Glioblastoma tumor cells release microvesicles (exosomes) containing mRNA, miRNA and angiogenic proteins. These microvesicles are taken up by normal host cells, such as brain microvascular endothelial cells. By incorporating an mRNA for a reporter protein into these microvesicles we demonstrate that microvesicle-delivered messages are translated by recipient cells. These microvesicles are also enriched in angiogenic proteins and elicit tubule formation by endothelial cells. Tumor-derived microvesicles therefore serve as a novel means of delivery of genetic information as well as proteins to recipient cells in the tumor environment. Glioblastoma microvesicles also stimulated proliferation of a human glioma cell line, indicating a self-promoting aspect. Messenger RNA mutant/variants and microRNAs characteristic of gliomas can be detected in serum microvesicles of glioblastoma patients. The tumor-specific EGFRvIII was detected in serum microvesicles from 7 out of 25 glioblastoma patients. Thus, tumor-derived microvesicles may provide diagnostic information and aid in therapeutic decisions for cancer patients through a blood test.
Glioblastomas are highly malignant brain tumors with a poor prognosis despite intensive research and clinical efforts1. These tumors as well as many others have a remarkable ability to mold their stromal environment to their own advantage. Tumor cells alter surrounding normal cells to facilitate tumor cell growth, invasion, chemoresistance, immune evasion and metastasis 2–4. The tumor cells also hijack the normal vasculature and stimulate rapid formation of new blood vessels to supply tumor nutrition 5. Although the immune system can initially suppress tumor growth, it is often progressively blunted by tumor activation of immunosuppressive pathways 6. Recent studies show the importance of communication between tumor cells and their environment through shedding of membrane microvesicles which can fuse to cells in the vicinity 7.
Microvesicles are 30–100 nm in diameter and shed from many different cell types under both normal and pathological conditions 8. These exosomes can be formed through inward budding of endosomal membranes giving rise to intracellular multivesicular bodies (MVB) that later fuse with the plasma membrane, releasing the exosomes to the exterior 8,9. They can also be shed directly by outward budding of the plasma membrane, as shown for Jurkat T-cells 10.
Microvesicles in Drosophila, termed argosomes, contain morphogens such as Wingless protein and move throughout the imaginal disc epithelium in the developing embryos 11. Microvesicles found in semen, known as prostasomes, can promote sperm motility, stabilize the acrosome reaction, facilitate immunosuppression and inhibit angiogenesis 12. On the other hand, prostasomes released by malignant prostate cells promote angiogenesis. It has been shown that microvesicles can transfer some of their contents to other cell types 13–16.
The content of microvesicles and their biological function depends on the cell of origin. Microvesicles derived from B-cells and dendritic cells have potent immuno-stimulatory and antitumor effects in vivo and have been used as antitumor vaccines 17. Dendritic cell-derived microvesicles contain co-stimulatory proteins necessary for T-cell activation, whereas most tumor cell-derived microvesicles do not. Instead they act to suppress the immune response and accelerate tumor growth and invasiveness 18–21. Breast cancer microvesicles stimulate angiogenesis, and platelet-derived microvesicles promote tumor progression and metastasis of lung cancer cells 22,23.
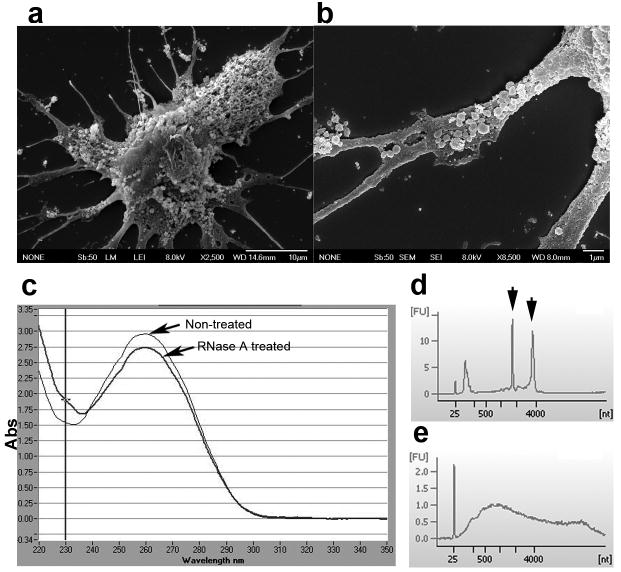
Human glioblastoma tissues were obtained from surgical resections and tumor cells were dissociated and cultured as monolayers in medium using fetal bovine serum (FBS) depleted for microvesicles (dFBS). Cultured primary cells obtained from three glioblastoma tumors were found to produce microvesicles at early and later passages (1–15 passages). Tumor cells were covered with microvesicles varying in size from about 50 – 500 nm (Fig. 1a and b). The microvesicles contained RNA and protein in an approximate ratio of 1:80. To evaluate whether the RNA was contained inside the microvesicles, they were either exposed to RNase A or left untreated before RNA extraction (Fig. 1c). There was always less than a 7% decrease in RNA content following RNase treatment. Thus, it appears that almost all of the RNA is contained within the vesicles and is thereby protected from external RNases by the surrounding membrane. Bioanalysis of RNA from microvesicles and their donor cells revealed that the microvesicles contain a broad range of RNA sizes consistent with a variety of mRNAs and miRNAs, but lack the ribosomal RNA peaks characteristic of cellular RNA (Fig. 1d and e).
Figure 1. Glioblastoma cells produce microvesicles containing RNA.
