Abstract
A 30-nucleotide DNA aptamer (5′-AGGAAGGCTTTAGGTCTGAGATCTCGGAAT-3′, denoted PF1) selected for high affinity to human immunodeficiency virus reverse transcriptase (HIV RT) using a primer-free SELEX (systematic evolution of ligands by exponential enrichment) method was characterized to determine features promoting tight binding. PF1's equilibrium dissociation constant for RT was ∼80 nM, over 10-fold lower than a random 30-mer. Changing the 2 terminal diguanosine repeats (underlined above) to diadenosine or dithymidine modestly decreased binding. Any changes to the 2 central diguanosines dramatically decreased binding. Binding was highly sensitive to length, with any truncations that deleted part of the 4 diguanosine motifs resulting in a 6-fold or more decrease in affinity. Even a construct with all the diguanosine motifs but lacking the 5′ terminal A and 3 nucleotides at the 3′ end showed ∼3-fold binding decrease. Changes to the nucleotides between the diguanosines, even those that did not alter PF1's low secondary structure (free energy of folding ΔG=−0.61 kcal/mol), dramatically decreased binding, suggesting sequence specificity. Despite the diguanosine motifs, circular dichroism (CD) spectra indicated that PF1 did not form a G-quartet. PF1 inhibited HIV RT synthesis with a half-maximal inhibitory value (IC50) of ∼60 nM. Larger, more structured RT DNA aptamers based on the HIV polypurine tract and those that formed G-quartets (denoted S4 and R1T) were more potent inhibitors, with IC50 values of ∼4 and ∼1 nM, respectively. An RNA pseudoknot aptamer (denoted 1.1) showed an IC50 near 4 nM. Competition binding assays with PF1 and several previously characterized RT aptamers indicated that they all bound at or near the primer–template pocket. These other more structured and typically larger aptamers bound more tightly than PF1 to RT based on filter binding assays. Results indicate that PF1 represents a new class of RT aptamers that are relatively small and have very low secondary structure, attributes that could be advantageous for further development as HIV inhibitors.
Introduction
Since its invention by 3 separate groups in the early 1990s (JOYCE, 1989; Ellington and Szostak, 1990; Tuerk and Gold, 1990), systematic evolution of ligands by exponential enrichment (SELEX) has been used for selecting small nucleic acids (RNA or DNA) that bind to specific targets with high affinity. SELEX starts with a large, random pool of nucleic acids incubated with a limiting amount of target. These specific targets are typically proteins, but they may also include nucleic acids, cultured cells, viruses, and other small molecules (Lang et al., 1989; Wolinsky et al., 1996; Wang et al., 2000; Hicke et al., 2001; Zhu et al., 2003; Wen and Gray, 2004; Zhang et al., 2004; Cerchia et al., 2005; Hicke et al., 2006; Missailidis and Perkins, 2007; Tang et al., 2007; Pieve et al., 2009). Nucleic acids that bind to the target with higher affinity will preferentially associate to form a complex. This complex can be isolated by several methods including gel shift, nitrocellulose filter binding, and capillary electrophoresis (Mosing et al., 2005). The selected pool is expanded by polymerase chain reaction amplification, and the new, enriched pool of nucleic acids is subjected to subsequent rounds of target binding. After several rounds nucleic acids with high affinity for the target (aptamers) are isolated. Aptamers have many potential uses in diagnostic and therapeutic applications, such as replacements of antibodies in biochemical assays (e.g., enzyme-linked immunosorbent assay), utilization as biosensors, as a tool for studying the molecular biology of virus replication, and in the development of antiviral drugs (GOLD, 1995; Brody and Gold, 2000; Joshi et al., 2003; Zhang et al., 2004; Nimjee et al., 2005; DEISINGH, 2006; Porschewski et al., 2006; JAMES, 2007; Mok and Li, 2008). Essentially, any technique that involves sensitive and stringent recognition of protein or nucleic acids could benefit from aptamer-based approaches.
Most aptamers are screened through standard SELEX procedures, which include a starting pool of random nucleic acids. These random oligonucleotides contain fixed flanking regions of known sequence at the 5′ and 3′ termini. After selection and modifications to enhance binding and reduce size, the selected aptamers often retain sequences from the fixed flanking regions, which are required to achieve tight binding. This approach poses a disadvantage, where the fixed sequences create biases on the selection by interacting with nucleotides in the random region of the selected oligonucleotides.
Newly developed methods that overcome this drawback have emerged. These include “primer-free” SELEX, where selection occurs in the absence of any fixed, flanking nucleotides (Wen and Gray, 2004; Pan and Clawson, 2010; Pan et al., 2008), and “minimal primer” SELEX, in which the fixed sequences are reduced to only a few nucleotides (Vater and Klussmann, 2003; Jarosch et al., 2006). A notable drawback of primer-free approaches is that they are more time consuming and include several additional steps. There are also additional steps at which bias may be introduced (Lai and Destefano, 2011).
Our lab has introduced a novel primer-free SELEX-based approach for selecting single-stranded DNA sequences (Lai and Destefano, 2011). Sequences that bound ∼10-fold tighter than the starting random sequences to human immunodeficiency virus reverse transcriptase (HIV RT) in gel-shift assays were isolated. All of the recovered 30-nucleotide aptamers had essentially the same sequence and were characterized by the regular arrangement of 4 separate guanosine doublets in the sequences (5′-AGGAAGGCTTTAGGTCTGAGATCTCGGAAT-3′, denoted PF1 in this report) and a negligible predicted secondary structure (free energy of folding ΔG=−0.61 kcal/mol, predicted using DNA mfold program; ZUKER, 2003). The repeated guanosine doublets were also observed in a 15-nucleotide-long aptamer that binds tightly to human thrombin (denoted as thrombin aptamer, or TBA) (Bock et al., 1992; Macaya et al., 1993). The TBA (5′-GGTTGGTGTGGTTGG-3′) folds into an anti-parallel G-quadruplex, with 2 G-quadruplex layers connected by a TGT and 2 TT loops. G-quadruplexes are formed by 4 guanine residues associated with one another by Hoogsteen bonds in a square planar configuration. Most monovalent cations stabilize G-quadruplex formation by the cation binding specifically to guanine O6 carbonyl groups between the planes of the G-quartets. The stability is highly dependent on the cation size, with K+ or Sr2+ having the most favorable ionic radius at 1.3–1.4 Å, while Na+ and Li+ destabilize G-quadruplex formation (Tohl and Eimer, 1997; SIMONSSON, 2001). These structures can assume both parallel and anti-parallel configurations and can form between 2 or more separate nucleic acid strands (intermolecular), or within the same strand (intramolecular). G-quadruplexes have been implicated in several cellular processes including telomerase regulation and gene expression (Hershman et al., 2008; Gonzalez and Hurley, 2010).
Several other aptamers to various HIV proteins area also predicted to form G-quadruplexes. These include aptamers that block HIV envelope glycoprotein gp120 by binding to the gp120 cationic V3 loop region (Hotoda et al., 1998). Various classes of G-quadruplex aptamers were found to inhibit RNase H activity (Andreola et al., 2001; de Soultrait et al., 2002), and integrase activity (Kelley et al., 2011). In previous studies, Schneider et al. and Michalowski et al. characterized a class of HIV RT aptamers with a bimodular structure consisting of a 3-tier G-quadruplex and a stem-loop helical element (Schneider et al., 1995; Michalowski et al., 2008). In addition to HIV proteins, G-quadruplex aptamers to several other viral and non-viral proteins have been selected, suggesting that G-quadruplexes are a common motif for high affinity protein–nucleic acid interactions (Sissi et al., 2011).
In this report, we characterized the interaction between HIV RT and the PF1 single-stranded DNA derived from primer-free SELEX as denoted above. The 2 sets of central diguanosines played a more pivotal role in tight binding than did the flanking ones. Altering spacing between the diguanosines or shortening PF1 by as few as 3 nucleotides significantly reduced binding to RT. Modifications that increased the level of secondary structure had a negative impact on binding. However, replacing the nucleotides between the diguanosines with runs of Ts, though retaining the low secondary structure, strongly inhibited binding, indicating a role for the specific non-diguanosine sequences of PF1. Comparisons with of other classes of HIV RT DNA and RNA aptamers on RT inhibition and binding were also made. These included (1) a 38-nucleotide primer–template flipback aptamer (termed 38 NT SELEX in this report) that mimics the polypurine tract at the 3′ end (DeStefano and Cristofaro, 2006; DeStefano and Nair, 2008); (2) 35 (S4) and 41 (R1T) nucleotide G-quadruplex aptamers (Michalowski et al., 2008); (3) a large 81-nucleotide DNA aptamer (RT1) (Schneider et al., 1995; Kissel et al., 2007); and (4) a 24-nucleotide RNA pseudoknot type aptamer (1.1) (Tuerk et al., 1992). All of these highly structured aptamers bound RT more tightly than PF1 and were in general better inhibitors of RT activity, although PF1 was also inhibitory. Interestingly, based on circular dichroism (CD) analysis, PF1 did not form G-quadruplexes.
Materials and Methods
Materials
Human immunodeficiency virus reverse transcriptase (HIV RT) was obtained from Worthington Biochemical Corporation. Taq polymerase, restriction enzymes, and T4 polynucleotide kinase (PNK) were obtained from New England Biolabs. Deoxyribonucleotide triphosphates (dNTPs) and RNase (DNase free) were from Roche Applied Sciences. RNase inhibitor (RNasin) was from Promega. Klenow DNA polymerase, dideoxyguanosine triphosphate (ddGTP), and DNase I (RNase free) were from United States Biochemical Corporation. Radiolabeled compounds were obtained from Perkin Elmer. Nitrocellulose filter disks (Protran BA 85, 0.45-μm pore size and 25-mm diameter) were from Whatman. Sephadex G-25 spin columns were from Harvard Apparatus. All oligonucleotides were commercially synthesized from Integrated DNA Technologies. Cell culture media (RPMI) was from Lonza and fetal bovine serum (FBS) was from Aleken Biologicals. All other chemicals were from Sigma-Aldrich Co., Thermo Fisher Scientific, Inc., or VWR Scientific, Inc.
End-labeling of oligonucleotides with T4 PNK
Reactions for labeling various oligos were done in a 50-μL volume containing 25 pmol of the oligo, 70 mM Tris-HCl (pH=7.6), 10 mM MgCl2, 5 mM dithiothreitol (DTT), 5 μL of γ-32P ATP (3,000 Ci/mmol, 10 μCi/μL), and 2 μL (20 units) of T4 polynucleotide kinase. The reaction mixture was incubated for 30 minutes at 37°C, and then the PNK was heat inactivated for 20 minutes at 70°C according to manufacturer's recommendation. The material was then run through a Sephadex G-25 spin column.
Determination of equilibrium dissociation constants with 5′ end-labeled substrates by gel-shift analysis
PF1 (2 nM) from the primer-free SELEX or other designed sequences (Table 1) were 5′ end labeled with γ-32P ATP and mixed with various amounts of HIV RT in 10 μL of buffer containing 50 mM Tris-HCl (pH=8), 1 mM DTT, 80 mM KCl, and 6 mM MgCl2, for 10 minutes at room temperature. Two microliters of 6×native gel loading buffer [40% sucrose, 0.25% (w/v) bromphenol blue, 0.25% (w/v) xylene cyanol] was added and the samples were run on a 6% native polyacrylamide gel (29:1 acrylamide:bisacrylamide) at ∼100 V until the blue dye marker was ∼2–3 inches below the wells (Sambrook and Russell, 2001). The amount of shifted vs. non-shifted material was quantified using a Fujifilm FLA-5100 or FLA-7000 phosphoimager. Values for equilibrium dissociation constants (Kd) were determined by plotting the concentration of shifted product (nM) versus the concentration of HIV RT and fitting the data by nonlinear least square fit to the quadratic equation: [ED]=0.5([E]t+[D]t+Kd)−0.5[([E]t+[D]t+Kd)2 – 4[E]t[D]t]1/2, where [E]t is the total enzyme concentration and [D]t is the total primer–template concentration (Hsieh et al., 1993). In cases where the affinity of HIV RT for the nucleic acid was low (Kd≥500 nM), a more precise Kd value was not determined.
