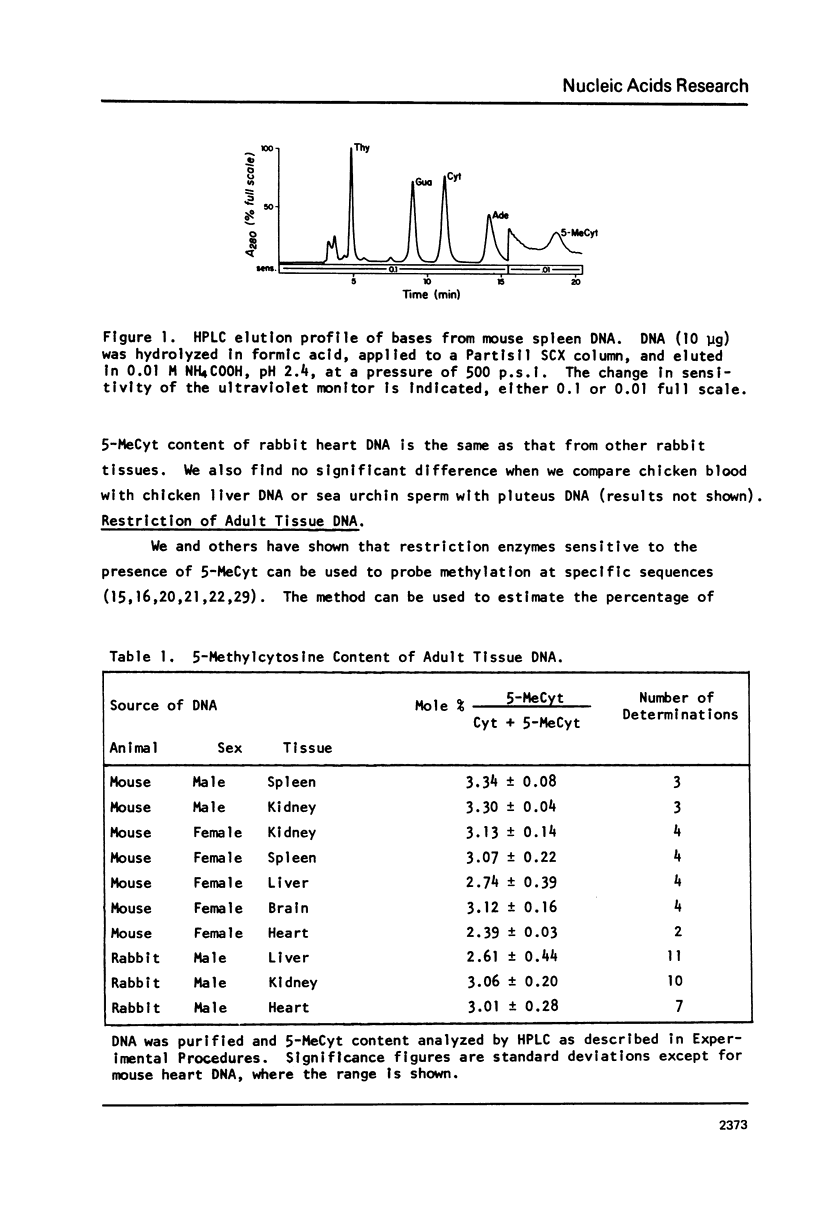
