Abstract
Bacterial plasmids propagate through microbial populations via the directed process of conjugative plasmid transfer (CPT). Because conjugative plasmids often encode antibiotic resistance genes and virulence factors, several approaches to inhibit CPT have been described. Bisphosphonates and structurally related compounds (BSRCs) were previously reported to disrupt conjugative transfer of the F (fertility) plasmid in E. coli. We have further investigated the effect of these compounds on the transfer of two additional conjugative plasmids, pCU1 and R100, between E. coli. The impact of BSRCs on E. coli survival and plasmid transfer was found to be dependent on the plasmid type, the length of time the E. coli were exposed to the compounds, and the ratio of plasmid donor to plasmid recipient cells. Therefore, these data indicate that BSRCs produce a range of effects on the conjugative transfer of bacterial plasmids in E. coli. Since their impact appears to be plasmid type-dependent, BSRCs are unlikely to be applicable as broad inhibitors of antibiotic resistance propagation.
Keywords: Conjugative Plasmid Transfer, F plasmid, Bisphosphonate, Antibiotic Resistance, Relaxase
Introduction
The horizontal transfer of genetic material between bacterial cells increases the genetic diversity of bacteria. For example, horizontal gene transfer (HGT) can lead to the rapid dissemination of antibiotic resistance genes and bacterial virulence factors among bacterial pathogens [1–3]. As a result, HGT is central to the emergence and spread of bacterial species harboring antibiotic resistance genes [4,5]. Patient morbidity and mortality drastically increases upon infection by antibiotic resistant bacteria because treatment options for these patients are limited [4–6]. Motivated by these facts, several approaches to inhibiting HGT have been pursued [7–13].
Conjugative plasmid transfer (CPT), a type of HGT, is a well-established route of antibiotic resistance propagation in both laboratory and clinical settings [1–3]. During CPT, the transferred (T) strand of a conjugative double stranded DNA (dsDNA) plasmid is unwound from the parent strand and then transported from a donor to a recipient bacterium. For successful DNA transfer, CPT relies upon two multi-protein complexes, the relaxosome and the type IV secretion system (T4SS), linked by a type IV coupling protein (T4CP) [14–19]. Within the relaxosome complex, a highly conserved relaxase enzyme initiates and terminates CPT using a series of metal-dependent DNA nicking steps [14,17,18]. Conjugative relaxases have been described as the anticipated targets of at least two classes of CPT-specific inhibitors [9,10]. In the Lujan et al. work, produced by the same group as the current study, the conjugative F plasmid relaxase was reported to be the target of bisphosphonates, which were shown to disrupt the transfer of the F plasmid in E. coli. This paper provided structural and in vitro DNA nicking activity data with the F plasmid relaxase, as well as plasmid transfer data in the presence of bisphosphonates. Under the published experimental conditions, inhibition of F plasmid transfer and selective killing of F plasmid-harboring cells was reported for a specific set of bisphosphonates [10].
While the initial reported impact of bisphosphonates on F plasmid transfer was promising, additional work was required to determine the range of plasmids and bacterial hosts that might be sensitive to these compounds under different experimental conditions. Subsequent structural data appeared to support the initial conclusion that relaxases were targeted by BSRCs. Citrate, a putative BSRC-type inhibitor, was observed by our group bound to the active site metal ion in the crystal structure of the plasmid pCU1 relaxase, an enzyme homologous to the F plasmid relaxase [20]. Here we report an expanded study that seeks to elucidate the impact of BSRCs on plasmid systems beyond the F plasmid. We found that the effects of BSRCs are variable and are dependent on plasmid type and experimental conditions.
Materials and Methods
In Cell Plasmid Transfer Experiments with the F, R100, and pCU1 Plasmids
Bacterial strains and plasmids are listed in Supplementary Table 1 along with the antibiotics that were used to select for each strain and their subsequent transconjugants. Concentrations of each antibiotic necessary to select for each strain were established by minimum inhibitory concentration (MIC) tests performed by serial dilution to determine the antibiotic resistance profiles of the strains used.