Table 1.
Affinity for Human Immunodeficiency Virus Reverse Transcriptase of PF1 Aptamer and Various Modifications
| Name | Sequence | Kd (nM) | ΔG (kcal/mol) |
|---|---|---|---|
| 5′-N30-3′ | 5′-NNNNNNNNNNNNNNNNNNNNNNNNNNNNN-3′ | ∼1000 | |
| PF1 | 5′-AGGAAGGCTTTAGGTCTGAGATCTCGGAAT-3′ | 82±7 | −0.61 |
| Diguanosine modifications of PF1 (Changes underlined) | |||
| PF1-GG>CC | 5′-ACCAACCCTTTACCTCTGAGATCTCCCAAT-3′ | >500 | >0 |
| PF1-GG>AA | 5′-AAAAAAACTTTAAATCTGAGATCTCAAAAT-3′ | >500 | −0.02 |
| PF1-terminal GG>CC | 5′-ACCAAGGCTTTAGGTCTGAGATCTCCCAAT-3′ | 140±10 | >0 |
| PF1-GCmid | 5′-AGGAACCCTTTACCTCTGAGATCTCGGAAT-3′ | ∼500 | −0.61 |
| PF1C6-7 | 5′-AGGAACCCTTTAGGTCTGAGATCTCGGAAT-3′ | ∼500 | −1.07 |
| PF1C13-14 | 5′-AGGAAGGCTTTACCTCTGAGATCTCGGAAT-3′ | >500 | −1.11 |
| PF1-C6 | 5′-AGGAACGCTTTAGGTCTGAGATCTCGGAAT-3′ | 420±70 | −0.61 |
| PF1-C14 | 5′-AGGAAGGCTTTAGCTCTGAGATCTCGGAAT-3′ | ∼500 | −0.73 |
| PF1-3G | 5′-AGGAAGGCTTTAGGGTCTGAGATCTCGGAAT-3′ | 310±90 | −0.61 |
| PF1-3G2 | 5′-AGGAAGGCTTTAGGGTCTGAGATCTCGGGAAT-3′ | 260±73 | −0.13 |
| PF1-4G | 5′-AGGAAGGCTTTAGGGGTCTGAGATCTCGGAAT-3′ | ∼500 | −0.61 |
| Truncated PF1 constructs (diguanosines and changes underlined) | |||
| PF1-25nt3′ cut | 5′-AGGAAGGCTTTAGGTCTGAGATCTC-3′ | ∼500 | >0 |
| PF1-25nt5′ cut | 5′-GGCTTTAGGTCTGAGATCTCGGAAT-3′ | >750 | −0.61 |
| PF1-20nt3′ cut | 5′-AGGAAGGCTTTAGGTCTGAG-3′ | >750 | >0 |
| PF1-20nt5′ cut | 5′-TAGGTCTGAGATCTCGGAAT-3′ | >750 | −0.61 |
| PF1-15nt3′ cut | 5′-AGGAAGGCTTTAGGT-3′ | >750 | >0 |
| PF1-15nt5′ cut | 5′-CTGAGATCTCGGAAT-3′ | >750 | −0.09 |
| PF1-27nt5′ cut | 5′-AAGGCTTTAGGTCTGAGATCTCGGAAT-3′ | ∼500 | −0.61 |
| PF1-26ntG | 5′-GGAAGGCTTTAGGTCTGAGATCTCGG-3′ | 233±29 | >0 |
| PF1-26ntC | 5′-CCAAGGCTTTAGGTCTGAGATCTCCC-3′ | ∼500 | >0 |
| PF1-26ntT | 5′-TTAAGGCTTTAGGTCTGAGATCTCTT-3′ | 131±24 | >0 |
| PF1-Δ (8–12) | 5′-AGGAAGGGGTCTGAGATCTCGGAAT-3′ | >750 | −0.61 |
| PF1-Δ (16–25) | 5′-AGGAAGGCTTTAGGTGGAAT-3′ | >750 | >0 |
| PF1-Δ (8–12/16–25) | 5′-AGGAAGGGGTGGAAT-3′ | >750 | >0 |
| PF1-15ntmidG | 5′-AAGGCTTTAGGTCTG-3′ | >750 | >0 |
| PF1-18ntTlinkG | 5′-AGGTTGGTTGGTTGGAAT-3′ | >750 | >0 |
| TBA (thrombin aptamer) | 5′-GGTTGGTGTGGTTGG-3′ | >750 | >0 |
| PF1-RNA | 5′-AGGAAGGCUUUAGGUCUGAGAUCUCGGAAU-3′ | >750 | −4.40 |
| Structural controls (diguanosines underlined) | |||
| PF1A | 5′-AGGAAGGCAAAAGGAAAAAAAAAAAGGAAA-3′ | >750 | 0 |
| PF1Flip | 5′-AGGAAGGCTTTAGGTTTCCTAAAGCCTTCC-3′ | 667±144 | −12.06 |
| PF1Loop | 5′-AGGAAGGCTTTAGGTCCGAGATCTCGGAAT-3′ | 558±3 | −3.38 |
| PF1N | 5′-NGGNNGGNNNNNGGNNNNNNNNNNNGGNNN-3′ | ∼500 | |
| PF1T | 5′-TGGTTGGTTTTTGGTTTTTTTTTTTGGTTT-3′ | 448±25 | −2.67 |
Repeated diguanosines are underlined in PF1, while changes to PF1 are underlined in the diguanosine-modified oligos. Both diguanosines and changes are underlined in the truncated oligos and diguanosines only in the structural controls.
Equilibrium dissociation constants (Kd) were determined using a gel-shift assay as described in Material and Methods. Results are an average of 3 experiments±standard deviation where indicated. Other results (denoted as ∼or>) were estimated based on at least 2 independent experiments.
Free energy of folding (ΔG) values were estimated using mfold (see Methods). If more than one structure was predicted, only the one with the lowest free energy is listed;>0 indicates a predicted positive value for ΔG.
Determination of Kd with 5′ end-labeled substrates by nitrocellulose filter binding
Experiments were conducted by mixing 5′ end-labeled aptamers (1 nM final concentration) with various amounts of HIV RT in 10 μL of buffer containing 50 mM Tris-HCl (pH 8.0), 80 mM KCl, 6 mM MgCl2, and 1 mM DTT. Enzyme amounts that flanked the approximate Kd value (estimated from initial experiments) were used. After 5 minutes at room temperature, the sample was vacuum filtered through a 25-mm nitrocellulose filter disk presoaked in reaction buffer. Filters were washed with 1 mL of buffer containing 25 mM Tris-HCl (pH 7.5) and 10 mM KCl at a rate of ∼200 μL per second, air dried, and the amount of bound material was determined by liquid scintillation. Kd determinations were obtained by fitting the data to a one site saturation equation in SigmaPlot. The equation is: Y=[Bmax(x)]/(Kd+x), –where Y is the total or relative amount of bound substrate (aptamer), x is the concentration of enzyme, and Bmax is the maximum amount of substrate that can be bound to the filter with saturating enzyme. In the approach used, Bmax was determined automatically by the program. This simple approach for calculating Kd works best when the concentration of substrate used is very low. This approach simplifies calculations by assuming that Bmax accounts for all the substrate in the reaction that can bind to the enzyme. This is not always the case, as interactions between various ligands on nitrocellulose filter can be complex. Regardless, the method provided a good way to compare a variety of closely related ligands.
Competition binding assay and determination of Kd using this assay
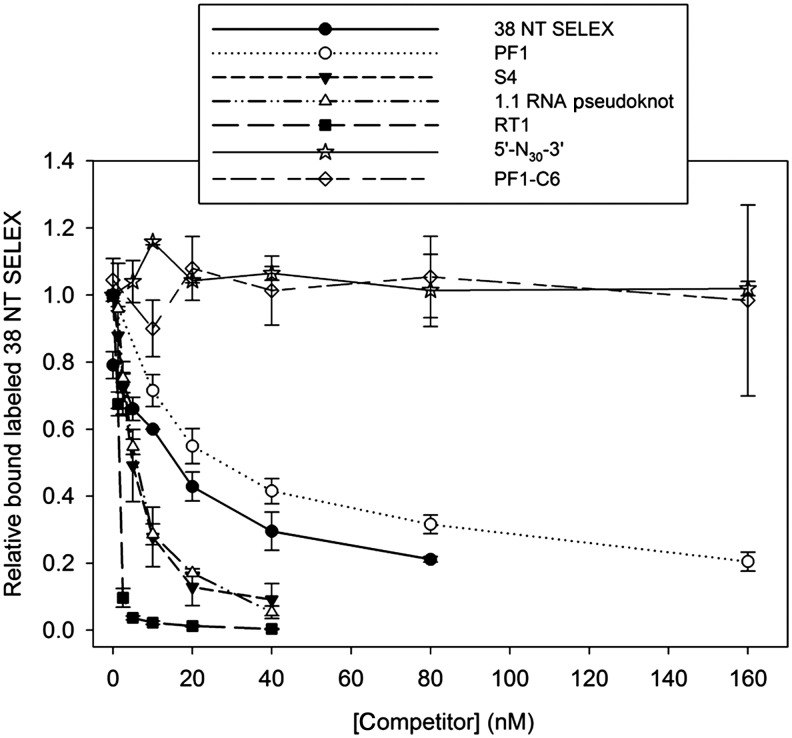
Experiments were conducted by mixing 5′ end-labeled 38 NT SELEX (10 nM final concentration) and various amounts of other aptamers (indicated in Fig. 3) in 8 μL of buffer. Two microliters of HIV RT (3 nM final concentration) were added and the material was incubated at room temperature for 1.5 hours. Final concentrations of buffer components were 50 mM Tris-HCl (pH 8.0), 80 mM KCl, 6 mM MgCl2, and 1 mM DTT. After incubation the sample was filtered through nitrocellulose and processed as described above. A graph with the amount of “hot” radiolabeled 38 NT SELEX bound to the filters at a given aptamer concentration, relative to the amount in the absence of added cold aptamer versus the concentration of cold aptamer added was constructed (Fig. 3). Plotted points are average values from 2–3 experiments, with error bars representing standard deviations. For some aptamers Kd values were calculated from the competition assay using the condition with 10 nM hot 38 NT SELEX and 10 nM cold competitor aptamer. Calculations were as described in Creighton et al., 1992.
FIG. 3.
Competition filter binding experiment. Unlabeled (cold) aptamers (5′-N30-3′, PF1, PF1-C6, 38 NT SELEX, S4, R1T, or 1.1 RNA pseudoknot, as indicated) were used as competitors with radiolabeled (hot) 38 NT SELEX (10 nM). Increasing amounts of cold aptamer (x-axis) were added to incubations with the hot aptamer and 3 nM human immunodeficiency virus reverse transcriptase (HIV RT) and incubated at room temperature for 1.5 hours. Samples were then filtered over nitrocellulose and counted by liquid scintillation. The y-axis shows the amount of hot 38 NT SELEX bound to the filters at a given aptamer concentration, relative to the amount in the absence of added cold aptamer. Plotted points are average values from 2–3 experiments, with error bars representing standard deviations.