Scanning EM image of a primary glioblastoma cell (bar = 10 μm). (b) Higher magnification showing the microvesicles on the cell surface. Vesicles can be binned into diameters of around 50 nm and 500 nm (bar = 1 μm). (c) Microvesicles were exposed to RNase A or mock-treated prior to RNA isolation and levels of RNA determined (n = 5). (d) Bioanalyzer data shows the size distribution of total RNA extracted from primary glioblastoma cells and (e) microvesicles isolated from them. The smallest peak represents an internal standard. The two prominent peaks in (d) (arrows) represent 18S (left) and 28S (right) ribosomal RNA, absent in microvesicles.
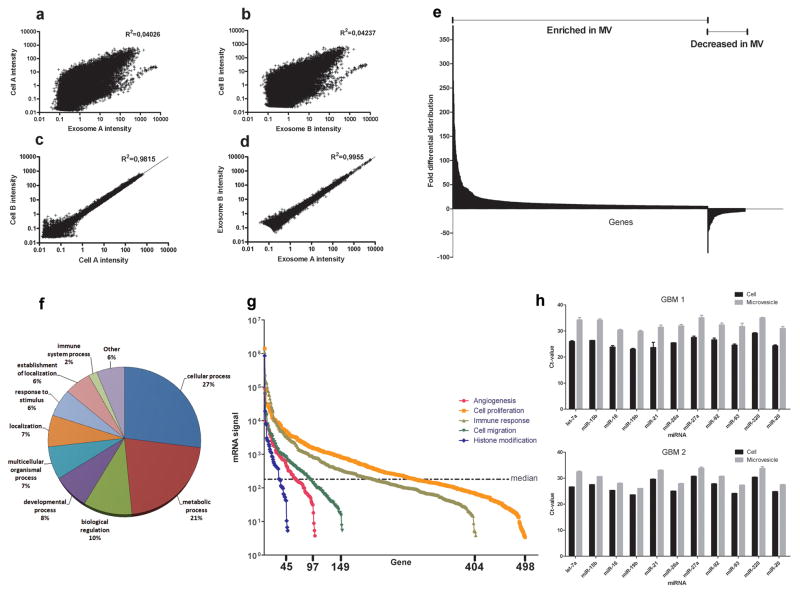
Microarray analysis of mRNA populations in microvesicles and their donor glioblastoma cells was performed using the Agilent 44K whole genome microarray. Approximately 22,000 gene transcripts were found in the cells and 27,000 transcripts in the microvesicles (detected at well above background levels, 99% confidence interval) on both arrays. Approximately 4,700 different mRNAs were detected exclusively in microvesicles on both arrays, indicating a selective enrichment process within the microvesicles (Supplementary Table 1). Consistent with this, there was a poor overall correlation in levels of mRNAs in the cells as compared to microvesicles from two tumor preparations (Fig. 2a and b), supporting selective enrichment of some cellular mRNAs in microvesicles. In contrast, a comparison of levels of specific mRNAs in different preparations of donor cells or of microvesicles showed a strong correlation, indicating a consistent distribution within these distinct cellular compartments (Fig. 2c and d). We found 3426 transcripts differentially distributed more than 5-fold (p-value <0.01). Of these, 2238 transcripts were enriched (up to 380-fold) and 1188 transcripts were less abundant than in the cells (up to 90-fold) (Fig. 2e). The intensities and ratios of all gene transcripts are shown in Supplementary Table 2. Ontologies of mRNA transcripts enriched or reduced more than 10-fold are listed in Supplementary Table 3.
Figure 2. Characterization of the microvesicle RNA.
(a, b) Scatterplots of mRNA levels in the microvesicles compared to donor cells from two different experiments. Linear regressions showed that levels in cells versus microvesicles were not well correlated. (c, d) In contrast, mRNA intensities in two different cell or two different microvesicle preparations were closely correlated. (e) 3426 genes were found to be more than 5-fold differentially distributed in the microvesicles as compared to the cells from which they were derived (p-value <0.01). (f) The biological process ontology of the 500 most abundant mRNA species in the microvesicles is displayed. (g) The intensity of microvesicle RNAs belonging to ontologies related to tumor growth is shown with the x-axis representing the number of mRNA transcripts present in the ontology. The median intensity levels on the arrays were 182. (h) Levels of mature miRNAs in microvesicles and glioblastoma cells from two different patients (GBM1 and GBM2) were analysed using quantitative miRNA RT-PCR. The cycle threshold (Ct) value is presented as the mean ± SEM (n = 4).
The mRNA transcripts that are highly enriched in microvesicles compared to cells are not always the most abundant in the microvesicles. The most abundant transcripts would be more likely to generate an effect in the recipient cell upon delivery. The 500 most abundant mRNA transcripts in microvesicles were divided into different biological processes based on their ontology descriptions and displayed in Fig. 2f. Glioblastoma microvesicle mRNAs belonging to ontologies such as angiogenesis, cell proliferation, immune response, cell migration and histone modification were plotted to compare their levels and contribution to the mRNA spectrum (Fig. 2g). These ontologies were selected as they represent functions that could be involved in remodelling the tumor stroma and enhancing tumor growth. All five ontologies contained mRNA with very high expression levels compared to the median signal intensity level of the array.
Mature miRNA in microvesicles and donor cells was detected using quantitative miRNA reverse transcription PCR. A subset of 11 miRNAs known to be abundant in gliomas (Krichevsky et al., in preparation) was readily detected in donor cells and microvesicles from two different primary glioblastomas (GBM 1 and GBM 2) (Fig. 2h). The levels were generally lower in microvesicles per μg total RNA than in parental cells (10%, corresponding to approximate 3 Ct-values), but correlated well with the tumor profile.
