvements in siRNA stability and functionality. Nucleic Acids Res. 2005;33:439–447. doi: 10.1093/nar/gki193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- FIRE A. XU S. MONTGOMERY M.K. KOSTAS S.A. DRIVER S.E. MELLO C.C. Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature. 1998;391:806–811. doi: 10.1038/35888. [DOI] [PubMed] [Google Scholar]
- FORD L.P. TOLOUE M.M. Delivery of RNAi mediators. Wiley Interdiscip. Rev. RNA. 2010;1:341–350. doi: 10.1002/wrna.12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- FOSTER D.J. BARROS S. DUNCAN R. SHAIKH S. CANTLEY W. DELL A., et al. Comprehensive evaluation of canonical versus Dicer-substrate siRNA in vitro and in vivo. RNA. 2012;18:557–568. doi: 10.1261/rna.031120.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- GUO P. The emerging field of RNA nanotechnology. Nat. Nanotechnol. 2010;5:833–842. doi: 10.1038/nnano.2010.231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- GUO S. HUANG F. GUO P. Construction of folate-conjugated pRNA of bacteriophage phi29 DNA packaging motor for delivery of chimeric siRNA to nasopharyngeal carcinoma cells. Gene Ther. 2006;13:814–820. doi: 10.1038/sj.gt.3302716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- HICKERSON R.P. FLORES M.A. LEAKE D. LARA M.F. CONTAG C.H. LEACHMAN S.A., et al. Use of self-delivery siRNAs to inhibit gene expression in an organotypic pachyonychia congenita model. J. Invest. Dermatol. 2011;131:1037–1044. doi: 10.1038/jid.2010.426. [DOI] [PubMed] [Google Scholar]
- HICKERSON R.P. SMITH F.J. REEVES R.E. CONTAG C.H. LEAKE D. LEACHMAN S.A., et al. Single-nucleotide-specific siRNA targeting in a dominant-negative skin model. J. Invest. Dermatol. 2008a;128:594–605. doi: 10.1038/sj.jid.5701060. [DOI] [PubMed] [Google Scholar]
- HICKERSON R.P. VLASSOV A.V. WANG Q. LEAKE D. ILVES H. GONZALEZ-GONZALEZ E., et al. Stability study of unmodified siRNA and relevance to clinical use. Oligonucleotides. 2008b;18:345–354. doi: 10.1089/oli.2008.0149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- HOERTER J.A.H. KRISHNAN V. LIONBERGER T.A. WALTER N.G. siRNA-like double-stranded RNAs are specifically protected against degradation in human cell extract. PLoS One. 2011;6:e20359. doi: 10.1371/journal.pone.0020359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- HOWARD K.A. PALUDAN S.R. BEHLKE M.A. BESENBACHER F. DELEURAN B. KJEMS J. Chitosan/siRNA nanoparticle-mediated TNF-alpha knockdown in peritoneal macrophages for anti-inflammatory treatment in a murine arthritis model. Mol. Ther. 2009;17:162–168. doi: 10.1038/mt.2008.220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- JACKSON A.L. BURCHARD J. LEAKE D. REYNOLDS A. SCHELTER J. GUO J., et al. Position-specific chemical modification of siRNAs reduces off-target transcript silencing. RNA. 2006;12:1197–1205. doi: 10.1261/rna.30706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- JACKSON A.L. LINSLEY P.S. Recognizing and avoiding siRNA off-target effects for target identification and therapeutic application. Nat. Rev. Drug Discov. 2010;9:57–67. doi: 10.1038/nrd3010. [DOI] [PubMed] [Google Scholar]