Preparation of substrate for RT inhibition assays
Sixty picomoles of 50-nucleotide template (5′-TTGTAATACGACTCACTATAGGGCGAATTCGAGCTCGGTACCCGGGGATC-3′) and 50 pmol of 33 nucleotide 5′ 32P end-labeled primer (5′- TTCCCCGGGTACCCGAGCTCGAAT TCGCCCTATAG-3′) were mixed in 20 μL of buffer containing 50 mM Tris-HCl (pH=8), 1 mM DTT, and 80 mM KCl. The mixture was heated to 80°C for 2 minutes, and then cooled at a rate of 1°C per minute to 30°C. This material was used directly in the assays.
Preparation of ddG-terminated 38 NT SELEX substrate
Fifty picomoles of 38 NT SELEX flipback DNA was incubated with 2 units of Klenow DNA polymerase in 50 μL of buffer containing 50 mM Tris-HCl (pH=8), 1 mM DTT, 50 mM KCl, 6 mM MgCl2, and 50 μM ddGTP for 30 minutes at 37°C. The material was extracted with phenol-chloroform and precipitated with ethanol. The precipitated material was run through a Sephadex G-25 spin column to remove any remaining unused ddGTP. The recovered material was not extendable by HIV RT in the presence of dNTPs (determined by 5′ end-labeling a portion of the material with γ-32P ATP), indicating that it was 3′ terminated with ddG.
RT inhibition assay
Reactions contained substrate (1:1.2 primer:template; final concentration in reactions was 50 nM in 5′ 32P end-labeled primer) in 30 μL of buffer containing 50 mM Tris-HCl (pH=8), 1 mM DTT, 80 mM KCl, 6 mM MgCl2, and 0.1 μg/μL bovine serum albumin. Various amounts of aptamer inhibitors were used in the assays: 0.1, 0.5, 1.0, or 5.0 nM for 38 NT SELEX (terminated with ddG), R1T, S4, RT1, and 1.1 RNA pseudoknot; 25, 50, 100, and 150 nM for PF1; and higher amounts for PF1 derivatives that bound with lower affinity. Five microliters of a supplement containing 800 μM dNTPs (100 μM final) in the above buffer were added to the reactions. The mixture was placed at 37°C for 2 minutes. Primer extension was initiated by adding 5 μL of HIV RT (0.25 nM final concentration in reactions). Five-microliter aliquots were removed at 2, 5, 10, 15, and 20 minutes and added to 5 μL of 2× formamide gel-loading buffer (90% formamide, 20 mM EDTA (pH=8), 0.25% xylene cyanol, 0.25% bromphenol blue). Samples were run on a 10% denaturing polyacrylamide gel (19:1 acrylamide:bisacrylamide, 7 M urea), dried, and then quantified using a phosphoimager as described above. A graph of the number of phosphoimager counts [photo-stimulated luminescence radiation units] versus time was plotted. Experiments were repeated at least once and typically 3 or more times. The RT inhibition was achieved by a decrease in the concentration of available enzyme, which was sequestered by the aptamer inhibitor. The half-maximal inhibitory values (IC50) were calculated from the relationship between the aptamer concentration and percent of inhibition achieved at the particular inhibitor concentration. The concentrations used for measurements were as stated above, and the 10-minute time point was used. A 4-parameter logistic equation was used for curve-fitting with SigmaPlot 10.0 software to obtain IC50 for the various aptamers: Y=min+(max–min) / [1+10(logIC50–x) Hillslope], where Y is the percent inhibition at a certain inhibitor concentration and x is the log of the inhibitor concentration. Error terms for reported IC50 values were standard deviations among triplicate assays.`
Assay for aptamer stability in cell culture media
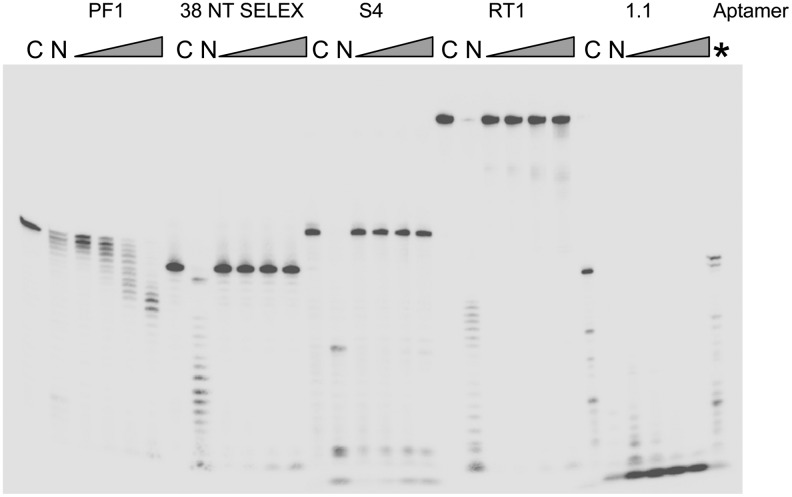
Aptamers (25 nM final concentration, 5′ end-labeled with 32P) were incubated in 200 μL of RPMI cell culture media containing 10% FBS at 37°C. Twenty-microliter aliquots were removed at 0 (control), 15, 30, 60, or 120 minutes and added to 20 μL of 2×formamide gel loading buffer. A nuclease digestion control was performed by adding 5 units of DNase I for DNA aptamers or 0.5 μg of RNase for RNA aptamer to 20 μL of aptamer containing media and incubating for 30 minutes before the addition of gel-loading buffer. For the 1.1 RNA pseudoknot aptamer, a 20-μL 15-minute incubation that included 20 units of RNasin was also performed. The samples were electrophoreses on a 12% denaturing gel as described above.
CD spectroscopy analysis
The DNA aptamer samples (see Tables 1 and 2) were heated to 90°C for 2 minutes then cooled at a rate of 2°C per minute to 30°C. The aptamer concentration was adjusted to 4 μM in potassium or sodium buffers [potassium buffer final concentration: 50 mM Tris-HCl (pH=8), 80 mM KCl, 6 mM MgCl2, and 1 mM DTT; sodium buffer final concentration: 50 mM Tris-HCl (pH=8), 80 mM NaCl, 6 mM MgCl2, and 1 mM DTT]. Near-ultraviolet CD spectra were acquired at 25°C using a Jasco J-810 spectropolarimeter at 1.0-nm intervals between 200 nm and 300 nm in a 1.0-mm quartz cuvette, at a scan speed of 50 nm per minute, response time at 8 seconds, and scan sensitivity at 100 mdegrees.
Table 2.
Equilibrium Dissociation Constant Determinations by Filter Binding and Competition Assay
| Name | Sequence | Kd (nM) |
|---|---|---|
| PF1 | 5′-AGGAAGGCTTTAGGTCTGAGATCTCGGAAT-3′ | 125±55 |
| (9.7±1.6) | ||
| Primer-template aptamer | ||
| 38 NT SELEX | 5′-TAATACCCCCCCTTCGGTGCAAAGCACCGAAGGGGGGG-3′ | 6.5±1.1 |
| G-quadruplex aptamers | ||
| R1T | 5′-CGCCTGATTAGCGATACTCAGGCGTTGGGTGGGTGGGTGGG-3′ | 1.1±0.2 |
| (0.4±0.1) | ||
| S4 | 5′-CGCCTGACCCTTCAGGCGTTGGGTGGGTGGGTGGG-3′ | 1.0±0.6 |
| (1.8±0.3) | ||
| Large DNA aptamer | ||
| RT1 | 5′-ATCCGCCTGATTAGCGATACTCAGAAGGATAAACTGTCCAGAACTTGGATATATACACTTGAGCAAAATCACCTGCAGGGG-3′ | 0.7±0.1 |
| RNA pseudoknot aptamer | ||
| 1.1 | 5′-UCCGUUUUCAGUCGGGAAAAACUG-3′ | 2.6±0.6 |
| (2.1±0.4) | ||
Names are as described in text. Other than PF1, the names designated are from the various manuscripts where the aptamers were reported are used.
Underlined nucleotides indicate the polypurine tract-like sequence in the primer–template aptamer and the region that forms the G-quadruplex in the G-quadruplex aptamers.
Secondary structure and ΔG determinations
All secondary structure and ΔG determinations were done using the mfold server (ZUKER, 2003). The conditions used were 37°C, 80 mM Na+ and 6 mM Mg2+. If more than one structure was predicted, only the one with the lowest ΔG is shown.
Results
The central diguanosine doublets are critical for tight binding of PF1 to HIV RT
In the previous study, Kd values were determined for the PF1 aptamer derived from the primer-free SELEX protocol (Lai and Destefano, 2011). Gel-shift analysis (Fig. 1) was used for these determinations as described in Materials and Methods. In the current experiments, PF1 had a Kd value of 82±7 nM (Table 1). In contrast, the random sequence material used to start the SELEX procedure (5′-N30-3′) bound with much lower affinity (Kd>1 μM).
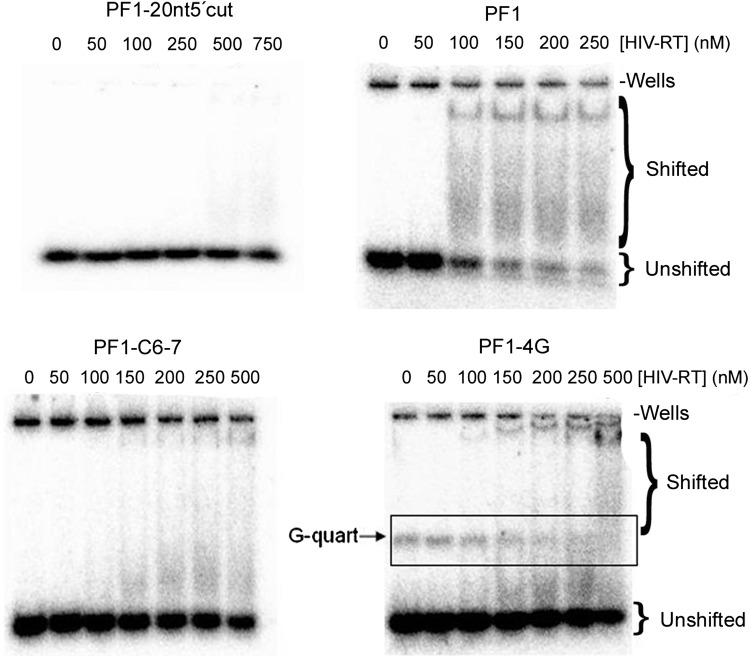
FIG. 1.
Representative gel-shift analysis of the PF1 aptamer and various derivatives. Increasing amounts of HIV RT (as indicated above wells; note different amounts with different constructs) were mixed with 5′ 32P end-labeled PF1 or other modified constructs (see Table 1). Samples were run on a native 6% polyacrylamide gel. Positions of shifted and unshifted material are indicated. For PF1-4G, G-quartets (G-quart, boxed lanes) were observed migrating above the unshifted DNA. A small portion of the material remained in the gel wells with some aptamers, but this occurred even when no RT was added, indicating it was not dependent on the added protein. In general, PF1 and the other derivatives tested did not form a single discrete band in gel-shift assays. A concentrated shifted band was observed just below the wells and a diffuse area of shifted material appeared underneath the band. This probably resulted from some complexes dissociating during the electrophoresis process.