A protocol for following transfer of plasmids F, R100, and pCU1 from their respective donor strain into their respective recipient strain was modified from that published for analysis of F plasmid transfer (Supplementary Figure 1) [10]. Modifications were necessary to obtain optimal plasmid transfer conditions for new conjugative systems as well as to examine the effect of BSRCs (Supplementary Figure 2, Sigma-Aldrich) on F plasmid transfer under conditions not previously studied. This protocol incorporates the use of Oxygen Biosensor (OBS) plates (BD Biosciences) to determine the size of each cell population (donor, D, recipient, R, transconjugant, T) at the end of the plasmid transfer assay. OBS plates contain an oxygen-sensitive fluorophore embedded in a gel that fluoresces (excitation wavelength of 485 nm, emission wavelength of 620 nm) in an anaerobic environment. Following the addition of a population of bacterial cells, oxygen consumption will cause an increase in the fluorescent signal of the gel. The time of rise in fluorescence signal is logarithmically related to the size of the cell population initially placed into the well [21].
Plasmid transfer between bacterial cells was analyzed using the following protocol. Cultures of donor (D) and recipient (R) were grown to saturation overnight under antibiotic selection in Luria Broth (LB, Fisher Scientific) at 37°C. Donors and recipients were then diluted into LB such that they reached an OD600 of 0.5 simultaneously when grown without selection at 37°C.
In the case of plasmid pCU1 transfer, donor and recipient subcultures were then mixed in a 2 to 5 donor to recipient volume:volume ratio and pelleted at 5000×g. Pelleted cells were resuspended in LB to increase the cell density by a factor of 4, and 250 μL of the resuspended mixture of cells was applied to each well of a 96-well filter-bottom plate (0.2 μm hydrophobic PVDF membrane, Corning). The plate was centrifuged for 2 min at 3000×g to adhere the cells onto the filters and remove the LB. LB or LB + BSRC (250 μL) was then applied to the 96-well filter-bottom plate to generate LB controls and experimental samples, respectively. Filter-bottom plates were then incubated at 37°C for a designated time period (2, 24, or 48 h). Following incubation, filter-bottom plates were centrifuged for 2 min at 3000×g to remove LB and LB + BSRC from the cells. Cells were resuspended in 250 μL LB and then diluted 4-fold in LB to generate the final samples and analyzed as described below.
In the case of F and R100 plasmid transfer, donor and recipient subcultures were then mixed at a ratio of 1:9 D:R, 1:1 D:R, or 9:1 D:R in the presence of LB or LB + BSRC to generate LB controls and experimental samples, respectively. Reactions were incubated at 37°C for 100 min. After 100 min, reactions were centrifuged for 5 min at 3000×g to pellet the cells; supernatant containing BSRC was removed. Pelleted cells were resuspended in an equal volume of LB and analyzed as described below.
For analysis, samples were transferred to OBS plates to determine the effect each BSRC had on the size of donor and recipient populations, and the number of transconjugants produced during the duration of the assay. In assays involving the R100 plasmid (all mating ratios) and the F plasmid (9:1 mating ratio), wells were covered with the minimal amount of heavy mineral oil necessary to limit the reabsorption of oxygen into the LB media after the time of rise in fluorescence. The addition of mineral oil to the wells did not alter the cell populations determined from the assays (data not shown). To determine the relationship between signal generated on an OBS plate and the absolute size of the cell population, LB controls, as defined above, were serially diluted and analyzed both by the OBS plate on a PHERAstar fluorescent plate reader (BMG Labtech) and by enumeration of colony forming units (CFUs) on selective media. These data were combined to correlate the time delay of the rise in fluorescence signal to a specific population size.
Results
Transfer of the Conjugative F and R100 Plasmids
The impact of BSRCs on CPT and bacterial cell survival was investigated using a series of plasmid transfer assays in E. coli (Supplementary Figure 1). First, the populations of F plasmid donor cells (D), recipient cells (R), and transconjugant cells (T) were examined over four experimental days using two BSRCs - citrate, which was not examined in the original study, and etidronate, which was previously examined (Figure 1) (Lujan et al., 2007). We found that each compound exhibited significant day-to-day variability in impacting D, R, and T populations in these assays. For example, on days 1 and 3, T populations were abolished at low citrate concentrations, while on days 2 and 4, this effect was only seen at a higher citrate concentration (Figure 1). As such, the standard error associated with the effective concentration required for 50% inhibition of cell survival (EC50) across six BSRCs were found to show significant variability for F plasmid studies using the experimental methods described previously [10] (Figure 2A). As outlined below for other plasmid systems (i.e., R100 and pCU1), a similar degree of dayto-day variability was also observed. Taken together, these results show that inconsistent results across experimental days confounded the ability to determine clear effects of BSRCs on plasmid-containing bacterial cells.