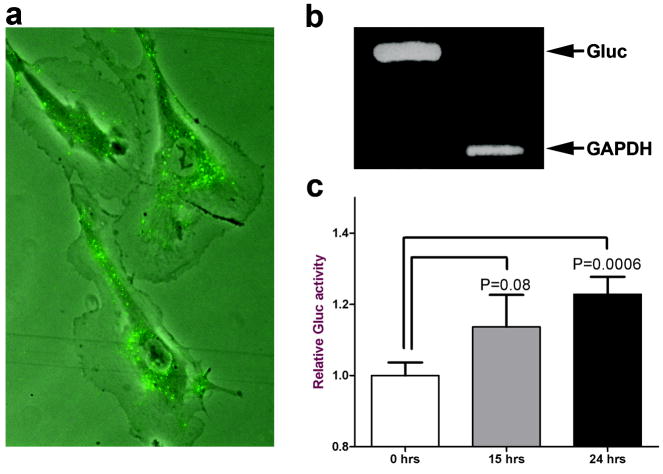
Glioblastoma microvesicles labelled with the fluorescent dye PKH67 were incubated with human brain microvascular endothelial cells (HBMVEC) in culture. The PKH67-labelled microvesicles were internalized into endosome-like structures by brain endothelial cells (Fig. 3a). Similar results were obtained when adding the fluorescently labelled microvesicles to primary glioblastoma cells (data not shown).
Figure 3. Glioblastoma microvesicles can deliver functional RNA to HBMVECs.
(a) Purified microvesicles were labelled with membrane dye PKH67 (green) and added to HBMVECs. The microvesicles were internalised into endosome-like structures within an hr. (b) Microvesicles were isolated from glioblastoma cells stably expressing Gluc. RNA extraction and RTPCR of Gluc and GAPDH mRNAs showed that both were incorporated into microvesicles. (c) Microvesicles were then added to HBMVECs and incubated for 24 hrs. The Gluc activity was measured in the medium at 0, 15 and 24 hrs after microvesicle addition and normalized to Gluc activity in microvesicles. The results are presented as the mean ± SEM (n = 4).
To determine if the mRNA delivered by glioblastoma-derived microvesicles could be expressed in recipient cells, glioblastoma cells were first transduced with a lentivirus vector encoding a secreted luciferase from Gaussia (Gluc) 24, and microvesicles produced by them were purified from conditioned medium. RT-PCR analysis showed that the mRNA for Gluc (555 bp product), as well as GAPDH (226 bp product), were present in the microvesicles (Fig. 3b). Purified microvesicles containing Gluc mRNA were added to HBMVEC cells and Gluc activity released into the medium by these endothelial cells was monitored over time (Fig. 3c). Gluc activity produced by recipient cells showed a continuing increase over 24 hrs, supporting ongoing translation of the Gluc mRNA. This novel method shows that mRNA incorporated into the tumor microvesicles can be delivered into recipient normal cells and generate a functional protein.
Nucleic acids are of high value as biomarkers because of highly sensitive PCR detection. We evaluated whether the RNA in microvesicles could be used as biomarkers for glioblastoma tumors. The epidermal growth factor receptor (EGFR) mRNA is particularly interesting since expression of the EGFRvIII mutant/variant is specific to some tumors and defines a clinical subtype of glioma 25. We used a nested RT-PCR to determine if EGFRvIII mRNA was found in resected glioma tissue and compared the result with microvesicles purified from a frozen serum sample from the same patient. The samples were coded and the PCRs were performed in a blind fashion. Fourteen of the 30 tumor samples (47%) contained the EGFRvIII transcript, which is consistent with the percentage of glioblastomas found to contain this mutant message in other studies 26. EGFRvIII could be amplified from microvesicles in seven of the 25 patients (28%) from whom serum was drawn around the time of surgery (Table 1; Supplementary Fig. 1). Interestingly, two patients with an EGFRvIII-negative tumor sample turned out to be positive in the serum microvesicles, supporting heterogeneous foci of EGFRvIII expression in the glioma tumor. EGFRvIII message was not detected in five serum samples drawn two weeks after extensive resection of the tumor, with four corresponding to EGFRvIIIpositive tumors, consistent with this tumor being the source of microvesicles. Furthermore, EGFRvIII was not found in serum exosomes from 30 normal control individuals (Supplementary Fig. 2). We also found that miRNA-21, known to be over-expressed in glioblastoma tumors27, was elevated in serum microvesicles from these patients as compared to controls (Supplementary Fig. 3). The identification of tumor-specific RNAs in serum microvesicles thus provides a window into somatic mutations and changes in gene expression in the tumor cells.
Table 1. RNA in glioblastoma microvesicles can be used as sensitive biomarkers.
Nested RT-PCR was used to monitor EGFRvIII mRNA in glioma tissue and exosomes purified from a frozen serum sample from the same patient. Samples from 30 patients were analysed in a blinded fashion and PCR reactions were repeated at least three times for each sample. No EGFRvIII mRNA was found in serum microvesicles from 30 normal controls.