PF1 had a striking arrangement of 4 diguanosine repeats. To test the role of these repeats in binding to HIV RT, several variations of this motif were made. Table 1 summarizes the modifications and lists the determined Kd values for binding to HIV RT. Changing the 4 guanosine doublets to cytidine (PF1-GG>CC) or adenosine (PF1-GG>AA) increased the Kd values to>500 nM, nearly equal to the starting material (5′-N30-3′). This demonstrated the importance of the diguanosine repeats for tight binding to RT. The effects of minor changes in the diguanosine runs were then tested. When the guanosine doublets at 5′ and 3′ termini were changed to dicytidine (PF1-terminal GG>CC), there was a small increase in the Kd (144±10 nM). In contrast, changing the 2 central diguanosines to cytidines (PF1-GCmid) led to a dramatic increase in the Kd values to near the value for 5′-N30-3′. Changing even one of the two central diguanosines to C (PF1C6-7 and PF1C13-14), or even replacing one G nucleotide with a C in one of the central doublets (PF1-C6 and PF1-C14) resulted in a significant loss in binding affinity. These results indicate that the 2 central G doublets are pivotal for tight binding to RT, while the terminal doublets play lesser roles. Interestingly, adding 1 or 2 extra guanosine(s) to the central doublet closer to the 3′ end also increased the Kd by 2- to 5-fold [PF1-3G, PF1-3G2 (extra G added to 3′ central and 3′ terminal diguanosines) and PF1-4G (2 Gs added to 3′ central diguanosine)]. The PF1-4G sequence showed G-quartet formation in gel-shift assay as was evidenced from a portion of the construct shifting upward on the gel (Fig. 1). This was presumably due to the formation of interstrand G-quartets. Finally, a construct that contained the 2 central diguanosine motifs but was truncated at the 5′ and 3′ ends such that the final length was only 15 nucleotides (PF1-15ntmidG) showed low affinity for RT. Overall the results highlight the importance of the central diguanosines in the context of the flanking sequences while showing that the central motif by itself does not enhance affinity for RT.
Truncations of the aptamer abolished its tight binding to HIV RT indicating a minimal length requirement
The length of the selected aptamer was also tested as a potential factor for tight binding. Table 1 shows the various oligos that were designed by progressively eliminating nucleotides from the termini of the 30-nucleotide PF1. As little as a 3-nucleotide deletion at the 5′ end (PF1-27nt5′cut) and 5-nucleotide deletion at the 3′ end [PF1-25nt3′ (fewer than 5 was not tested at this end)] dramatically decreased binding to a Kd of ∼500 nM. Further truncations at either end (PF1-25nt5′cut, PF1-20nt5′cut, PF1-15nt5′cut, PF1-20nt3′, and PF1-15nt3′cut) led to an even more severe decrease in binding to essentially the level of 5′-N30-3′ (Kd>750 nM). Similarly, when the nucleotides in between the guanosine doublets were eliminated [PF1-Δ(8–12), PF1-Δ(16–25), PF1-Δ(8–12/16–25), and PF1-18ntTlinkG (closely resembles TBA aptamer)], the shortened oligos lost the ability to bind RT tightly. These results indicate that PF1's affinity for RT is highly dependent on the length of the oligo, as the binding of RT significantly decreases even when 3 nucleotides were trimmed off, as in PF1-27nt5′cut. However, it is important to point out that the truncations discussed above eliminate at least 1 of the 4 diguanosine motifs. Therefore they do not completely separate the role of the diguanosine repeats versus nucleotide length in achieving strong binding to HIV RT. To test this further, a construct with 1 and 3 nucleotides eliminated from the 5′ and 3′ termini, respectively, was tested (PF1-26ntG). This construct retains all 4 diguanosines but was shortened by 4 nucleotides. This resulted in about a 3-fold increase in the Kd in comparison to PF1. The role of the terminal diguanosines was further evaluated by changing the diguanosine in PF1-26ntG to either dicytidines (PF1-26ntC) or dithymidines (PF1-26ntT). While dicytidine had a negative effect on binding in comparison to diguanosine, dithymidine had a modestly positive effect. Overall, these results suggest a complex interaction between RT and the constructs and a critical role for the central diguanosines. Beyond this, length and the nucleotide sequence at the termini play some role in high affinity binding but are less vital.
An RNA version of PF1 or the TBA aptamer does not confer tight binding to HIV RT
An RNA analog of the 30-nucleotide PF1 aptamer (PF1-RNA) was tested for RT binding. PF1-RNA did not bind with high affinity to RT, indicating that the high affinity interaction was specific to DNA (Table 1). Also, the 15-nucleotide TBA aptamer that forms a G-quadruplex and binds tightly to thrombin (see Introduction) did not bind tightly to HIV RT.
The diguanosine repeats require the specific sequences in PF1 to retain HIV RT tight binding
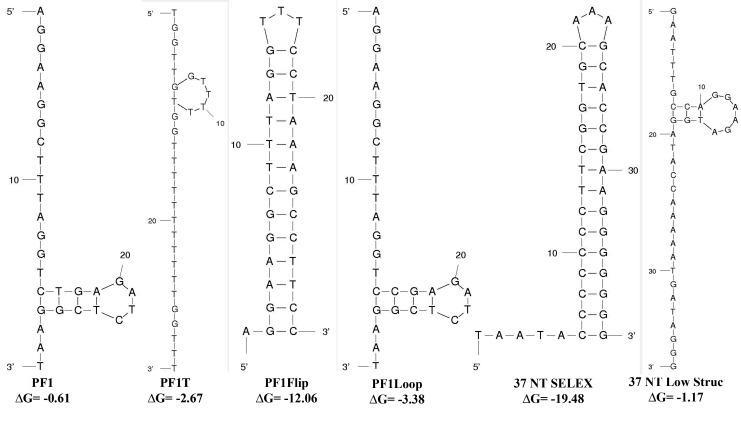
As was noted above, PF1 has very low predicted secondary structure (ΔG=–0.61 kcal/mol). It was possible that the intervening sequences between the diguanosines served as “spacers” that allowed the repeats to be optimally positioned for tight binding to RT. The low secondary structure could reduce the rigidity of PF1, allowing the diguanosines greater flexibility to be positioned for maximal binding to RT. To test this, the potential to form secondary structure for the bases between the diguanosine repeats was eliminated by changing them to all As or all Ts [PF1A (ΔG=0), PF1T (ΔG=–2.67 kcal/mol), respectively; see Fig. 2 for predicted structures of various constructs]. A third oligo in which the intervening bases were randomized (PF1N) was also tested. Two constructs with greater secondary structure were also tested: PF1Flip (ΔG=–12.06 kcal/mol) contained a predicted flipback structure in the presence of 3 of the diguanosine repeats; and PF1Loop (ΔG=–3.83 kcal/mol) had a predicted hairpin-loop structure in between the middle diguanosines. Despite the structural differences, all of the constructs had Kd values that increased approximately 6-fold or even more in comparison to PF1 (Table 1). The results with the PF1A and PF1T low secondary structure construct indicated that the intervening sequences do not simply serve as low secondary structure spacers; for example, PF1A had a Kd value over 750 nM, while the Kd for PF1T was ∼450 nM, each binding much more poorly than PF1. Since the more structured constructs also decreased binding, it is tempting to conclude that both low secondary structure and specific nucleotide sequences are required for tight binding. However, interpretations are complicated by the sequence changes that were required to induce more structure in PF1.
FIG. 2.
Predicted structures of some aptamers and derivatives. Structures and free energy of folding values (ΔG, in kcal/mol) were obtained using mfold as described in Materials and Methods.
Kd determinations for PF1 and other aptamers by filter binding
Nitrocellulose filter binding assays were used to compare the binding of several aptamers to HIV RT. Although this method, like the gel-shift analysis used above, is not an equilibrium binding method and therefore does not necessary yield a highly accurate Kd value (Hall and Kranz, 1999), it is the simplest and most cost effective approach available for comparative binding. In addition to PF1, we used a class of G-quadruplex RT aptamers studied by Michalowski et al. that were potent inhibitors of HIV RT (Michalowski et al., 2008). The primary G-quadruplex aptamer, R1T, was a 41-nucleotide oligo with a 3-tier parallel G-quadruplex (Michalowski et al., 2008). We also evaluated S4, a 35-nucleotide DNA similar to R1T with some modifications to the stem-loop component while retaining the G-quadruplex structure (Michalowski et al., 2008). Our lab developed a unique primer–template flipback aptamer, 38 NT SELEX, which is a 38-nucleotide-long single-strand DNA and a potent inhibitor of HIV RT (DeStefano and Cristofaro, 2006; DeStefano and Nair, 2008) (see Introduction). Another previously characterized DNA aptamer, RT1 (Schneider et al., 1995; Kissel et al., 2007), was much larger than the other aptamers (81 nucleotides), and finally, a small previously characterized 24-nucleotide RNA pseudoknot type aptamer, 1.1 (Tuerk et al., 1992) was also examined. The results along with the sequences of the various aptamers are shown in Table 2. The 2 G-quartet type inhibitors, S4 and R1T, along with RT1 all bound very tightly, with Kd values of 1.1 nM or less. The RNA pseudoknot 1.1 bound slightly less tightly (Kd=2.6±0.6 nM), while the primer–template based aptamer 38 NT SELEX bound even less tightly (6.5±1.1 nM). Results were in general consistent with previous characterizations of these aptamers with the exception of 38 NT SELEX, which bound RT about an order of magnitude more tightly in previous reports (DeStefano and Nair, 2008). However, binding was measured by an RT nucleotide-extension assay in the previous work, so the numbers are difficult to compare with those obtained here. Aptamer PF1 was by far the weakest binding, with a Kd value of 125±55 nM. The average value was also higher than, though not statistically different from, the value calculated by gel-shift analysis (82±7 nM).