Figure 1.
Survival of donor, recipient, and transconjugant cell populations during F plasmid transfer in the presence of citrate or etidronate. The fraction or percentage of each cell line which survived, relative to the unexposed control, is provided for eight concentrations of citrate and etidronate. Donor is shown in blue, recipient in red, and transconjugant in green. Etidronate data are from Lujan et al. (2007). Error bars present on the plot of the averaged data represent the standard error.
Figure 2.
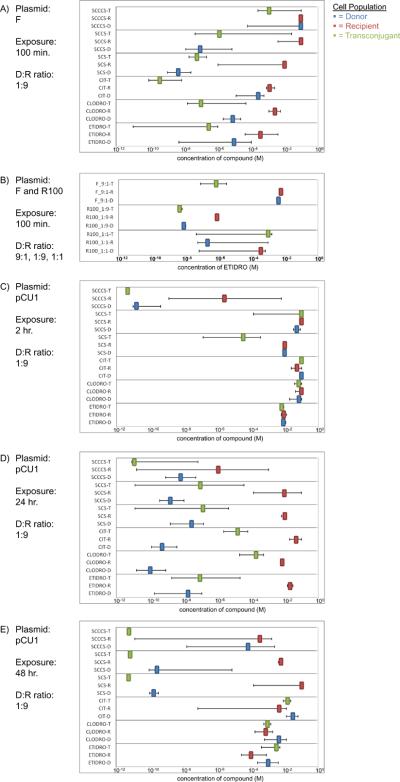
EC50 values for plasmid transfer assays. The effective molar concentration required to kill 50% (the EC50) of donor (D), recipient (R), or transconjugant (T) cell populations for a selection of bisphosphonates and structurally related compounds(BSRCs) during F (A,B), R100 (B), and pCU1 (C,D,E) plasmid transfer assays. For each plot, the plasmid, time of exposure to the compound(s), and mating ratio of donor to recipient cells (D:R) are provided. Error bars represent the standard error associated with each measurement. Etidronate and clodronate data in (A) are from Lujan et al. (2007).
We next examined R100, a conjugative plasmid closely related to the F plasmid [22]. Experimental conditions were altered slightly relative to those published for the F plasmid [10] to enhance R100 transfer. Adjusting the conditions of transfer also allowed us to determine the effects changes in experimental design would have on BSRC efficacy. In particular, the ratio of R100 donors (D) to R100 recipients (R) was set at both the original 1D:9R ratio used when following F plasmid transfer and a new 1D:1R ratio. An exposure time of 100 minutes was maintained, identical to that used when characterizing F plasmid transfer. As shown in Figures 2B and 3A–B, the ratio of donor to recipient bacterial cells had a significant impact on BSRC efficacy. For example, at a 1D:9R R100 ratio, etidronate spared the R population relative to the D and T populations; at a 1D:1R ratio, etidronate's impact on the three populations was statistically identical (Figure 2B).
Figure 3.
Survival of donor, recipient, and transconjugant cell populations during plasmid transfer. Cell survival during F, R100, and pCU1 plasmid transfer assays. The percentage of each cell line which survived, relative to the unexposed control, is provided for eight concentrations of etidronate (A) , PCP (B), and citrate or etidronate (C). Donor is shown in blue, recipient in red, and transconjugant in green. Error bars represent the standard error of each measurement. The impact of etidronate and PCP (A and B) on cell survival was dependent on the ratio of donor and recipient cell populations exposed to each compound. The impact of citrate and etidronate (C) on cell survival was dependent on the time the mating cells were exposed to each compound.