| Patient# | Time of serum collection* | Serum volume | Biopsy EGFRvIII | Serum exosome EGFRvIII |
|---|---|---|---|---|
| 1 | 0 | 3 ml | Yes | Yes |
| 2 | 0 | 2 ml | No | No |
| 3 | 0 | 2.5 ml | No | No |
| 4 | 0 | 1 ml | Yes | No |
| 5 | 0 | 1 ml | Yes | No |
| 6 | 0 | 1 ml | No | No |
| 7 | 0 | 0.6 ml | Yes | Yes |
| 8 | 0 | 1 ml | No | No |
| 9 | 0 | 1 ml | Yes | Yes |
| 10 | 0 | 1 ml | No | Yes |
| 11 | 0 | 2 ml | Yes | No |
| 12 | 0 | 2 ml | Yes | Yes |
| 13 | 0 | 2 ml | No | Yes |
| 14 | 0 | 2 ml | Yes | Yes |
| 15 | 0 | 2 ml | No | No |
| 16 | 0 | 2 ml | No | No |
| 17 | 0 | 1 ml | Yes | No |
| 18 | 0 | 0.8 ml | Yes | No |
| 19 | 0 | 1 ml | No | No |
| 20 | 0 | 1 ml | No | No |
| 21 | 0 | 1 ml | No | No |
| 22 | 0 | 1 ml | No | No |
| 23 | 0 | 1 ml | No | No |
| 24 | 0 | 1 ml | No | No |
| 25 | 0 | 1 ml | No | No |
| 26 | 14 | 0.6 ml | Yes | No |
| 27 | 14 | 1.2 ml | No | No |
| 28 | 14 | 0.8 ml | Yes | No |
| 29 | 14 | 0.9 ml | Yes | No |
| 30 | 14 | 0.6 ml | Yes | No |
Days post-surgery
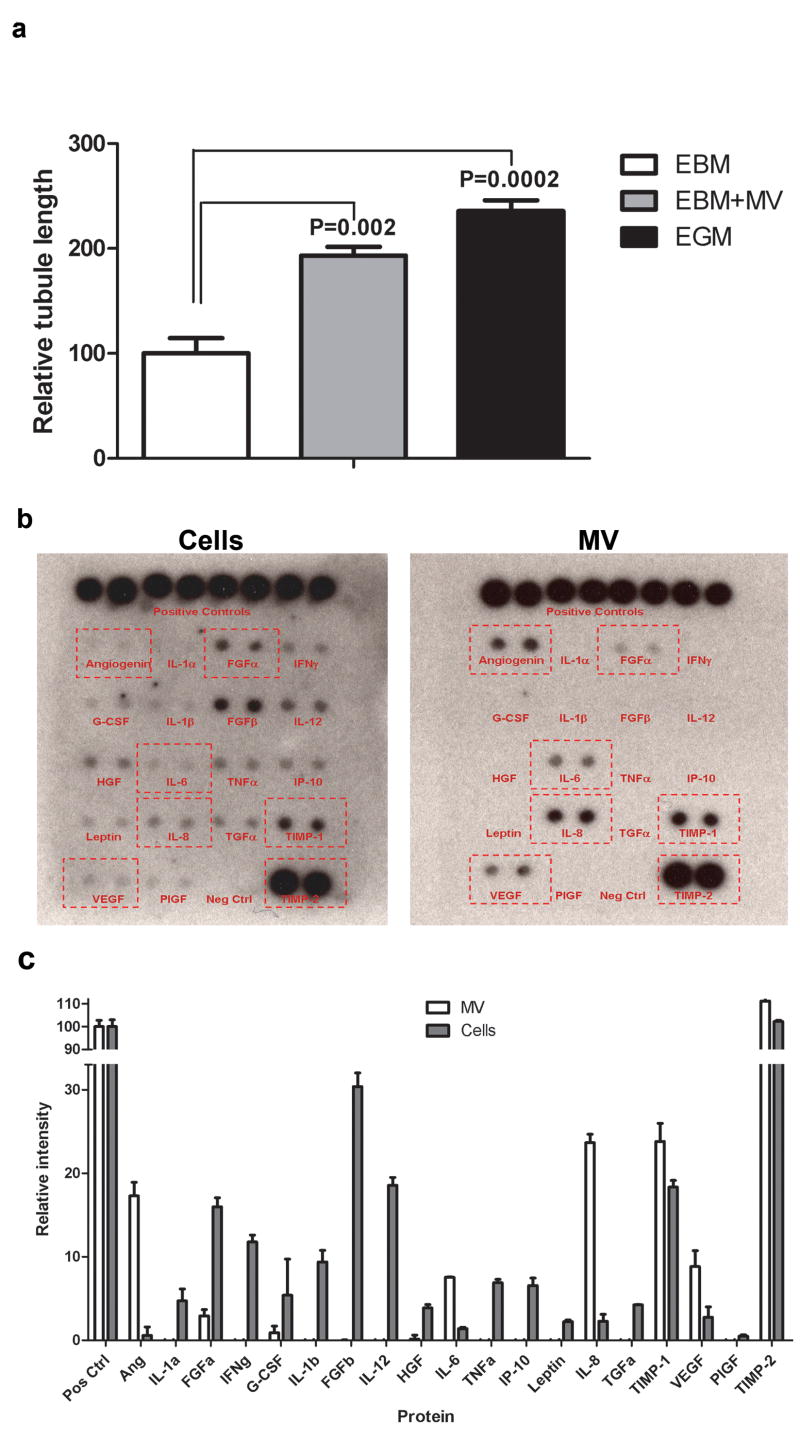
To address whether glioblastoma microvesicles could contribute to angiogenesis, we used an in vitro angiogenesis assay. HBMVECs were cultured in matrigel-coated plates in endothelial basal medium (EBM), EBM supplemented purified glioblastoma microvesicles, or EBM plus angiogenic growth factors (EGM). In the presence of microvesicles there was a doubling of tubule length by the HBMVECs within 16 hrs, comparable to when exposed to angiogenic factors (Fig. 4a). This finding supports a role for glioblastoma-derived microvesicles in initiating angiogenesis in brain endothelial cells.
Figure 4. Glioblastoma microvesicles stimulate angiogenesis in vitro and contain angiogenic proteins.
a) HBMVECs were cultured on Matrigel™ in basal medium (EBM) alone, or supplemented with GBM microvesicles (EBM+MV) or angiogenic factors (EGM). Tubule formation was measured after 16 hrs as average tubule length ± SEM compared to cells grown in EBM (n = 6). (b) Total protein from primary glioblastoma cells and microvesicles (MV) from them (1 mg each) was analysed on a human angiogenesis antibody array. (c) The arrays were scanned and the intensities analysed with the ImageJ software (n = 4).