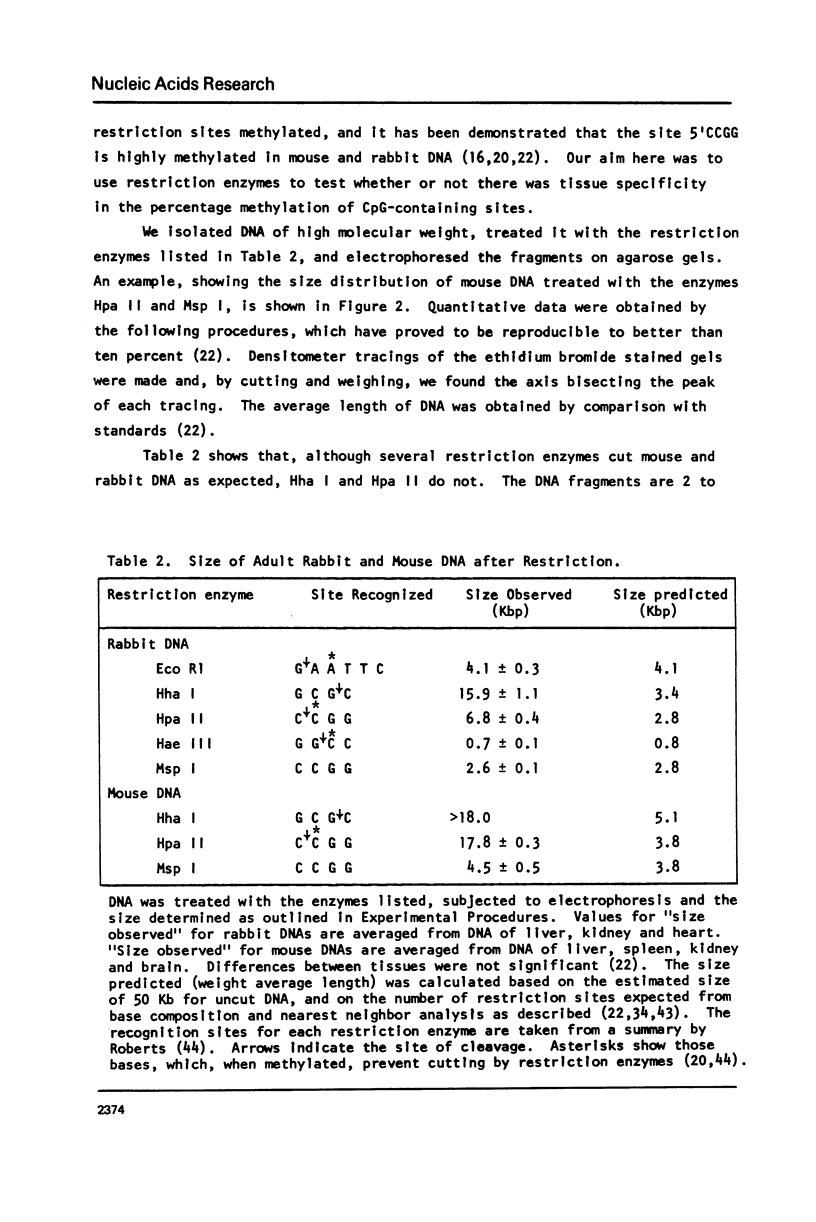
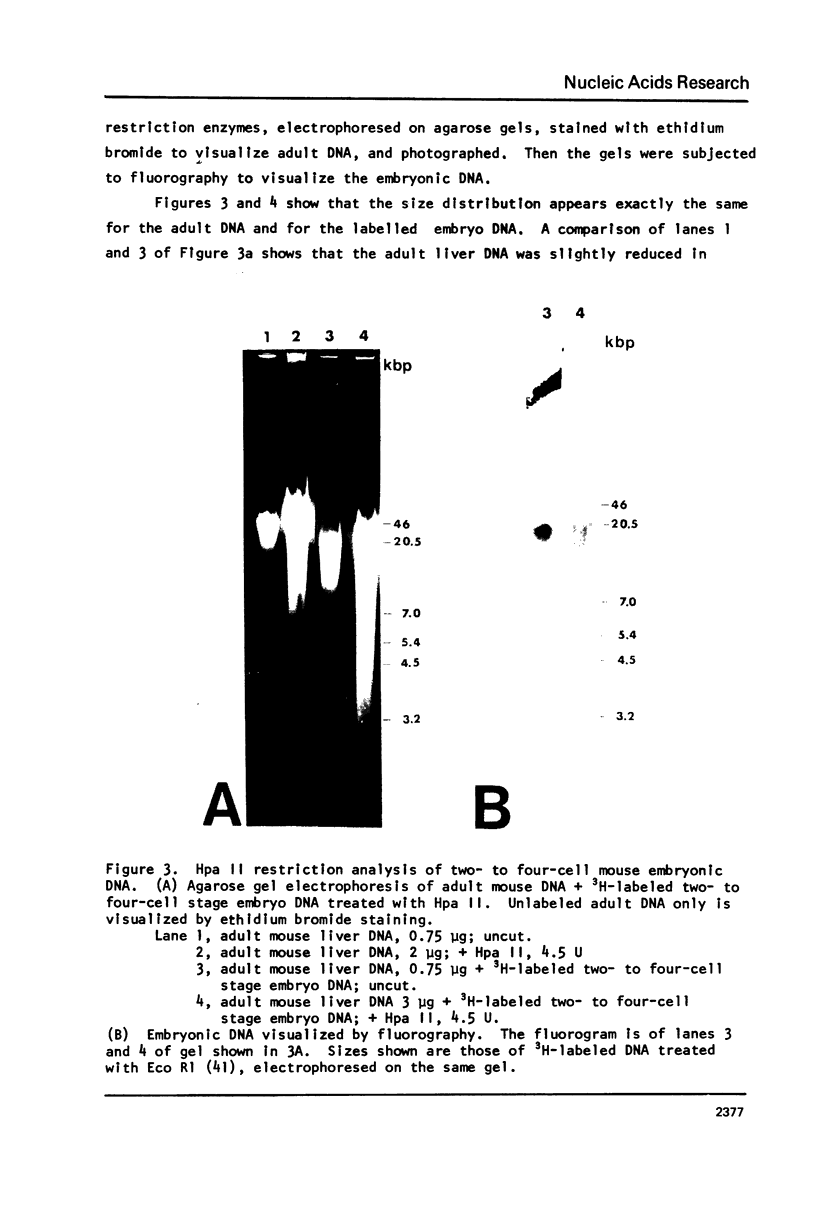