We then tested the effects of plasmid donor (D) to plasmid recipient (R) ratio on the F plasmid system. In particular, the efficacy of BSRCs in the presence of a 1D:9R ratio was compared to their efficacy in the presence of a 9D:1R ratio. The results of these experiments mirrored those generated for the R100 studies (Figure 2A–B, Figures 3A–B). Slight differences in EC50 values of etidronate observed at 9D:1R ratios (Fig. 2B) disappeared when 1D:9R ratios were used(Fig. 2A). Taken together, these F and R100 plasmid studies highlight the effect bacterial cell population ratios have on BSRC efficacy.
Transfer of the Conjugative Plasmid pCU1
To expand the analysis of BSRCs beyond that of the F-like plasmid family, the conjugative plasmid pCU1, isolated from Salmonella typhimurium [23,24], was also examined. The rate of plasmid pCU1 transfer was found to be significantly slower than that of the F plasmid; pCU1 plasmid transfer is known to be drastically enhanced when transfer occurs on a solid medium [25]. Therefore, the previously published experimental conditions used to study the transfer of the F plasmid [10] were modified to optimize pCU1 plasmid transfer, and to explore the impact of these modifications on BSRC efficacy. In particular, the ratio of donor to recipient cells was shifted from 1D:9R to 2D:5R, and the mating experiment was performed on a filter, as opposed to liquid (see Materials and Methods). The timeline of the experiment was also extended in an attempt to compensate for the slow rate of pCU1 plasmid transfer, and the concentration of donor and recipient cells was increased 4-fold during the experiment to increase the local concentration of potential mating pairs. In particular, donors and recipients were allowed to mate (undergo plasmid transfer) in the presence of each compound of interest for 2, 24, and 48 hours.
As shown in Figures 2C–E and 3C, these experiments revealed a time-dependence of BSRC efficacy on cells containing pCU1. The impact on cell survival and plasmid transfer were most dramatic when the cells were exposed to putative inhibitors for 24 hours (Figures 2C–E). After 2 or 48 hours, in contrast, either a weak effect or no effect was observed (Figures 2C–E). For example, clodronate showed little efficacy against all three cell populations at 2 and 48 hours of exposure, but exhibited efficacy versus donor and, to a lesser extent, transconjugant populations at 24 hours of exposure (see Figures 2C–E). Furthermore, it was also noted that compound efficacy differed between plasmid types even at roughly the same exposure time. Clodronate demonstrated some efficacy for F plasmid donor cells at 100 minutes of exposure (Figure 2A); however, the same compound at 2 hours of exposure demonstrated no effect on any cell populations containing plasmid pCU1 (Figure 2C). Taken together, these results establish significant variability between exposure times and plasmid types for any effect by BSRCs on plasmid transfer and cell survival.
Impact of Metal Chelation during Plasmid Transfer Experiments
Finally, bisphosphonates are known metal chelators, and their role in the treatment of osteoporosis relies upon this activity [26–28]. Thus, we compared the effects of the metal chelator EDTA with those of etidronate on bacterial cell survival and plasmid transfer for E coli containing either the F or R100 plasmids (Figures 4A and 4B, respectively). We find little difference between these two compounds for both F- or R100-containing cells, indicating that metal chelation may be the mechanism by which BSRCs elicit effects on bacterial cell survival and plasmid transfer. In summary, the broader plasmid transfer experiments presented here demonstrate time-, bacterial cell line-, and plasmid-specific variability in the impact of BSRCs on bacterial cells, and point to a general metal chelation effect rather than a relaxase-specific inhibition.
Figure 4.
Survival of donor, recipient, and transconjugant cell populations during F and R100 plasmid transfer in the presence of etidronate or EDTA. Cell survival during F (A) and R100 (B) plasmid transfer assays. The percentage of each cell line which survived, relative to the unexposed control, is provided for eight concentrations of EDTA. For comparison, survival curves following exposure of plasmids F and R100 to etidronate are provided. Donor is shown in blue, recipient in red, and transconjugant in green. Error bars represent the standard error of each measurement.