To further characterise the angiogenic capability of microvesicles we analyzed levels of angiogenic proteins in microvesicles and compared them with levels in glioblastoma donor cells using a human angiogenesis antibody array (Fig. 4b). Seven of the 19 angiogenic proteins were readily detected in the microvesicles, with 6 of them (angiogenin, IL-6, IL-8, TIMP-1, VEGF and TIMP-2) being at higher levels on a total protein basis than in glioblastoma cells (Fig. 4c). The three most enriched angiogenic proteins were angiogenin, IL-6 and IL-8, all of which have been implicated in glioma angiogenesis and increased malignancy 28–30. This indicates that the angiogenic effect of microvesicles is mediated at least in part by angiogenic proteins.
Human U87 glioma cells were incubated in normal growth medium or medium supplemented with microvesicles isolated from primary glioblastoma cells. After three days, untreated U87 cells had increased 5-fold in number whereas the microvesicle supplemented cells had increased 8-fold (Supplementary Fig. 4). Thus, glioblastoma microvesicles appear to stimulate proliferation of other glioma cells.
These studies support the ability of microvesicles shed from tumor cells to serve as a means whereby tumors can manipulate their environment in order to make it more permissive to tumor growth and invasion. In this study we document the abundant shedding of microvesicles by primary human glioblastoma cells. We characterized mRNA and miRNA profiles present in these cells and microvesicles and showed that particular mRNAs are highly enriched in these microvesicles as compared to donor cells. In fact, more mRNA transcripts were detected well above background in microvesicles as compared to cells. This difference could be due in part to the large amount of ribosomal RNA in the cells compared to the microvesicles, increasing the relative amount of mRNA/μg total RNA in the microvesicles. Ontology analysis showed that a number of mRNA transcripts associated with cell migration, angiogenesis, cell proliferation, immune response and histone modification are present in high levels in the microvesicles. The miRNAs in microvesicles appeared to parallel their distribution in the glioblastoma cells.
We have shown that glioblastoma microvesicles can enter HMVECs and translate a reporter mRNA carried by the microvesicles. This suggests that the tumor-derived microvesicles can modify the surrounding normal cells by changing their translational profile. Further, we have shown that glioblastoma microvesicles can stimulate an angiogenic phenotype in normal brain endothelial cells and can stimulate the proliferation of other glioma cells. In addition to the potential role of mRNAs in these processes, microvesicles also contain a number of angiogenic proteins, such as angiogenin, FGF., IL-6, IL-8, TIMP-1, VEGF and TIMP-2. Most of these presumably interact with cognate receptors on the surface of endothelial cells to promote angiogenesis, and may require extracellular lysis of the microvesicles with release of proteins contained within them. It has been proposed that the acidic environment in established tumors can promote lysis of some of the microvesicles, making intravesicular proteins bioavailable 31. On the other hand, angiogenic proteins like angiogenin require transportation across the membrane for biological effect 32, which could be facilitated by the microvesicles. Tumor microvesicles thus act as a multicomponent delivery vehicle for mRNA, miRNA and proteins to communicate genetic information as well as signalling proteins to cells in their environs.
This study presents a thorough analysis of mRNAs that are enriched in the microvesicles versus donor cells, suggesting that there may be a cellular mechanism for localizing these messages into microvesicles, possibly via a zip code in the 3′UTR as described for mRNAs translated in specific cellular locations, e.g. beta actin 33. The conformation of the mRNAs in the microvesicles is not known, but they may be present as ribonuclear particles (RNPs) 34. Evidence suggests that retroviruses, like HIV, can utilize the endogenous microvesicle machinery for budding and generation of new virus particles 10. Interestingly, several endogenous retrovirus RNA sequences were found to be highly enriched in the microvesicles (Supplementary Table 2).
The RNA found in the microvesicles contains a “snapshot” of a substantial array of the cellular transcriptome at any given time. Among the mRNAs found in glioblastoma-derived microvesicles, the EGFR mRNA is of specific interest since the EGFRvIII mutant splice variant is found specifically in many glioblastomas 26. Using nested RT-PCR we were able to detect the EGFRvIII message in tumor biopsies and serum microvesicles from glioblastoma patients, but not in any of 30 normal control sera. We showed that brain tumors release microvesicles into the bloodstream and that we can genetically type EGFRvIII status of glioblastoma tumors by nested RT PCR of RNA in microvesicles isolated from a small amount of patient serum as compared to other methods that require invasive brain surgery. The sensitivity of this assay may depend on factors as tumor size, tumor location and serum volume, as well as the method of RNA extraction, cDNA conversion and PCR used. Information about EGFRvIII status of glioblastoma patients could be useful in the ongoing EGFRvIII vaccine and other therapeutic clinical trials 35. One study showed that EGFRvIII-positive gliomas are over 50 times more likely to respond to treatment with EGFR-inhibitors like erlotinib (Tarceva®) or gefitinib (Iressa®) 36. Thus, we propose a new way of looking at molecular determinants of cancer, including but not limited to EGFRvIII, by isolating microvesicles from serum and extracting the RNA for profiling and detection of mutations, splice variants and levels of mRNAs and miRNAs characteristic of tumor formation, progression and response to therapy.
Microvesicles may provide a means of detecting evolving genetic changes relative to tumor progression using serum samples drawn over time, presumably no matter what type of cancer or where the tumor foci are situated in the individual. Further, knowledge of tumor genotype and phenotype gained through microvesicle analysis may help in designing tailored therapies to curtail tumor growth. And lastly, microvesicles may prove useful as a delivery vehicle for therapeutic RNAs and proteins.