Competition binding assays indicate that all aptamers compete with the primer–template for binding and most likely bind in or near the substrate binding pocket
Some of the aptamers studied in the filter binding assays were used in a competition filter binding assay to further understand how they interacted with RT. In this assay, all of the aptamers were competed against 38 NT SELEX. This aptamer was used because it is essentially a primer–template and therefore would bind to RT like the normal nucleic acid substrate. An ability to displace the aptamer would suggest that the competitor binds in or near the same pocket. Displacement based on allosteric binding is much less likely but cannot be completely ruled out. All assays contained 10 nM radiolabeled (hot) 38 NT SELEX and 3 nM HIV RT. Increasing amounts of unlabeled (cold) aptamers were added and allowed to incubate for 1.5 hours before passing the material over nitrocellulose filter disks. Cold 38 NT SELEX was able to compete the hot material off of RT with 10 nM cold material resulting in about a 40% decrease in binding (Fig. 3), which is approximately what would be predicted based on the Kd for 38 NT SELEX determined above (Table 2). In contrast, both 5′-N30-3′ and PF1-C6 were essentially uncompetitive even when added at a 16-fold molar excess. Note that PF1-C6 has only a single nucleotide change of a G at position 6 to a C, and this change resulted in ∼5-fold reduction in binding in the gel-shift assay (Table 1). Apparently even this level of change results in a complete inability to displace a strong binding primer–template. It is also possible that some binding of either 5′-N30-3′ or PF1-C6 occurred, but at a site that did not displace 38 NT SELEX. The other aptamers displaced 38 NT SELEX and yielded results that were consistent with the determined Kd values (Table 2), at least from a qualitative perspective. PF1 was the least competitive, while the S4 G-quartet and 1.1 RNA pseudoknot were modestly better competitors than 38 NT SELEX, and RT1 was a much better competitor. Still, both PF1 and RT1 were more competitive than would be predicted based on the differences in the Kd values between them and 38 NT SELEX. Another way to analyze this is to use the determined Kd value from above of 38 NT SELEX (6.5 nM) to calculate the Kd values for the aptamers used as competitors. Since the amount of enzyme used for binding was limiting, Kd values relative to 38 NT SELEX can be approximated from assays that contained 10 nM hot 38 NT SELEX and 10 nM of the cold competitor (Creighton et al., 1992). These values are also listed in Table 2 (in parentheses in the Kd column, a value for R1T is included although results are not shown on Fig. 3). The values for S4 and 1.1 were close to the values determined by filter binding in the absence of a competitor, while the value for R1T was modestly lower. A reliable value could not be obtained for RT1 due to displacement of labeled 38 NT SELEX to near background levels when 10 nM cold RT1 was added. Even 2.5 nM RT1 almost completely displaced RT from 38 NT SELEX. RT1 is much larger than the other aptamers. It is possible that it has more than 1 binding site for RT or forms a complex that dissociates extremely slowly. If the latter were the case, obtaining equilibrium may require more than the 1.5-hour incubation time used for these assays. The 9.7±1.6 nM value for PF1 is much lower than the 125±55 nM determined value. The reason for this discrepancy is unclear but may have to do with a direct interaction between PF1 and 38 NT SELEX or synergistic binding of PF1 in the presence of 38 NT SELEX. It could also result of the PF1-RT complex being more susceptible to dissociation during the filter washing or binding steps, an effect that would artificially lower the Kd value when measured directly. Interesting, a several-fold lowering of the determined Kd value was also observed when PF1 was competed against the 1.1 RNA pseudoknot, but to a much lesser extent when it was competed against the S4 G-quartet (data not shown). We were unable to test binding using PF1 as the labeled aptamer and other aptamers as competitors because too much enzyme was required to get enough radioactive material bound to the filters. This was in part due to a high background level of binding in the absence of enzyme with PF1.
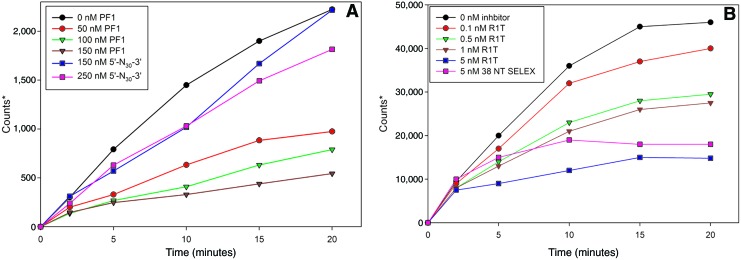
The selected aptamer PF1 can inhibit HIV RT primer extension, but not as effectively as some other previously characterized RT aptamers
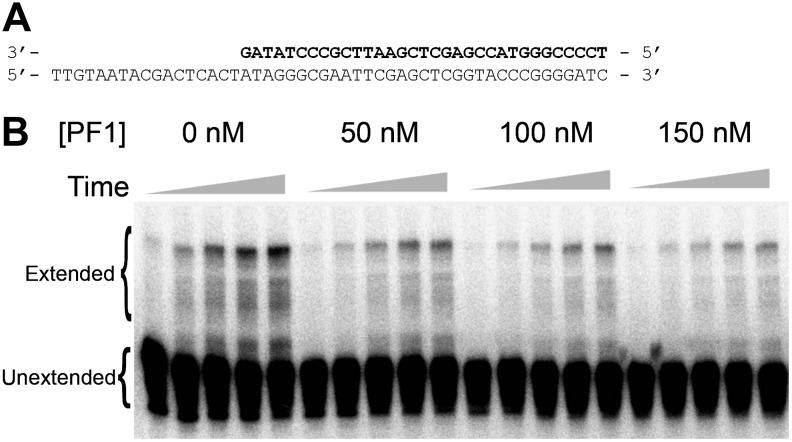
An HIV RT primer extension assay was performed to determine whether PF1 could effectively inhibit HIV RT in vitro. A 33-nucleotide 5′ 32P end-labeled DNA primer was hybridized to a 50-nucleotide template (50 nM final concentration; complex shown in Fig. 4A). HIV RT utilizes this primer–template to extend the primer portion toward the 5′ end of the template. Assays were performed using 50 nM primer–template and 0.25 nM HIV RT, and the amount of extended primer was measured over a time course from 2 to 20 minutes (Fig. 4B). Results were plotted (relative counts vs. time) and used to calculate IC50 as described in Material and Methods (Fig. 5A, B). The IC50 of PF1 was 63±24 nM (mean±standard deviations from 3 independent experiments) (Fig. 5A; Table 3). This concentration is comparable to the primer–template concentration used in the experiment, suggesting that PF1 binds approximately as well as the primer–template. The IC50 for 5′-N30-3′ was>500 nM (Table 3; Fig. 5A). Interestingly, even those oligos that showed only modestly lower affinities for RT in the gel-shift assays (PF1-26ntT and PF1-terminal GG>CC) were poor inhibitors of RT extension relative to PF1.
FIG. 4.
A representative autoradiogram using various concentrations of PF1 in the RT inhibition assay. (A) Schematic diagram of the primer (black)-template (gray) used to conduct the reverse transcriptase (RT) inhibition assay. The primer strand was radiolabeled with 32P at the 5′ end. (B) The assay was conducted by incubating the prehybridized primer–template (50 nM) with 100 μM dNTPs and various concentrations of aptamers as indicated. Reactions were initiated with HIV RT (0.25 nM), and aliquots were removed at 2, 5, 10, 15, and 20 minutes. The samples were run on a denaturing 10% polyacrylamide gel and quantified as described in Materials and Methods.
FIG. 5.
Inhibition of HIV RT primer extension in the presence of various amounts of PF1 and starting material (5′-N30-3′) (5A), or R1T and 38 NT SELEX ddG (5B). *Graphs of counts (photo-stimulated luminescence radiation units from phosphoimager) versus time derived from experiment of the type shown in Fig. 4. The difference in magnitude of the y-axis in the experiment in A and B is not meaningful, as the counts are dependent on the specific activity of the primer and the time of exposure. The assay was conducted by incubating the primer-template (5′ γ-32P end-labeled primer, 50 nM) with various concentrations of aptamers. Reactions were initiated with HIV RT (0.25 nM), and aliquots were removed at the indicated times. The samples were run on a denaturing 10% polyacrylamide gel and quantified as described in Materials and Methods. In Fig. 5B, the 38 NT SELEX aptamer contained a 3′ terminal dideoxy G residue in place of the normal dG to prevent extension of the aptamer by RT. Refer to Tables 1 and 2 for information on the specific oligonucleotides used as inhibitors. Color images available online at www.liebertonline.com/nat
Table 3.
Inhibition of Human Immunodeficiency Virus Reverse Transcriptase Primer Extension by Various Aptamers
| Name | Sequence | IC50 (nM) |
|---|---|---|
| Starting material: 5′-N30-3′ | 5′-NNNNNNNNNNNNNNNNNNNNNNNNNNNNN-3′ | >500 |
| PF1 | 5′-AGGAAGGCTTTAGGTCTGAGATCTCGGAAT-3′ | 63±24 |
| Primer-template aptamer | ||
| 38 NT SELEX (3′ ddG) | 5′-TAATACCCCCCCTTCGGTGCAAAGCACCGAAGGGGGGG-3′ | 3.7±1.1 |
| G-quadruplex aptamers | ||
| R1T | 5′-CGCCTGATTAGCGATACTCAGGCGTTGGGTGGGTGGGTGGG-3′ | 0.9±0.3 |
| S4 | 5′-CGCCTGACCCTTCAGGCGTTGGGTGGGTGGGTGGG-3′ | 0.5±0.1 |
| Large DNA aptamer | ||
| RT1 | 5′-ATCCGCCTGATTAGCGATACTCAGAAGGATAAACTGTCCAGAACTTGGATATATACACTTGAGCAAAATCACCTGCAGGGG-3′ | 0.3±0.1 |
| RNA pseudoknot aptamer | ||
| 1.1 | 5′-UCCGUUUUCAGUCGGGAAAAACUG-3′ | 4.7±0.2 |
| Modified constructs | ||
| PF1-terminal GG>CC | 5′-ACCAAGGCTTTAGGTCTGAGATCTCCCAAT-3′ | ∼250 |
| PF1-26ntG | 5′-GGAAGGCTTTAGGTCTGAGATCTCGG-3′ | ∼250 |
| PF1-26ntT | 5′-TTAAGGCTTTAGGTCTGAGATCTCTT-3′ | >250 |
Underlined nucleotides indicate the polypurine tract-like sequence in the primer-template aptamer and the region that forms the G-quadruplex in the G-quadruplex aptamers.
The half-maximal inhibitory value (IC50) values were determined using a reverse transcriptase inhibition assay as described under Material and Methods. Results for the experimental and control oligonucleotides were an average of 3 experiments±standard deviation. The results for the starting material and modified constructs were estimates based on 2–3 independent experiments.
Consistent with their lower Kd values in filter binding assays (Table 2), the 1.1 RNA pseudoknot, RT1, R1T (Fig. 5B), S4, and 38 NT SELEX (Fig. 5B) aptamers were all much more potent inhibitors in the RT extension assay (Table 3). Aptamers 38 NT SELEX and 1.1 had comparable IC50 values in the ∼4 nM range, while S4, R1T, and RT1 were 1 nM or less. For the RT inhibition experiments, 38 NT SELEX had the terminal 3′ guanosine residue replaced with a ddG to prevent extension by HIV RT.
The G-quartet and primer–template aptamers are highly stable in serum containing media, while PF1 is less stable and 1.1 RNA pseudoknot is highly unstable
The ability of an aptamer to resist degradation in the host would be an important property for any aptamer that was being considered as a model for future drug design. We used incubation in cell media (RPMI) with 10% FBS to examine stability for various aptamers (Fig. 6). This is obviously not a direct test of stability in a host model but can still yield useful comparative information. Of the 5 aptamers tested, both 38 NT SELEX and the S4 G-quartet aptamer were highly stable, showing minimal breakdown over the 2-hour time course. This is consistent with the high degree of structure for these aptamers. RT1, the 81-nucleotide DNA aptamer, was also quite stable, although some breakdown was observed. Again, this aptamer is highly structured (Schneider et al., 1995; Kissel et al., 2007). PF1 was less stable than the other DNA aptamers and showed a slow but progressive shortening over the time course. Degradation appeared to result from removal of nucleotides from the 3′ end of the 5′ end-labeled aptamer, as the amount of label did not decrease while the length of the products did. Still, breakdown was modest considering the very low secondary structure of the aptamer and the fact that no attempts to protect it from degradation (by using phosphorothioate nucleotides, for example) were made. In contrast to the DNA aptamers, the 1.1 RNA pseudoknot aptamer was highly unstable and was nearly completely degraded after only 15 minutes in the media. Degradation clearly resulted from RNase, as addition of the RNase A-type inhibitor, RNasin, stabilized the aptamer (Fig. 6, furthest lane on the right, 15 minutes incubation with RNasin included). Overall the experiment suggested that DNA aptamers may be much more stable than RNAs, giving them a clear advantage as potential drug models.
FIG. 6.