- KENSKI D.M. COOPER A.J. LI J.J. WILLINGHAM A.T. HARINGSMA H.J. YOUNG T.A., et al. Analysis of acyclic nucleoside modifications in siRNAs finds sensitivity at position 1 that is restored by 5′-terminal phosphorylation both in vitro and in vivo. Nucleic Acids Res. 2010;38:660–671. doi: 10.1093/nar/gkp913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- KENSKI D.M. WILLINGHAM A.T. HARINGSMA H.J. LI J.J. FLANAGAN W.M. In vivo activity and duration of short interfering RNAs containing a synthetic 5′-phosphate. Nucleic Acid Ther. 2012;22:90–95. doi: 10.1089/nat.2011.0333. [DOI] [PubMed] [Google Scholar]
- KHVOROVA A. REYNOLDS A. JAYASENA S.D. Functional siRNAs and miRNAs exhibit strand bias. Cell. 2003;115:209–216. doi: 10.1016/s0092-8674(03)00801-8. [DOI] [PubMed] [Google Scholar]
- KIM D.H. BEHLKE M.A. ROSE S.D. CHANG M.S. CHOI S. ROSSI J.J. Synthetic dsRNA Dicer substrates enhance RNAi potency and efficacy. Nat. Biotech. 2005;23:222–226. doi: 10.1038/nbt1051. [DOI] [PubMed] [Google Scholar]
- KIM D.H. ROSSI J.J. Strategies for silencing human disease using RNA interference. Nat. Rev. Genet. 2007;8:173–184. doi: 10.1038/nrg2006. [DOI] [PubMed] [Google Scholar]
- KLEINMAN M.E. KANEKO H. CHO W.G. DRIDI S. FOWLER B.J. BLANDFORD A.D., et al. Short-interfering RNAs induce retinal degeneration via TLR3 and IRF3. Mol. Ther. 2012;20:101–108. doi: 10.1038/mt.2011.212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- KLEINMAN M.E. YAMADA K. TAKEDA A. CHANDRASEKARAN V. NOZAKI M. BAFFI J.Z., et al. Sequence- and target-independent angiogenesis suppression by siRNA via TLR3. Nature. 2008;452:591–597. doi: 10.1038/nature06765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- KUBO T. TAKEI Y. MIHARA K. YANAGIHARA K. SEYAMA T. Amino-modified and lipid-conjugated dicer-substrate siRNA enhances RNAi efficacy. Bioconjug. Chem. 2012;23:164–173. doi: 10.1021/bc200333w. [DOI] [PubMed] [Google Scholar]
- KUBO T. ZHELEV Z. OHBA H. BAKALOVA R. Modified 27-nt dsRNAs with dramatically enhanced stability in serum and long-term RNAi activity. Oligonucleotides. 2007;17:445–464. doi: 10.1089/oli.2007.0096. [DOI] [PubMed] [Google Scholar]
- KUBO T. ZHELEV Z. OHBA H. BAKALOVA R. Chemically modified symmetric and asymmetric duplex RNAs: an enhanced stability to nuclease degradation and gene silencing effect. Biochem. Biophys. Res. Commun. 2008;365:54–61. doi: 10.1016/j.bbrc.2007.10.116. [DOI] [PubMed] [Google Scholar]
- LARA M.F. GONZALEZ-GONZALEZ E. SPEAKER T.J. HICKERSON R.P. LEAKE D. MILSTONE L.M., et al. Inhibition of CD44 gene expression in human skin models using self-delivery siRNA administered by dissolvable microneedle arrays. Hum. Gene Ther. 2012 doi: 10.1089/hum.2011.211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LAYZER J.M. MCCAFFREY A.P. TANNER A.K. HUANG Z.A.N. KAY M.A. SULLENGER B.A. In vivo activity of nuclease-resistant siRNAs. RNA. 2004;10:766–771. doi: 10.1261/rna.5239604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LEACHMAN S.A. HICKERSON R.P. SCHWARTZ M.E. BULLOUGH E.E. HUTCHERSON S.L. BOUCHER K.M., et al. First-in-human mutation-targeted siRNA phase Ib