Materials and methods
Collection of tumor samples and serum from glioblastoma patients
For cell culture, brain tumor specimens from patients diagnosed by a neuropathologist as glioblastoma multiforme were taken directly from surgery and placed in cold sterile Neurobasal media (Invitrogen, Carlsbad, CA, USA). The specimens were dissociated into single cells within 1 hr from the time of surgery using a Neural Tissue Dissociation Kit (Miltenyi Biotech, Bergisch Gladbach, Germany) and plated in DMEM 5% microvesicle-depleted dFBS (prepared by ultracentrifugation at 110,000 × g for 16 hrs to remove bovine microvesicles) supplemented with penicillin-streptomycin (10 IU ml−1 and 10 μg ml−1, respectively, Sigma-Aldrich, St Louis, MO, USA). Matched de-identified frozen tumor and serum samples from confirmed glioblastoma patients were obtained from the Department of Neurosurgery (Massachusetts General Hospital, Boston, USA and the Cancer Research Center; VU Medical Center, Amsterdam, The Netherlands). These samples were kept at −80°C until use.
Scanning EM
Human glioblastoma cells were placed on ornithine-coated coverslips, fixed in 0.5 × Karnovskys fixative and then washed 2×5 min with PBS. The cells were dehydrated in 35% EtOH 10 min, 50% EtOH 2×10 min, 70% EtOH 2×10 min, 95% EtOH 2×10 min, 100% EtOH 4×10 min and then transferred to a Tousimis SAMDRI-795 semi-automatic Critical Point Dryer followed by coating with chromium in a GATAN Model 681 High Resolution Ion Beam Coater.
Microvesicle isolation
Glioblastoma cells at passage 1–15 were cultured in microvesicle-free media (DMEM containing 5% dFBS). The conditioned medium from 40 million cells was harvested after 48 hrs. The microvesicles were purified by differential centrifugation 15. In brief, glioblastomaconditioned medium was centrifuged for 10 min at 300 × g to eliminate cell contamination. Supernatants were further centrifuged for 20 min at 16,500 × g and filtered through a 0.22 μm filter. Microvesicles were pelleted by ultracentrifugation at 110,000 × g for 70 min. The microvesicle pellets were washed in 13 ml PBS, pelleted again and resuspended in PBS. Exosomes were measured for their protein content using DC Protein Assay (Bio-Rad, Hercules, CA, USA). Serum exosomes from healthy controls and glioblastoma patients were diluted up to 13 ml in PBS and sterile filtered before centrifugation.
RNA isolation
To evaluate whether RNA was present inside the microvesicles, RNase A (Fermentas, Glen Burnie, MD, USA) was added to suspensions of microvesicles at a final concentration of 100 μg/ml and incubated for 15 min at 37°C. Total RNA was then purified using the MirVana RNA isolation kit (Ambion, Austin TX, USA) according to the manufacturer’s protocol. The RNA was quantified using a nanodrop ND-1000 (Thermo Fischer Scientific, Wilmington, DE, USA). Snap frozen tumor biopsies were thawed on RNAlater ICE (Ambion, Austin TX, USA) according to manufacturer’s recommendation followed by RNA extraction using the MirVana RNA isolation kit.
Microarray analysis
The microarray experiments were performed by Miltenyi Biotech (Auburn, CA, USA) using the Agilent Whole Human Genome Microarray, 4×44K, two color array. The array was performed on two different RNA preparations from primary glioblastoma cells and their microvesicles. The data was analysed using the GeneSifter software (Vizxlabs, Seattle, WA, USA). The Intersector software (Vizxlabs) was used to extract the genes readily detected on both arrays.
Quantitative miRNA RT-PCR
Total RNA was isolated using the mirVana RNA isolation kit. Total RNA (30 ng) was converted into cDNA using specific miR-primers (Applied Biosystems, Foster City, CA, USA) and further amplified according to the manufacturer’s protocol.
HBMVEC in vitro angiogenesis assay
HBMVECs (30,000) (Cell Systems, Catalogue #ACBRI-376, Kirkland, WA, USA) were cultured on Matrigel-coated wells in a 24-well plate in basal medium (Lonza Biologics Inc., Portsmouth, NH, USA) only, or supplemented with glioblastoma microvesicles (7 μg/well) or a cocktail of angiogenic factors (EGM; hydrocortisone, EGF, FGF, VEGF, IGF, ascorbic acid, FBS, and heparin; Singlequots from Lonza). Tubule formation was measured after 16 hrs and analyzed with the ImageJ software (NIH).
Gluc mRNA translation assay
Primary human glioblastoma cells were infected with a selfinactivating lentivirus vector expressing secreted Gluc under a cytomegalovirus promoter 37 to achieve an infection efficiency of >95%. The cells were stably transduced and microvesicles generated during the subsequent passages (2–10) were isolated and purified as above. Microvesicles (50 μg) were added to 50,000 HBMVEC and incubated for 24 hrs. The Gluc activity in the supernatant was measured directly after microvesicle addition (0 hrs), and 15 hrs and 24 hrs later and normalised to the Gluc activity in the microvesicles. The results are presented as the mean ± SEM (n = 4).
PKH67 labelled microvesicle
Purified glioblastoma microvesicles were labelled with PKH67 Green Fluorescent labelling kit (Sigma-Aldrich, St Louis, MO, USA) as described 21. The labelled microvesicles were incubated with HBMVEC in culture (5 μg/50,000 cells). Microvesicles were allowed to bind for 20 min at 4°C and cells were then washed and incubated at 37°C for 1 hr.