Stability of various aptamers in cell culture media (RPMI+10% fetal bovine serum), shown in an autoradiogram of a gel with various 5′ end-labeled aptamers (as indicated above lanes) incubated in culture medium for increasing times. Each set has a control (C) time 0 sample, followed by a 30-minute incubation in cell media that included nuclease (N). Either DNase I (5 units) for the DNA aptamers or RNase (0.25 μgs) for the 1.1 RNA aptamer was used. This lane is followed by samples incubated for 15, 30, 60, and 120 minutes. *For the 1.1 RNA pseudoknot aptamer, an extra lane where 20 units of RNasin RNase inhibitor was added to the media and incubated for 15 minutes is shown. Note that 38 NT SELEX [38 nucleotides (nts)] runs faster on the gel than either PF1 (30 nts) or S4 (35 nts), despite being longer. This is likely do to it being highly structured.
Circular dichroism spectroscopy indicates that PF1 does not form a G-quadruplex
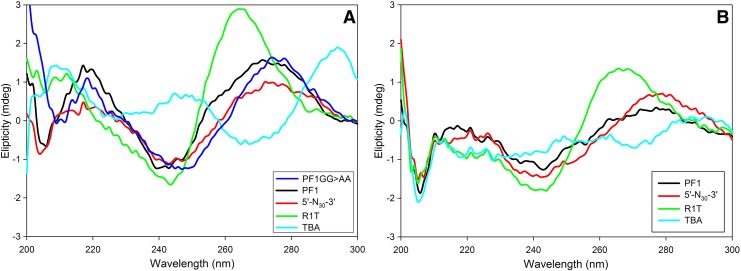
CD spectroscopy is an analytical tool for detecting secondary structures and G-quadruplexes in nucleic acids. The characteristic feature of a G-quadruplex is a distinctive positive band at 210 nm, and between 260 and 265 nm, and a negative peak at ∼240 nm. For antiparallel quadruplexes, the negative peak is at ∼260 nm, and a positive peak at ∼290 nm. A regular B-DNA has a negative peak at ∼210 nm and ∼240 nm, plus positive peaks at ∼220 nm and between 270 and 280 nm (Kypr et al., 2009). For single-stranded DNA sequences, the CD spectrum is similar to the spectrum for the double-stranded DNA with the same coding strand sequence (Hung et al., 1994). A hairpin loop like 38 NT SELEX DNA flips back onto itself to form an intramolecular B-DNA (Kypr et al., 2009).
Consistent with the previous results, the CD spectrum for the TBA aptamer (Fig. 7A) contained positive peaks at 210 and 295 nm, and a negative peak at 265 nm, confirming its antiparallel G-quadruplex nature (Macaya et al., 1993). R1T showed positive peaks at 210 and 260 nm, and a negative peak at 240 nm at room temperature in the presence of K+, consistent with a parallel G-quadruplex. While K+ stabilizes G-quadruplex formation, Na+ disfavors formation (Tohl and Eimer, 1997; SIMONSSON, 2001). Despite this, R1T retained its G-quadruplex structure (although the spectrum is slightly flattened) in the presence of 80 mM NaCl, while the TBA G-quadruplex structure, consistent with previous results (Tohl and Eimer, 1997; SIMONSSON, 2001; Risitano and Fox, 2004), was disrupted under these conditions (Fig. 7B). The retention of quadruplex structure by R1T in the presence of Na+ is also consistent with previous results and likely reflects the high stability of this particular quadruplex (Michalowski et al., 2008).
FIG. 7.
CD spectra of various aptamers and derivatives. Spectra were acquired as described in Materials and Methods. The various aptamers and derivatives used are shown in Tables 1 and 2. (A) Spectra acquired in potassium buffer: 50 mM Tris-HCl (pH 8), 80 mM KCl, 6 mM MgCl2, and 1 mM DTT; (B) Spectra acquired in sodium buffer: 50 mM Tris-HCl (pH 8), 80 mM NaCl, 6 mM MgCl2, and 1 mM DTT. Color images available online at www.liebertonline.com/nat
In the presence of K+, PF1 showed a positive peak at ∼270 nm and a negative peak at ∼240 nm, as well as a positive peak at 220 nm and negative peak at about 210 nm (Fig. 7A). This spectrum was consistent with B-form DNA and indicates that PF1 was unlikely to form a G-quadruplex. A construct with the guanosine doublets substituted (PF1-GG>AA; see Table 1) yielded a spectrum similar to PF1 as did the starting material (5′-N30-3′). Neither PF1 nor 5′-N30-3′ showed significant alterations (other than modest flattening) in their spectra in the presence of Na+ (Fig. 7B), again suggesting that PF1 does not form a G-quadruplex. Spectra for 38 NT SELEX and a 38-nucleotide low secondary structure oligonucleotide (38 NT Low Struc; see Fig. 2) as well as PF1-GG>CC were also consistent with B-form DNA (data not shown).
Discussion
Effects of the guanosine repeats on HIV RT binding and possible structures of PF1
In this study, we characterized a group of single-stranded DNA aptamers selected for binding to HIV RT that were isolated using a novel primer-free SELEX technique (Lai and Destefano, 2011) and defined whether the aptamer's affinity for RT was dependent on its length, sequence, or structure. We speculated that the regular arrangement of 4 separate diguanosines in the sequences affected the tight binding of the selected aptamer to HIV RT. This prediction was based on this arrangement appearing in aptamers isolated from 2 separate independent SELEX experiments (Lai and Destefano, 2011). The likelihood of such a specific sequence arrangement occurring arbitrarily in 2 separate SELEX experiments, let alone in a single selection, was very low. Although it is possible that the diguanosine array resulted from the unique SELEX protocol used to select the aptamers, the fact that the selected aptamers bind much more tightly to HIV RT than to the SELEX experiment starting material strongly suggests that their selection was based on affinity for RT. It could also be argued that the low predicted secondary structure of PF1 was at least in part due to the thermostable RNA ligase step in the SELEX protocol favoring low secondary structure and therefore excluding structured aptamers from being selected. However, results showed that only strong secondary structure led to inefficient ligation (Lai and Destefano, 2011). The low secondary structure of PF1 may permit it to more easily conform to RT and maximize binding. The low secondary structure could provide flexibility that allows the diguanosines to be positioned for optimal binding to RT or formation of a G-quadruplex. However, low secondary structure alone clearly cannot account for PF1's affinity to RT as oligos with low (PF1T) or no predicted structure (PF1A) and identical diguanosine spacing bound RT poorly (Table 1). Therefore, PF1's strong binding results at least in part from the specific sequence between the diguanosine repeats, even though those sequences presumably generate little structure. This point is further supported by the fact that the 2 separate SELEX experiments produced aptamers not only with identical diguanosine spacing, but also with closely related intervening sequences (Lai and Destefano, 2011). Taken together with other results that suggested a potential role for low secondary structure (see Results), the interaction between PF1 and HIV RT is likely complex and may be driven by diguanosine repeats optimally spaced by specific sequences that generate little structure.
The diguanosine repeats in PF1 were similar to those found in intramolecular G-quadruplex forming aptamers that bind tightly to other proteins (Macaya et al., 1993; Michalowski et al., 2008). In addition, G-quadruplex forming aptamers have been shown to bind HIV RT and many other proteins with high affinity (see Introduction). Surprisingly, CD spectroscopy indicated PF1 did not form a G-quadruplex (Fig. 7A). This does not necessarily rule out the possibility of PF1 forming other higher order structures. Since both the low and high structure single-stranded constructs all showed similar CD spectra (see Results), this technique, as applied here, does not clearly differentiate the presence of structure. Although PF1 by itself does not appear to form a classical G-quadruplex that can be identified by CD spectrum, it is still possible that interaction between HIV RT and PF1 may induce a change in the conformation of the aptamer leading to G-quadruplex or other structural formation. If this were the case, and assuming that quadruplex formation is required for tight binding, then changes in any of the 4 guanosine doublets would be expected to be highly detrimental to RT binding. Changing any doublets did decrease binding, but the 2 central doublets were clearly more important, as changes in these had a profound effect on RT affinity (Table 1). PF1-terminal GG>CC, which changes the diguanosines at the termini to Cs showed just a modest decrease in binding. PF1-26ntT, which changed the terminal Gs to Ts and also deleted the flanking terminal nucleotides, similarly showed a small change in binding. It is important to reiterate, however, that neither gel-shift nor filter binding are true equilibrium binding methods for measuring affinity (Hall and Kranz, 1999). Therefore the extent to which they are highly quantitative is dependent on the behavior and stability of the particular protein-ligand interaction under gel-shift of filter binding conditions. Consistent with this, PF1-26ntT and PF1-terminal GG>CC were much weaker inhibitors than PF1 in the RT extension assay (Table 3) despite the small changes observed by gel-shift analysis. To clearly determine whether G-quadruplex formation is important to PF1's interaction with RT, structural analysis using nuclear magnetic resonance or X-ray crystallography would be required.
The role of aptamer length to HIV RT tight binding
Although PF1 demonstrated ∼10-fold tighter binding to RT than the starting material, deletion of just a few nucleotides caused a significant decrease in binding (Table 1). Even PF1-26ntG, which retains all of the diguanosines and intervening sequences but lacked 1 and 3 nucleotides from the 5′ and 3′ ends, respectively, showed a modest loss in binding. Any changes that eliminated or changed the spacing between diguanosine runs essentially eliminated tight binding. From these results it appears that there may be length limitations for tight binding to RT as well as specific sequence and spacing requirement for the diguanosine. With respect to length, DNA aptamers to HIV RT reported in the literature are typically larger than PF1. The 38-nucleotide and 35-nucleotide aptamers (38 NT SELEX and S4, respectively) used in the binding and inhibition studies here are among the shortest reported for aptamers that bind in the nM range (Tables 2 and 3). Perhaps 30 nucleotides is approaching the limit required to obtain low nM binding for a DNA aptamer. In contrast, RNA aptamers of fewer than 30 nucleotides that form pseudoknots can bind RT with very high affinity (pM binding constants have been reported) (Kensch et al., 2000). The 24-nucleotide aptamer we used here (1.1) bound just slightly less tightly than the G-quadruplex and large RT1 aptamers in filter binding experiments and was a potent competitor for RT binding and inhibition (Tables 2 and 3; Fig. 3). We are currently attempting to select larger and smaller DNA aptamers than PF1 using the primer-free SELEX method. This should help determine the importance and limitations of aptamer length.
Comparison between various classes of aptamers in HIV RT binding and inhibition assays
HIV RT and other DNA polymerases utilize a primer–template duplex as their natural substrate. In an RT extension assay, the HIV RT aptamers can compete with the primer–template complex, as has been shown for all of the previously characterized aptamers used here (Kissel et al., 2007; DeStefano and Nair, 2008; Michalowski et al., 2008). The G-quadruplex and RT1 inhibitors were the most potent of those tested here, with IC50 values that were ∼5- to 10-fold lower than 38 NT SELEX and 1.1, which were also potent inhibitors (Table 3). It is notable that the G-quadruplex inhibitors and RT1 seem to bind even more tightly than an optimized primer–template (38 NT SELEX), the natural substrate for HIV RT (Table 2; Fig. 3). Even though PF1 did not inhibit as well as the other aptamers, the determined IC50 value (63±24 nM) indicated that it still bound approximately as tightly as the primer–template used in the assays. Such tight binding would not be predicted for a single-stranded B-form 30-mer with very low predicted structure. The possibility that PF1 was forming a loop-back type structure or dimerizing to form a primer–template was investigated. Folding and hybridization programs did not predict any of these structures and no extension of PF1 was observed in assays with HIV RT and dNTPs (data not shown). This suggests that no 3′ recessed termini were formed.