trial of an inherited skin disorder. Mol. Ther. 2010;18:442–446. doi: 10.1038/mt.2009.273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LU P.Y. WOODLE M.C. Delivering small interfering RNA for novel therapeutics. Methods Mol. Biol. 2008;437:93–107. doi: 10.1007/978-1-59745-210-6_3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LUNDBERG P. YANG H.J. JUNG S.J. BEHLKE M.A. ROSE S.D. CANTIN E.M. Protection against TNFalpha-dependent liver toxicity by intraperitoneal liposome delivered DsiRNA targeting TNFalpha in vivo. J. Control Release. 2011. http://dx.doi.org/10.1016/j.jconrel.2011.10.034. http://dx.doi.org/10.1016/j.jconrel.2011.10.034 [DOI] [PMC free article] [PubMed]
- MATRANGA C. TOMARI Y. SHIN C. BARTEL D.P. ZAMORE P. D. Passenger-strand cleavage facilitates assembly of siRNA into Ago2-containing RNAi enzyme complexes. Cell. 2005;123:607–620. doi: 10.1016/j.cell.2005.08.044. [DOI] [PubMed] [Google Scholar]
- MOSCHOS S.A. WILLIAMS A.E. LINDSAY M.A. Cell-penetrating-peptide-mediated siRNA lung delivery. Biochem. Soc. Trans. 2007;35:807–810. doi: 10.1042/BST0350807. [DOI] [PubMed] [Google Scholar]
- OCAMPO S.M. ROMERO C. AVINO A. BURGUENO J. GASSULL M.A. BERMUDEZ J., et al. Functionally enhanced siRNA targeting TNFalpha attenuates DSS-induced colitis and TLR-mediated immunostimulation in mice. Mol. Ther. 2012;20:382–390. doi: 10.1038/mt.2011.236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- PETRI S. DUECK A. LEHMANN G. PUTZ N. RUDEL S. KREMMER E., et al. Increased siRNA duplex stability correlates with reduced off-target and elevated on-target effects. RNA. 2011;17:737–749. doi: 10.1261/rna.2348111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- POECK H. BESCH R. MAIHOEFER C. RENN M. TORMO D. MORSKAYA S. S., et al. 5′-Triphosphate-siRNA: turning gene silencing and Rig-I activation against melanoma. Nat. Med. 2008;14:1256–63. doi: 10.1038/nm.1887. [DOI] [PubMed] [Google Scholar]
- REYNOLDS A. LEAKE D. BOESE Q. SCARINGE S. MARSHALL W.S. KHVOROVA A. Rational siRNA design for RNA interference. Nat. Biotechnol. 2004;22:326–330. doi: 10.1038/nbt936. [DOI] [PubMed] [Google Scholar]
- ROSE S.D. KIM D.H. AMARZGUIOUI M. HEIDEL J.D. COLLINGWOOD M.A. DAVIS M.E., et al. Functional polarity is introduced by Dicer processing of short substrate RNAs. Nucleic Acids Res. 2005;33:4140–4156. doi: 10.1093/nar/gki732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- SANO M. SIERANT M. MIYAGISHI M. NAKANISHI M. TAKAGI Y. SUTOU S. Effect of asymmetric terminal structures of short RNA duplexes on the RNA interference activity and strand selection. Nucleic Acids Res. 2008;36:5812–5821. doi: 10.1093/nar/gkn584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- SANTEL A. ALEKU M. KEIL O. ENDRUSCHAT J. ESCHE V. DURIEUX B., et al. RNA interference in the mouse vascular endothelium by systemic administration of siRNA-lipoplexes for cancer therapy. Gene Ther. 2006a;13:1360–1370. doi: 10.1038/sj.gt.3302778. [DOI] [PubMed] [Google Scholar]
- SANTEL A. ALEKU M. KEIL O. ENDRUSCHAT J. ESCHE V. FISCH G., et al. A novel siRNA-lipoplex technology for RNA interference in the mouse vascular endothelium. Gene Ther. 2006b;13:1222–1234. doi: 10.1038/sj.gt.3302777. [DOI] [PubMed] [Google Scholar]