RT PCR and nested PCR
RNA was extracted using the MirVana RNA isolation kit. RNA was converted to cDNA using the Omniscript RT kit (if starting material was >50 ng) or Sensiscript RT kit (if starting material was <50 ng) (Qiagen Inc., Valencia, CA, USA) using a mix of oligo dT and random hexamer primer according to manufacturer’s recommendation. The following PCR primers were used: GAPDH primers; Forw 5′-GAA GGT GAA GGT CGG AGT C-3′, Reverse 5′-GAA GAT GGT GAT GGG ATT TC-3′. EGFR/EGFRvIII PCR1; Forw 5′-CCAGTATTGATCGGGAGAGC-3′, Reverse 5′-TCAGAATATCCAGTTCCTGTGG-3′, EGFR/EGFRvIII PCR2; Forw 5′-ATG CGA CCC TCC GGG ACG-3′, Reverse 5′-GAG TAT GTG TGA AGG AGT-3′. The Gluc primers have been described previously 24. PCR protocol: 94°C 3 min; 94°C 45 s, 60°C 45 s, 72°C 2 min × 35 cycles; 72°C 7 min.
Angiogenesis antibody array
One mg total protein from either primary glioblastoma cells or purified microvesicles isolated from the same cells were lysed in Promega lysis buffer (Promega, Madison, WI, USA) and then added to the human angiogenesis antibody array (Panomics, Fremont, CA, USA) according to manufacturer’s recommendations. The arrays were scanned and analysed with the ImageJ software (NIH).
Statistics
The statistical analyses were performed using Students t-test.
Supplementary Material
Acknowledgments
We wish to express our gratitude to Dr. B. Tannous for supplying the Gluc lentivirus construct, Drs. C. Maguire, M. Broekman, K. Miranda, L. Russo and O. Saydam for fruitful discussions. We also would like to thank Applied Biosystems for supplying the miRNA qRT-PCR primers and Dr. Idema (Neuro-oncology Research Group, Cancer Center Amsterdam) for supplying some of the serum/biopsy samples. This work was kindly supported by the Wenner-Gren Foundation (JS) Stiftelsen Olle Engkvist Byggmästare (JS), NCI CA86355 (XOB & MSE), NCI CA69246 (XOB, MSE & RSC), the Brain Tumor Society (AMK) and the American Brain Tumor Association (TW).
References
- 1.Stupp R, et al. Radiotherapy plus concomitant and adjuvant temozolomide for glioblastoma. The New England journal of medicine. 2005;352:987–996. doi: 10.1056/NEJMoa043330. [DOI] [PubMed] [Google Scholar]
- 2.Mazzocca A, et al. Cancer research. 2005;65:4728–4738. doi: 10.1158/0008-5472.CAN-04-4449. [DOI] [PubMed] [Google Scholar]
- 3.Muerkoster S, et al. Tumor stroma interactions induce chemoresistance in pancreatic ductal carcinoma cells involving increased secretion and paracrine effects of nitric oxide and interleukin-1beta. Cancer research. 2004;64:1331–1337. doi: 10.1158/0008-5472.can-03-1860. [DOI] [PubMed] [Google Scholar]
- 4.Singer CF, et al. Differential gene expression profile in breast cancer-derived stromal fibroblasts. Breast Cancer Res Treat. 2007 doi: 10.1007/s10549-007-9725-2. [DOI] [PubMed] [Google Scholar]
- 5.Carmeliet P, Jain RK. Angiogenesis in cancer and other diseases. Nature. 2000;407:249–257. doi: 10.1038/35025220. [DOI] [PubMed] [Google Scholar]
- 6.Gabrilovich DI. Molecular mechanisms and therapeutic reversal of immune suppression in cancer. Current cancer drug targets. 2007;7:1. [PubMed] [Google Scholar]
- 7.Ratajczak J, Wysoczynski M, Hayek F, Janowska-Wieczorek A, Ratajczak MZ. Membrane-derived microvesicles: important and underappreciated mediators of cell-to-cell communication. Leukemia. 2006;20:1487–1495. doi: 10.1038/sj.leu.2404296. [DOI] [PubMed] [Google Scholar]
- 8.Thery C, Zitvogel L, Amigorena S. Exosomes: composition, biogenesis and function. Nature reviews. 2002;2:569–579. doi: 10.1038/nri855. [DOI] [PubMed] [Google Scholar]
- 9.Pan BT, Johnstone RM. Fate of the transferrin receptor during maturation of sheep reticulocytes in vitro: selective externalization of the receptor. Cell. 1983;33:967–978. doi: 10.1016/0092-8674(83)90040-5. [DOI] [PubMed] [Google Scholar]
- 10.Booth AM, et al. Exosomes and HIV Gag bud from endosome-like domains of the T cell plasma membrane. The Journal of cell biology. 2006;172:923–935. doi: 10.1083/jcb.200508014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Greco V, Hannus M, Eaton S. Argosomes: a potential vehicle for the spread of morphogens through epithelia. Cell. 2001;106:633–645. doi: 10.1016/s0092-8674(01)00484-6. [DOI] [PubMed] [Google Scholar]
- 12.Delves GH, Stewart AB, Cooper AJ, Lwaleed BA. Prostasomes, angiogenesis, and tissue factor. Seminars in thrombosis and hemostasis. 2007;33:75–79. doi: 10.1055/s-2006-958465. [DOI] [PubMed] [Google Scholar]
- 13.Mack M, et al. Nature medicine. 2000;6:769–775. doi: 10.1038/77498. [DOI] [PubMed] [Google Scholar]
- 14.Al-Nedawi K, et al. Nature cell biology. 2008;10:619–624. doi: 10.1038/ncb1725. [DOI] [PubMed] [Google Scholar]
- 15.Valadi H, et al. Exosome-mediated transfer of mRNAs and microRNAs is a novel mechanism of genetic exchange between cells. Nature cell biology. 2007;9:654–659. doi: 10.1038/ncb1596. [DOI] [PubMed] [Google Scholar]