The stronger inhibition observed here and tighter binding for the previously characterized aptamers indicates that these aptamers bind with significantly greater affinity to RT than PF1. This was confirmed using filter binding and competition binding assays (Table 2; and Fig. 3). There are several possible reasons for this, including the different SELEX protocols used, the length of the aptamers, and the extensive post-SELEX modifications of the 38 NT SELEX and the G-quadruplex aptamers. For example, the 38 NT SELEX binds ∼10-fold better than the aptamers selected from the original SELEX (DeStefano and Cristofaro, 2006; DeStefano and Nair, 2008), while the G-quadruplex aptamers used here are about twice as potent in inhibition assays as the parent aptamers from which they were derived (Michalowski et al., 2008). With respect to length, when 38 NT SELEX was reduced to only 27 nucleotides, its Kd increased by 100-fold, even though the run of 7 Gs was still present (DeStefano and Nair, 2008). Similarly, the shortest G-quadruplex aptamer was 35 nucleotides, and it required the stem-loop component to achieve tight binding to HIV RT (Schneider et al., 1995; Michalowski et al., 2008). It will be interesting to see if PF1 can also be modified for tighter binding, for example, by increasing it length.
Also notable was the ability of all the aptamers to compete with the primer–template aptamer in the competition assay (Fig. 3). This suggests that all of these and perhaps all current RT aptamers bind at or near the primer–template binding pocket. It appears that this site has by far the strongest affinity for nucleic acids and obtaining aptamers that bind elsewhere (if possible) would require blocking or mutating the primer–template binding pocket.
Applications and future work
The HIV RT aptamers tested here were derived from various SELEX protocols including a traditional protocol for the G-quadruplex, DNA, and RNA pseudoknot aptamers, and 2 protocols developed in this lab: one that selects primer–templates for 38 NT SELEX (DeStefano and Cristofaro, 2006), and a second primer-free protocol for PF1 (Lai and Destefano, 2011). Aptamers derived from each protocol had unique properties with respect to sequence and structure. In particular the primer-free SELEX generated aptamers with low secondary structure, although it was not entirely clear if this is a general property of this protocol or was specific to HIV RT. The protocol includes a step with thermostable RNA ligase that was sensitive to structure, but only when the substrates for ligation were highly structured. Presumably this step could be modulated to help regulate the structure of the aptamers that are selected. This could be done by lowering the ligation temperature (60°C in the standard protocol) or shortening the time allowed for ligation, both of which would favor aptamers with lower structure. The low structure aptamers might not bind as tightly to their target, as aptamer binding is often structure-dependent; however, they might have other advantages, particularly as models for the development of cellular inhibitors. It is known that highly structured nucleic acids tend to elicit innate immune responses in cells (Hornung and Latz, 2010). Low structure aptamers may help overcome this problem. Both G-rich oligonucleotides and those that can form G-quartets have effects on cells ranging from induction of senescence and aging (Riou et al., 2002) to inhibition of proliferation (Scaggiante et al., 1998; Cogoi et al., 2004a; Cogoi et al., 2004b). G-quadruplexes are also associated with inhibitory cellular properties, via the oligos presumably binding to nucleolin (Dempsey et al., 1999; Hanakahi et al., 1999), elongation factor 1A (eIF1A) (Dapas et al., 2003; Bernardi et al., 2006; Goodchild et al., 2007), and STAT3 (Tong et al., 1997). A clear drawback of the RNA aptamer in comparison with the DNAs was the much lower stability in cell culture media (Fig. 6). Highly stable aptamers that do not elicit unwanted cell responses could be valuable tools for the development of oligonucleotide therapies in the future.
Acknowledgments
This work was supported by National Institute of General Medicine grant number GM051140.
Author Disclosure Statement
No competing financial interests exist.
References
- ANDREOLA M.L. PILEUR F. CALMELS C. VENTURA M. TARRAGO-LITVAK L. TOULME J.J. LITVAK S. DNA aptamers selected against the HIV-1 RNase H display in vitro antiviral activity. Biochemistry. 2001;40:10087–10094. doi: 10.1021/bi0108599. [DOI] [PubMed] [Google Scholar]
- BERNARDI F. GAGGELLI E. MOLTENI E. PORCIATTI E. VALENSIN D. VALENSIN G. 1H and 13C-NMR and molecular dynamics studies of cyclosporin a interacting with magnesium(II) or cerium(III) in acetonitrile. Conformational changes and cis-trans conversion of peptide bonds. Biophys. J. 2006;90:1350–1361. doi: 10.1529/biophysj.105.074245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- BOCK L.C. GRIFFIN L.C. LATHAM J.A. VERMAAS E.H. TOOLE J.J. Selection of single-stranded DNA molecules that bind and inhibit human thrombin. Nature. 1992;355:564–566. doi: 10.1038/355564a0. [DOI] [PubMed] [Google Scholar]
- BRODY E.N. GOLD L. Aptamers as therapeutic and diagnostic agents. J. Biotechnol. 2000;74:5–13. doi: 10.1016/s1389-0352(99)00004-5. [DOI] [PubMed] [Google Scholar]
- CERCHIA L. DUCONGE F. PESTOURIE C. BOULAY J. AISSOUNI Y. GOMBERT K. TAVITIAN B. DE FRANCISCIS V. LIBRI D. Neutralizing aptamers from whole-cell SELEX inhibit the RET receptor tyrosine kinase. PLoS Biol. 2005;3:e123. doi: 10.1371/journal.pbio.0030123. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- COGOI S. BALLICO M. BONORA G.M. XODO L.E. Antiproliferative activity of a triplex-forming oligonucleotide recognizing a Ki-ras polypurine/polypyrimidine motif correlates with protein binding. Cancer Gene Ther. 2004a;11:465–476. doi: 10.1038/sj.cgt.7700722. [DOI] [PubMed] [Google Scholar]
- COGOI S. QUADRIFOGLIO F. XODO L.E. G-rich oligonucleotide inhibits the binding of a nuclear protein to the Ki-ras promoter and strongly reduces cell growth in human carcinoma pancreatic cells. Biochemistry. 2004b;43:2512–2523. doi: 10.1021/bi035754f. [DOI] [PubMed] [Google Scholar]
- CREIGHTON S. HUANG S.M. CAI H. ARNHEIM N. GOODMAN N.F. Base mispair extension kinetics. J. Biol. Chem. 1992;267:2633–2639. [PubMed] [Google Scholar]
- DAPAS B. TELL G. SCALONI A. PINES A. FERRARA L. QUADRIFOGLIO F. SCAGGIANTE B. Identification of different isoforms of eEF1A in the nuclear fraction of human T-lymphoblastic cancer cell line specifically binding to aptameric cytotoxic GT oligomers. Eur. J. Biochem. 2003;270:3251–3262. doi: 10.1046/j.1432-1033.2003.03713.x. [DOI] [PubMed] [Google Scholar]
- DE SOULTRAIT V.R. LOZACH P.Y. ALTMEYER R. TARRAGO-LITVAK L. LITVAK S. ANDREOLA M.L. DNA aptamers derived from HIV-1 RNase H inhibitors are strong anti-integrase agents. J. Mol. Biol. 2002;324:195–203. doi: 10.1016/s0022-2836(02)01064-1. [DOI] [PubMed] [Google Scholar]
- DEISINGH A.K. Aptamer-based biosensors: biomedical applications. Handb. Exp. Pharmacol. 2006:341–357. doi: 10.1007/3-540-27262-3_17. [DOI] [PubMed] [Google Scholar]
- DEMPSEY L.A. SUN H. HANAKAHI L.A. MAIZELS N. G4 DNA binding by LR1 and its subunits, nucleolin and hnRNP D, A role for G-G pairing in immunoglobulin switch recombination. J. Biol. Chem. 1999;274:1066–1071. doi: 10.1074/jbc.274.2.1066. [DOI] [PubMed] [Google Scholar]
- DESTEFANO J.J. CRISTOFARO J.V. Selection of primer-template sequences that bind human immunodeficiency virus reverse transcriptase with high affinity. Nucleic Acids Res. 2006;34:130–139. doi: 10.1093/nar/gkj426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DESTEFANO J.J. NAIR G.R. Novel aptamer inhibitors of human immunodeficiency virus reverse transcriptase. Oligonucleotides. 2008;18:133–144. doi: 10.1089/oli.2008.0103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ELLINGTON A.D. SZOSTAK J.W. In vitro selection of RNA molecules that bind specific ligands. Nature. 1990;346:818–822. doi: 10.1038/346818a0. [DOI] [PubMed] [Google Scholar]
- GOLD L. The SELEX process: a surprising source of therapeutic and diagnostic compounds. Harvey Lect. 1995;91:47–57. [PubMed] [Google Scholar]
- GONZALEZ V. HURLEY L.H. The c-MYC NHE III(1): function and regulation. Annu. Rev. Pharmacol. Toxicol. 2010;50:111–129. doi: 10.1146/annurev.pharmtox.48.113006.094649. [DOI] [PubMed] [Google Scholar]
- GOODCHILD A. KING A. GOZAR M.M. PASSIOURA T. TUCKER C. RIVORY L. Cytotoxic G-rich oligodeoxynucleotides: putative protein targets and required sequence motif. Nucleic Acids Res. 2007;35:4562–4572. doi: 10.1093/nar/gkm465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- HALL K.B. KRANZ J.K. Nitrocellulose filter binding for determination of dissociation constants. In RNA-protein interaction protocols. In: Haynes S.R., editor. Humana Press; Totowa, New Jersey: 1999. pp. 105–128. [DOI] [PubMed] [Google Scholar]
- HANAKAHI L.A. SUN H. MAIZELS N. High affinity interactions of nucleolin with G-G-paired rDNA. J. Biol. Chem. 1999;274:15908–15912. doi: 10.1074/jbc.274.22.15908. [DOI] [PubMed] [Google Scholar]
- HERSHMAN S.G. CHEN Q. LEE J.Y. KOZAK M.L. YUE P. WANG L.S. JOHNSON F.B. Genomic distribution and functional analyses of potential G-quadruplex-forming sequences in Saccharomyces cerevisiae. Nucleic Acids Res. 2008;36:144–156. doi: 10.1093/nar/gkm986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- HICKE B.J. MARION C. CHANG Y.F. GOULD T. LYNOTT C.K. PARMA D. SCHMIDT P.G. WARREN S. Tenascin-C aptamers are generated using tumor cells and purified protein. J. Biol. Chem. 2001;276:48644–48654. doi: 10.1074/jbc.M104651200. [DOI] [PubMed] [Google Scholar]
- HICKE B.J. STEPHENS A.W. GOULD T. CHANG Y.F. LYNOTT C.K. HEIL J. BORKOWSKI S. HILGER C.S. COOK G. WARREN S. SCHMIDT P.G. Tumor targeting by an aptamer. J. Nucl. Med. 2006;47:668–678. [PubMed] [Google Scholar]
- HORNUNG V. LATZ E. Intracellular DNA recognition. Nat. Rev. Immunol. 2010;10:123–130. doi: 10.1038/nri2690. [DOI] [PubMed] [Google Scholar]