- SANTEL A. ALEKU M. RODER N. MOPERT K. DURIEUX B. JANKE O., et al. Atu027 prevents pulmonary metastasis in experimental and spontaneous mouse metastasis models. Clin. Cancer Res. 2010;16:5469–5480. doi: 10.1158/1078-0432.CCR-10-1994. [DOI] [PubMed] [Google Scholar]
- SCHWARZ D.S. HUTVAGNER G. DU T. XU Z. ARONIN N. ZAMORE P. D. Asymmetry in the assembly of the RNAi enzyme complex. Cell. 2003;115:199–208. doi: 10.1016/s0092-8674(03)00759-1. [DOI] [PubMed] [Google Scholar]
- SEMPLE S.C. AKINC A. CHEN J. SANDHU A.P. MUI B.L. CHO C.K., et al. Rational design of cationic lipids for siRNA delivery. Nat. Biotechnol. 2010;28:172–176. doi: 10.1038/nbt.1602. [DOI] [PubMed] [Google Scholar]
- SIERANT M. KAZMIERCZAK-BARANSKA J. PADUSZYNSKA A. SOBCZAK M. PIETKIEWICZ A. NAWROT B. Longer 19-base pair short interfering RNA duplexes rather than shorter duplexes trigger RNA interference. Oligonucleotides. 2010;20:199–206. doi: 10.1089/oli.2010.0239. [DOI] [PubMed] [Google Scholar]
- SIOLAS D. LERNER C. BURCHARD J. GE W. LINSLEY P. S. PADDISON P. J., et al. Synthetic shRNAs as potent RNAi triggers. Nat. Biotechnol. 2005;23:227–31. doi: 10.1038/nbt1052. [DOI] [PubMed] [Google Scholar]
- SOUTSCHEK J. AKINC A. BRAMLAGE B. CHARISSE K. CONSTIEN R. DONOGHUE M., et al. Therapeutic silencing of an endogenous gene by systemic administration of modified siRNAs. Nature. 2004;432:173–178. doi: 10.1038/nature03121. [DOI] [PubMed] [Google Scholar]
- STRUMBERG D. SCHULTHEIS B. TRAUGOTT U. VANK C. SANTEL A. KEIL O., et al. Phase I clinical development of Atu027, a siRNA formulation targeting PKN3 in patients with advanced solid tumors. Int. J. Clin. Pharmacol. Ther. 2012;50:76–8. doi: 10.5414/cpp50076. [DOI] [PubMed] [Google Scholar]
- SUN X. ROGOFF H.A. LI C.J. Asymmetric RNA duplexes mediate RNA interference in mammalian cells. Nat. Biotechnol. 2008;26:1379–1782. doi: 10.1038/nbt.1512. [DOI] [PubMed] [Google Scholar]
- UI-TEI K. NAITO Y. ZENNO S. NISHI K. YAMATO K. TAKAHASHI F., et al. Functional dissection of siRNA sequence by systematic DNA substitution: modified siRNA with a DNA seed arm is a powerful tool for mammalian gene silencing with significantly reduced off-target effect. Nucleic Acids Res. 2008;36:2136–2151. doi: 10.1093/nar/gkn042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- VAISH N. CHEN F. SETH S. FOSNAUGH K. LIU Y. ADAMI R., et al. Improved specificity of gene silencing by siRNAs containing unlocked nucleobase analogs. Nucleic Acids Res. 2011;39:1823–1832. doi: 10.1093/nar/gkq961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- WEITZER S. MARTINEZ J. The human RNA kinase hClp1 is active on 3′ transfer RNA exons and short interfering RNAs. Nature. 2007;447:222–226. doi: 10.1038/nature05777. [DOI] [PubMed] [Google Scholar]
- WOLFRUM C. SHI S. JAYAPRAKASH K.N. JAYARAMAN M. WANG G. PANDEY R.K., et al. Mechanisms and optimization of in vivo delivery of lipophilic siRNAs. Nat. Biotechnol. 2007;25:1149–1157. doi: 10.1038/nbt1339. [DOI] [PubMed] [Google Scholar]