- 16.Baj-Krzyworzeka M, et al. Tumour-derived microvesicles carry several surface determinants and mRNA of tumour cells and transfer some of these determinants to monocytes. Cancer Immunol Immunother. 2006;55:808–818. doi: 10.1007/s00262-005-0075-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chaput N, Taieb J, Andre F, Zitvogel L. The potential of exosomes in immunotherapy. Expert opinion on biological therapy. 2005;5:737–747. doi: 10.1517/14712598.5.6.737. [DOI] [PubMed] [Google Scholar]
- 18.Wieckowski E, Whiteside TL. Human tumor-derived vs dendritic cell-derived exosomes have distinct biologic roles and molecular profiles. Immunologic research. 2006;36:247–254. doi: 10.1385/IR:36:1:247. [DOI] [PubMed] [Google Scholar]
- 19.Clayton A, Mitchell JP, Court J, Mason MD, Tabi Z. Human tumor-derived exosomes selectively impair lymphocyte responses to interleukin-2. Cancer research. 2007;67:7458–7466. doi: 10.1158/0008-5472.CAN-06-3456. [DOI] [PubMed] [Google Scholar]
- 20.Ginestra A, et al. The amount and proteolytic content of vesicles shed by human cancer cell lines correlates with their in vitro invasiveness. Anticancer research. 1998;18:3433–3437. [PubMed] [Google Scholar]
- 21.Liu C, et al. J Immunol. 2006;176:1375–1385. doi: 10.4049/jimmunol.176.3.1375. [DOI] [PubMed] [Google Scholar]
- 22.Janowska-Wieczorek A, et al. Microvesicles derived from activated platelets induce metastasis and angiogenesis in lung cancer. International journal of cancer. 2005;113:752–760. doi: 10.1002/ijc.20657. [DOI] [PubMed] [Google Scholar]
- 23.Millimaggi D, et al. Tumor vesicle-associated CD147 modulates the angiogenic capability of endothelial cells. Neoplasia New York, NY. 2007;9:349–357. doi: 10.1593/neo.07133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tannous BA, Kim DE, Fernandez JL, Weissleder R, Breakefield XO. Codon-optimized Gaussia luciferase cDNA for mammalian gene expression in culture and in vivo. Mol Ther. 2005;11:435–443. doi: 10.1016/j.ymthe.2004.10.016. [DOI] [PubMed] [Google Scholar]
- 25.Pelloski CE, et al. J Clin Oncol. 2007;25:2288–2294. doi: 10.1200/JCO.2006.08.0705. [DOI] [PubMed] [Google Scholar]
- 26.Nishikawa R, et al. Immunohistochemical analysis of the mutant epidermal growth factor, deltaEGFR, in glioblastoma. Brain tumor pathology. 2004;21:53–56. doi: 10.1007/BF02484510. [DOI] [PubMed] [Google Scholar]
- 27.Chan JA, Krichevsky AM, Kosik KS. MicroRNA-21 is an antiapoptotic factor in human glioblastoma cells. Cancer research. 2005;65:6029–6033. doi: 10.1158/0008-5472.CAN-05-0137. [DOI] [PubMed] [Google Scholar]
- 28.Brat DJ, Bellail AC, Van Meir EG. The role of interleukin-8 and its receptors in gliomagenesis and tumoral angiogenesis. Neuro-oncology. 2005;7:122–133. doi: 10.1215/S1152851704001061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Eberle K, et al. The expression of angiogenin in tissue samples of different brain tumours and cultured glioma cells. Anticancer research. 2000;20:1679–1684. [PubMed] [Google Scholar]
- 30.Rolhion C, et al. Journal of neurosurgery. 2001;94:97–101. doi: 10.3171/jns.2001.94.1.0097. [DOI] [PubMed] [Google Scholar]
- 31.Taraboletti G, et al. Bioavailability of VEGF in tumor-shed vesicles depends on vesicle burst induced by acidic pH. Neoplasia New York, NY. 2006;8:96–103. doi: 10.1593/neo.05583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Xu ZP, Tsuji T, Riordan JF, Hu GF. Identification and characterization of an angiogeninbinding DNA sequence that stimulates luciferase reporter gene expression. Biochemistry. 2003;42:121–128. doi: 10.1021/bi020465x. [DOI] [PubMed] [Google Scholar]
- 33.Kislauskis EH, Zhu X, Singer RH. Sequences responsible for intracellular localization of beta-actin messenger RNA also affect cell phenotype. The Journal of cell biology. 1994;127:441–451. doi: 10.1083/jcb.127.2.441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mallardo M, et al. Isolation and characterization of Staufen-containing ribonucleoprotein particles from rat brain. Proceedings of the National Academy of Sciences of the United States of America. 2003;100:2100–2105. doi: 10.1073/pnas.0334355100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sonabend AM, Dana K, Lesniak MS. Targeting epidermal growth factor receptor variant III: a novel strategy for the therapy of malignant glioma. Expert review of anticancer therapy. 2007;7:S45–50. doi: 10.1586/14737140.7.12s.S45. [DOI] [PubMed] [Google Scholar]
- 36.Mellinghoff IK, et al. Molecular determinants of the response of glioblastomas to EGFR kinase inhibitors. The New England journal of medicine. 2005;353:2012–2024. doi: 10.1056/NEJMoa051918. [DOI] [PubMed] [Google Scholar]
- 37.Badr CE, Hewett JW, Breakefield XO, Tannous BA. A highly sensitive assay for monitoring the secretory pathway and ER stress. PLoS ONE. 2007;2:e571. doi: 10.1371/journal.pone.0000571. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.