Discussion
We present an extended analysis of the potential impact of bisphosphonates and structurally related compounds (BSRCs) on conjugative plasmid transfer in bacteria and on the survival of bacterial cells containing conjugative plasmids. This follows a previous report from our group, in which we indicated that BSRCs selectively disrupted bacterial conjugation by targeting the relaxase enzyme [10]. The current study expands the scope of the investigation by including additional BSRCs, two additional bacterial plasmids, and a range of new experimental conditions. Data outlined in Figure 4 suggest that BSRCs exert their variable effects via metal ion chelation, as EDTA appeared to act in a manner similar to etidronate. EDTA was not examined in the previous study, although another metal chelating BSRC, pamidronate, showed little impact in that study on F-plasmid presence in E. coli [10]. Regardless, bacterial cell survival is closely linked to the intra- and extracellular concentration of metal ions, which are required cofactors for a range of processes but can be cytotoxic at high concentrations [29–31]. The integrity of the outer membrane (OM) of gram negative bacteria is maintained by the presence of divalent cations, which serve to stabilize and organize lipopolysaccharide (LPS) in the OM [32]. As a result, EDTA is commonly used to permeabilize the OM of gram-negative bacteria by chelating divalent cations, and the efficiency of gram-negative cell lysis by lysozyme is enhanced by the presence of EDTA [33–35]. Millimolar concentrations of citrate can also permeabilize the OM of gram-negative bacteria by metal chelation, though citrate's cytotoxic effect is prevented by an excess of MgCl2 [33,35].
The data presented here further show that BSRCs exhibit variable effects on bacterial cell survival and conjugative plasmid transfer (see Figures 1–3). Factors involved in this variability include plasmid type, ratio of plasmid donor to plasmid recipient cell types, and the time over which cells are exposed to BSRCs. Based on these results, we conclude that BSRCs will fail to exert a consistent effect on the transfer of conjugative plasmids, which often contain antibiotic resistance genes, in a clinical setting. Indeed, while in some conditions BSRCs can appear potent (e.g., Figure 2C–E), they lack potency if a key variable is changed (e.g., exposure time of bacteria to BSRCs). While the results presented here are disappointing from the perspective of limiting the spread of antibiotic resistance, they clarify the effect of BSRCs on the conjugation of additional plasmids in E. coli.
Supplementary Material
Highlights
Bisphosphonates and related compounds variably affect conjugative plasmid transfer.
Conjugation of plasmids R100, pCU1 and F are differentially impacted by BSRCs.
Changes to mating protocols alter the observed effect of BSRCs on plasmid transfer.
Metal chelation may be the mechanism by which BSRCs exert their effects.
Acknowledgments
We thank Suzanne Paterson for the gift of the plasmid pCU1, and Laura Frost for the gift of the plasmid R100-1. Supported by NIH grant AI78924. In addition, D.E.M., W.K.B. and M.R.R. were funded in part by Exigent Pharmaceuticals, a former commercial venture focused on the disruption of antibiotic resistance propagation. R.P.N. and S.W.M. received no funding from Exigent.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- [1].de la Cruz F, Davies J. Horizontal gene transfer and the origin of species: lessons from bacteria. Trends in Microbiol. 2000;8:128–133. doi: 10.1016/s0966-842x(00)01703-0. [DOI] [PubMed] [Google Scholar]