- HOTODA H. KOIZUMI M. KOGA R. KANEKO M. MOMOTA K. OHMINE T. FURUKAWA H. AGATSUMA T. NISHIGAKI T., et al. Biologically active oligodeoxyribonucleotides. 5. 5'-End-substituted d(TGGGAG) possesses anti-human immunodeficiency virus type 1 activity by forming a G-quadruplex structure. J. Med. Chem. 1998;41:3655–3663. doi: 10.1021/jm970658w. [DOI] [PubMed] [Google Scholar]
- HSIEH J.C. ZINNEN S. MODRICH P. Kinetic mechanism of the DNA-dependent DNA polymerase activity of human immunodeficiency virus reverse transcriptase. J. Biol. Chem. 1993;268:24607–24613. [PubMed] [Google Scholar]
- HUNG S.H. YU Q. GRAY D.M. RATLIFF R.L. Evidence from CD spectra that d(purine).r(pyrimidine) and r(purine).d(pyrimidine) hybrids are in different structural classes. Nucleic Acids Res. 1994;22:4326–4334. doi: 10.1093/nar/22.20.4326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- JAMES W. Aptamers in the virologists' toolkit. J. Gen. Virol. 2007;88:351–364. doi: 10.1099/vir.0.82442-0. [DOI] [PubMed] [Google Scholar]
- JAROSCH F. BUCHNER K. KLUSSMANN S. In vitro selection using a dual RNA library that allows primerless selection. Nucleic Acids Res. 2006;34:e86. doi: 10.1093/nar/gkl463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- JOSHI P.J. FISHER T.S. PRASAD V.R. Anti-HIV inhibitors based on nucleic acids: emergence of aptamers as potent antivirals. Curr. Drug Targets Infect. Disord. 2003;3:383–400. doi: 10.2174/1568005033481060. [DOI] [PubMed] [Google Scholar]
- JOYCE G.F. Amplification, mutation, and selection of catalytic RNA. Gene. 1989;82:83–87. doi: 10.1016/0378-1119(89)90033-4. [DOI] [PubMed] [Google Scholar]
- KELLEY S. BORODA S. MUSIER-FORSYTH K. KANKIA B.I. HIV-integrase aptamer folds into a parallel quadruplex: a thermodynamic study. Biophys. Chem. 2011;155:82–88. doi: 10.1016/j.bpc.2011.03.004. [DOI] [PubMed] [Google Scholar]
- KENSCH O. CONNOLLY B.A. STEINHOFF H.J. MCGREGOR A. GOODY R.S. RESTLE T. HIV-1 reverse transcriptase-pseudoknot RNA aptamer interaction has a binding affinity in the low picomolar range coupled with high specificity. J. Biol. Chem. 2000;275:18271–18278. doi: 10.1074/jbc.M001309200. [DOI] [PubMed] [Google Scholar]
- KISSEL J.D. HELD D.M. HARDY R.W. BURKE D.H. Active site binding and sequence requirements for inhibition of HIV-1 reverse transcriptase by the RT1 family of single-stranded DNA aptamers. Nucleic Acids Res. 2007;35:5039–5050. doi: 10.1093/nar/gkm420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- KYPR J. KEJNOVSKA I. RENCIUK D. VORLICKOVA M. Circular dichroism and conformational polymorphism of DNA. Nucleic Acids Res. 2009;37:1713–1725. doi: 10.1093/nar/gkp026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LAI Y.T. DESTEFANO J.J. A primer-free method that selects high-affinity single-stranded DNA aptamers using thermostable RNA ligase. Anal. Biochem. 2011;414:246–253. doi: 10.1016/j.ab.2011.03.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LANG W. PERKINS H. ANDERSON R.E. ROYCE R. JEWELL N. WINKELSTEIN W., JR. Patterns of T lymphocyte changes with human immunodeficiency virus infection: from seroconversion to the development of AIDS. J. Acquir. Immune Defic. Syndr. 1989;2:63–69. [PubMed] [Google Scholar]
- MACAYA R.F. SCHULTZE P. SMITH F.W. ROE J.A. FEIGON J. Thrombin-binding DNA aptamer forms a unimolecular quadruplex structure in solution. Proc. Natl. Acad. Sci. U. S. A. 1993;90:3745–3749. doi: 10.1073/pnas.90.8.3745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MICHALOWSKI D. CHITIMA-MATSIGA R. HELD D.M. BURKE D.H. Novel bimodular DNA aptamers with guanosine quadruplexes inhibit phylogenetically diverse HIV-1 reverse transcriptases. Nucleic Acids Res. 2008;36:7124–7135. doi: 10.1093/nar/gkn891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MISSAILIDIS S. PERKINS A. Update: aptamers as novel radiopharmaceuticals: their applications and future prospects in diagnosis and therapy. Cancer Biother. Radiopharm. 2007;22:453–468. doi: 10.1089/cbr.2007.357. [DOI] [PubMed] [Google Scholar]
- MOK W. LI Y. Recent progress in nucleic acid aptamer-based biosensors and bioassays. Sensors. 2008;8:7050–7084. doi: 10.3390/s8117050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MOSING R.K. MENDONSA S.D. BOWSER M.T. Capillary electrophoresis-SELEX selection of aptamers with affinity for HIV-1 reverse transcriptase. Anal. Chem. 2005;77:6107–6112. doi: 10.1021/ac050836q. [DOI] [PubMed] [Google Scholar]
- NIMJEE S.M. RUSCONI C.P. SULLENGER B.A. Aptamers: an emerging class of therapeutics. Annu. Rev. Med. 2005;56:555–583. doi: 10.1146/annurev.med.56.062904.144915. [DOI] [PubMed] [Google Scholar]
- PAN W. CLAWSON G.A. Primer-free aptamer selection using a random DNA library. Methods Mol. Biol. 2010;629:369–385. doi: 10.1007/978-1-60761-657-3_24. [DOI] [PubMed] [Google Scholar]
- PAN W. XIN P. CLAWSON G.A. Minimal primer and primer-free SELEX protocols for selection of aptamers from random DNA libraries. BioTechniques. 2008;44:351–360. doi: 10.2144/000112689. [DOI] [PubMed] [Google Scholar]
- PIEVE C.D. PERKINS A.C. MISSAILIDIS S. Anti-MUC1 aptamers: radiolabelling with (99m)Tc and biodistribution in MCF-7 tumour-bearing mice. Nucl. Med. Biol. 2009;36:703–710. doi: 10.1016/j.nucmedbio.2009.04.004. [DOI] [PubMed] [Google Scholar]
- PORSCHEWSKI P. GRATTINGER M.A. KLENZKE K. ERPENBACH A. BLIND M.R. SCHAFER F. Using aptamers as capture reagents in bead-based assay systems for diagnostics and hit identification. J. Biomol. Screen. 2006;11:773–781. doi: 10.1177/1087057106292138. [DOI] [PubMed] [Google Scholar]
- RIOU J.F. GUITTAT L. MAILLIET P. LAOUI A. RENOU E. PETITGENET O. MEGNIN-CHANET F. HELENE C. MERGNY J.L. Cell senescence and telomere shortening induced by a new series of specific G-quadruplex DNA ligands. Proc. Natl. Acad. Sci. U. S. A. 2002;99:2672–2677. doi: 10.1073/pnas.052698099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- RISITANO A. FOX K.R. Influence of loop size on the stability of intramolecular DNA quadruplexes. Nucleic Acids Res. 2004;32:2598–2606. doi: 10.1093/nar/gkh598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- SAMBROOK J. RUSSELL D.W. Molecular Cloning: A Laboratory Manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor; New York: 2001. pp. 5.40–5.46. 12.74, 12.78. [Google Scholar]
- SCAGGIANTE B. MORASSUTTI C. DAPAS B. TOLAZZI G. USTULIN F. QUADRIFOGLIO F. Human cancer cell lines growth inhibition by GTn oligodeoxyribonucleotides recognizing single-stranded DNA-binding proteins. Eur. J. Biochem. 1998;252:207–215. doi: 10.1046/j.1432-1327.1998.2520207.x. [DOI] [PubMed] [Google Scholar]
- SCHNEIDER D.J. FEIGON J. HOSTOMSKY Z. GOLD L. High-affinity ssDNA inhibitors of the reverse transcriptase of type 1 human immunodeficiency virus. Biochemistry. 1995;34:9599–9610. doi: 10.1021/bi00029a037. [DOI] [PubMed] [Google Scholar]
- SIMONSSON T. G-quadruplex DNA structures: variations on a theme. Biol. Chem. 2001;382:621–628. doi: 10.1515/BC.2001.073. [DOI] [PubMed] [Google Scholar]
- SISSI C. GATTO B. PALUMBO M. The evolving world of protein-G-quadruplex recognition: a medicinal chemist's perspective. Biochimie. 2011;93:1219–1230. doi: 10.1016/j.biochi.2011.04.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- TANG Z. SHANGGUAN D. WANG K. SHI H. SEFAH K. MALLIKRATCHY P. CHEN H.W. LI Y. TAN W. Selection of aptamers for molecular recognition and characterization of cancer cells. Anal. Chem. 2007;79:4900–4907. doi: 10.1021/ac070189y. [DOI] [PubMed] [Google Scholar]
- TOHL J. EIMER W. Interaction of a G-DNA quadruplex with mono- and divalent cations. A force field calculation. Biophys. Chem. 1997;67:177–186. doi: 10.1016/s0301-4622(97)00037-9. [DOI] [PubMed] [Google Scholar]
- TONG W. LU C.-D. SHARMA S.K. MATSUURA S. SO A.G. SCOTT W.A. Nucleotide-Induced Stable Complex Formation by HIV-1 Reverse Transcriptase. Biochemistry. 1997;36:5749–5757. doi: 10.1021/bi962410z. [DOI] [PubMed] [Google Scholar]
- TUERK C. GOLD L. Systematic evolution of ligands by exponential enrichment: RNA ligands to bacteriophage T4 DNA polymerase. Science. 1990;249:505–510. doi: 10.1126/science.2200121. [DOI] [PubMed] [Google Scholar]
- TUERK C. MACDOUGAL S. GOLD L. RNA pseudoknots that inhibit human immunodeficiency virus type 1 reverse transcriptase. Proc. Natl. Acad. Sci. U. S. A. 1992;89:6988–6992. doi: 10.1073/pnas.89.15.6988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- VATER A. KLUSSMANN S. Toward third-generation aptamers: Spiegelmers and their therapeutic prospects. Curr. Opin. Drug Discov. Devel. 2003;6:253–261. [PubMed] [Google Scholar]
- WANG J. JIANG H. LIU F. In vitro selection of novel RNA ligands that bind human cytomegalovirus and block viral infection. RNA. 2000;6:571–583. doi: 10.1017/s1355838200992215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- WEN J.D. GRAY D.M. Ff gene 5 single-stranded DNA-binding protein assembles on nucleotides constrained by a DNA hairpin. Biochemistry. 2004;43:2622–2634. doi: 10.1021/bi030177g. [DOI] [PubMed] [Google Scholar]
- WOLINSKY S.M. KORBER B.T. NEUMANN A.U. DANIELS M. KUNSTMAN K.J. WHETSELL A.J. FURTADO M.R. CAO Y. HO D.D. SAFRIT J.T. Adaptive evolution of human immunodeficiency virus-type 1 during the natural course of infection [see comments] Science. 1996;272:537–542. doi: 10.1126/science.272.5261.537. [DOI] [PubMed] [Google Scholar]
- ZHANG Z. BLANK M. SCHLUESENER H.J. Nucleic acid aptamers in human viral disease. Arch. Immunol. Ther. Exp. (Warsz) 2004;52:307–315. [PubMed] [Google Scholar]
- ZHU T. COREY L. HWANGBO Y. LEE J.M. LEARN G.H. MULLINS J.I. MCELRATH M.J. Persistence of extraordinarily low levels of genetically homogeneous human immunodeficiency virus type 1 in exposed seronegative individuals. J. Virol. 2003;77:6108–6116. doi: 10.1128/JVI.77.11.6108-6116.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ZUKER M. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 2003;31:3406–3415. doi: 10.1093/nar/gkg595. [DOI] [PMC free article] [PubMed] [Google Scholar]