- [2].Mazel D, Davies J. Antibiotic resistance in microbes. Cell. Mol. Life Sci. 1999;56:742–754. doi: 10.1007/s000180050021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Waters VL. Conjugative transfer in the dissemination of beta-lactam and aminoglycoside resistance. Front. Biosci. 1999;4:433–456. doi: 10.2741/waters. [DOI] [PubMed] [Google Scholar]
- [4].CDC National Nosocomial Infections Surveillance (NNIS) System Report, data summary from January 1992 through June 2004, issued October 2004. Am. J. Infect. Control. 2004;32:470–485. doi: 10.1016/S0196655304005425. [DOI] [PubMed] [Google Scholar]
- [5].Fish DN, Ohlinger MJ. Antimicrobial resistance: factors and outcomes. Crit. Care Clin. 2006;22:291–311. doi: 10.1016/j.ccc.2006.02.006. [DOI] [PubMed] [Google Scholar]
- [6].Hawkey PM, Jones AM. The changing epidemiology of resistance. J. Antimicrob. Chemother. 2009;64:i3–i10. doi: 10.1093/jac/dkp256. [DOI] [PubMed] [Google Scholar]
- [7].DeNap JCB, Thomas JR, Musk DJ, Hergenrother PJ. Combating drug-resistant bacteria: small molecule mimics of plasmid incompatibility as antiplasmid compounds. J. Am. Chem. Soc. 2004;126:15402–15404. doi: 10.1021/ja044207u. [DOI] [PubMed] [Google Scholar]
- [8].Fernandez-Lopez R, Machon C, Longshaw C, Martin S, Molin S, Zechner E, Espinosa M, Lanka E, de la Cruz F. Unsaturated fatty acids are inhibitors of bacterial conjugation. Microbiology. 2005;151:3517–3626. doi: 10.1099/mic.0.28216-0. [DOI] [PubMed] [Google Scholar]
- [9].Garcillan-Barcia M, Jurado P, Gonzalez-Perez B, Moncalian G, Fernandez L, de la Cruz F. Conjugative transfer can be inhibited by blocking relaxase activity within recipient cells with intrabodies. Mol. Microbiol. 2007;63:404–416. doi: 10.1111/j.1365-2958.2006.05523.x. [DOI] [PubMed] [Google Scholar]
- [10].Lujan SA, Guogas LM, Ragonese H, Matson SW, Redinbo MR. Disrupting antibiotic resistance propagation by inhibiting the conjugative DNA relaxase. Proc. Natl. Acad. Sci. U.S.A. 2007;104:12282–12287. doi: 10.1073/pnas.0702760104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Marraffini LA, Sontheimer EJ. CRISPR interference limits horizontal gene transfer in staphylococci by targeting DNA. Science. 2008;322:1843–1845. doi: 10.1126/science.1165771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Potts RG, Lujan SA, Redinbo MR. Winning the asymmetric war: new strategies for combating antibacterial resistance. Future Microbiol. 2008;3:119–123. doi: 10.2217/17460913.3.2.119. [DOI] [PubMed] [Google Scholar]
- [13].Williams JJ, Hergenrother PJ. Exposing plasmids as the Achilles' heel of drug-resistant bacteria. Curr. Opin. Chem. Biol. 2008;12:389–399. doi: 10.1016/j.cbpa.2008.06.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Byrd DR, Matson SW. Nicking by transesterification: the reaction catalysed by a relaxase. Mol. Microbiol. 1997;25:1011–1022. doi: 10.1046/j.1365-2958.1997.5241885.x. [DOI] [PubMed] [Google Scholar]
- [15].Francia MV, Varsaki A, Garcillan-Barcia MP, Latorre A, Drainas C, de la Cruz F. A classification scheme for mobilization regions of bacterial plasmids. FEMS Microbiol. Rev. 2004;28:79–100. doi: 10.1016/j.femsre.2003.09.001. [DOI] [PubMed] [Google Scholar]
- [16].Garcillan-Barcia MP, Francia MV, de la Cruz F. The diversity of conjugative relaxases and its application in plasmid classification. FEMS Microbiol. Rev. 2009;33:657–687. doi: 10.1111/j.1574-6976.2009.00168.x. [DOI] [PubMed] [Google Scholar]
- [17].Lanka E, Wilkins BM. DNA processing reactions in bacterial conjugation. Annu. Rev. Biochem. 1995;64:141–169. doi: 10.1146/annurev.bi.64.070195.001041. [DOI] [PubMed] [Google Scholar]
- [18].Llosa M, Gomis-Ruth F, Coll M, de la Cruz F. Bacterial conjugation: a two-step mechanism for DNA transport. Mol. Microbiol. 2002;45:1–8. doi: 10.1046/j.1365-2958.2002.03014.x. [DOI] [PubMed] [Google Scholar]
- [19].Willetts N, Wilkins B. Processing of plasmid DNA during bacterial conjugation. Microbiol. Rev. 1984;48:24–41. doi: 10.1128/mr.48.1.24-41.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Nash RP, Habibi S, Cheng Y, Lujan SA, Redinbo MR. The mechanism and control of DNA transfer by the conjugative relaxase of resistance plasmid pCU1. Nucleic Acids Res. 2010;38:5929–5943. doi: 10.1093/nar/gkq303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Stitt DT, Nagar MS, Haq TA, Timmins MR. Determination of growth rate of microorganisms in broth from oxygen-sensitive fluorescence plate reader measurements. Biotechniques. 2002;32:684–689. doi: 10.2144/02323dd06. [DOI] [PubMed] [Google Scholar]
- [22].Anthony KG, Klimke WA, Manchak J, Frost LS. Comparison of proteins involved in pilus synthesis and mating pair stabilization from the related plasmids F and R100-1: insights into the mechanism of conjugation. J. Bacteriol. 1999;181:5149–5159. doi: 10.1128/jb.181.17.5149-5159.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Khatoon H, Iyer RV, Iyer VN. A new filamentous bacteriophage with sex-factor specificity. Virology. 1972;48:145–155. doi: 10.1016/0042-6822(72)90122-5. [DOI] [PubMed] [Google Scholar]
- [24].Konarska-Kozlowska M, Iyer VN. Physical and genetic organization of the IncN-group plasmid pCU1. Gene. 1981;14:195–204. doi: 10.1016/0378-1119(81)90115-3. [DOI] [PubMed] [Google Scholar]
- [25].Thatte V, Bradley D, Iyer V. N conjugative transfer system of plasmid pCU1. J. Bacteriol. 1985;163:1229–1236. doi: 10.1128/jb.163.3.1229-1236.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Hu Y-Y, Rawal A, Schmidt-Rohr K. Strongly bound citrate stabilizes the apatite nanocrystals in bone. Proc. Natl. Acad. Sci. U.S.A. 2010;107:22425–22429. doi: 10.1073/pnas.1009219107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Russell RGG, Rogers MJ. Bisphosphonate: from the laboratory to the clinic and back again. Bone. 1999;25:97–106. doi: 10.1016/s8756-3282(99)00116-7. [DOI] [PubMed] [Google Scholar]
- [28].Russell RGG, Xia Z, Dunford JE, Oppermann U, Kwaasi A, Hulley PA, Kavanagh KL, Triffitt JT, Lundy MW, Phips RJ, Barnett BL, Coxon FP, Rogers MJ, Watts NB, Ebetino FH. Bisphosphonates: an update on mechanisms of action and how these relate to clinical efficacy. Ann. N. Y. Acad. Sci. 2007;1117:209–257. doi: 10.1196/annals.1402.089. [DOI] [PubMed] [Google Scholar]
- [29].Finney LA, O'Halloran TV. Transition metal speciation in the cell: insights from the chemistry of metal ion receptors. Science. 2003;300:931–936. doi: 10.1126/science.1085049. [DOI] [PubMed] [Google Scholar]
- [30].Outten CE, O'Halloran TV. Femtomolar sensitivity of metalloregulatory proteins controlling zinc homeostasis. Science. 2001;292:2488–2492. doi: 10.1126/science.1060331. [DOI] [PubMed] [Google Scholar]
- [31].Waldron KJ, Robinson NJ. How do bacterial cells ensure that metalloproteins get the correct metal? Nat. Rev. Microbiol. 2009;6:25–35. doi: 10.1038/nrmicro2057. [DOI] [PubMed] [Google Scholar]
- [32].Nikaido H, Vaara M. Molecular basis of bacterial outer membrane permeability. Microbiol. Rev. 1985;49:1–32. doi: 10.1128/mr.49.1.1-32.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Helander IM, Mattlia-Sandholm T. Fluorometric assessment of gram-negative bacterial permeabilization. J. Appl. Microbiol. 2000;88:213–219. doi: 10.1046/j.1365-2672.2000.00971.x. [DOI] [PubMed] [Google Scholar]
- [34].Leive L. The barrier function of the gram-negative envelope. Ann. N. Y. Acad. Sci. 1974;235:109–129. doi: 10.1111/j.1749-6632.1974.tb43261.x. [DOI] [PubMed] [Google Scholar]
- [35].Vaara M, Jaakkola J. Sodium hexametaphosphate sensitizes Pseudomonas aeruginosa, several other species of Pseudomonas, and Escherichia coli to hydrophobic drugs. Antimicrob. Agents Chemother. 1989;33:1741–1747. doi: 10.1128/aac.33.10.1741. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.