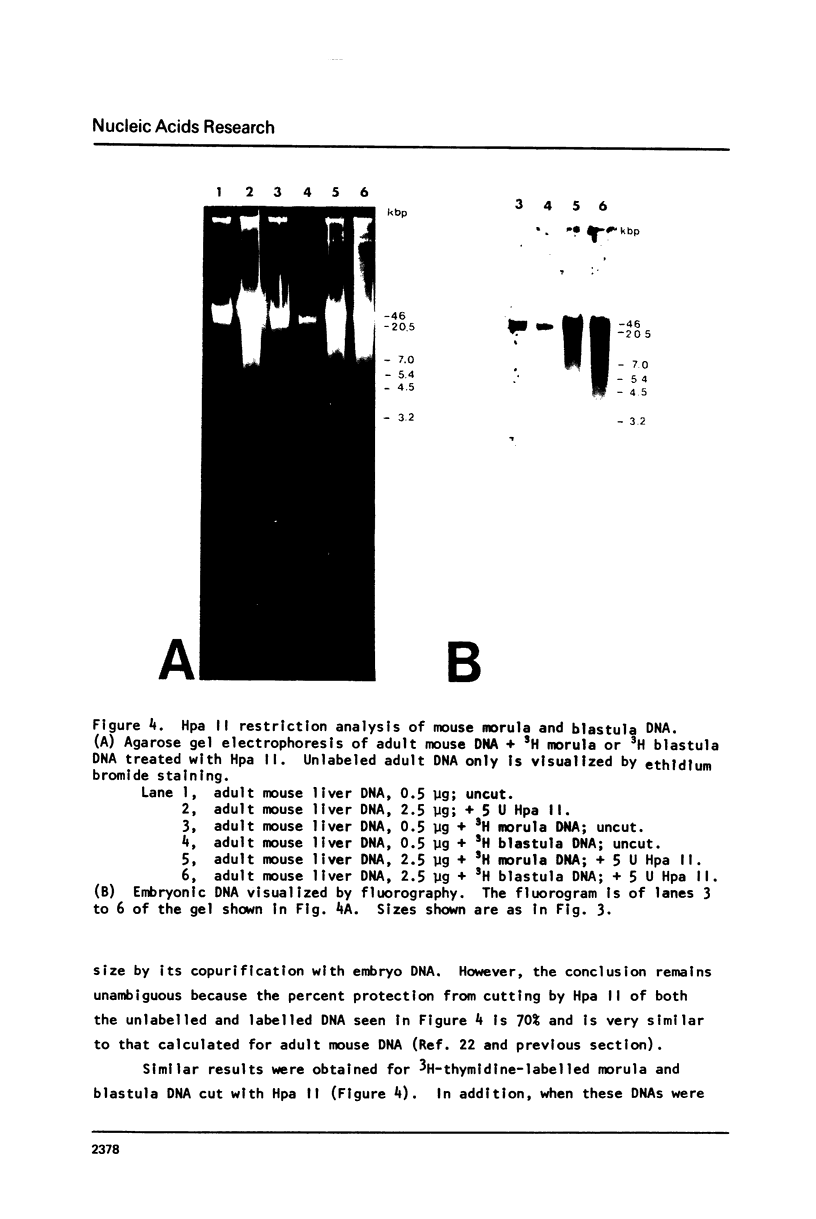
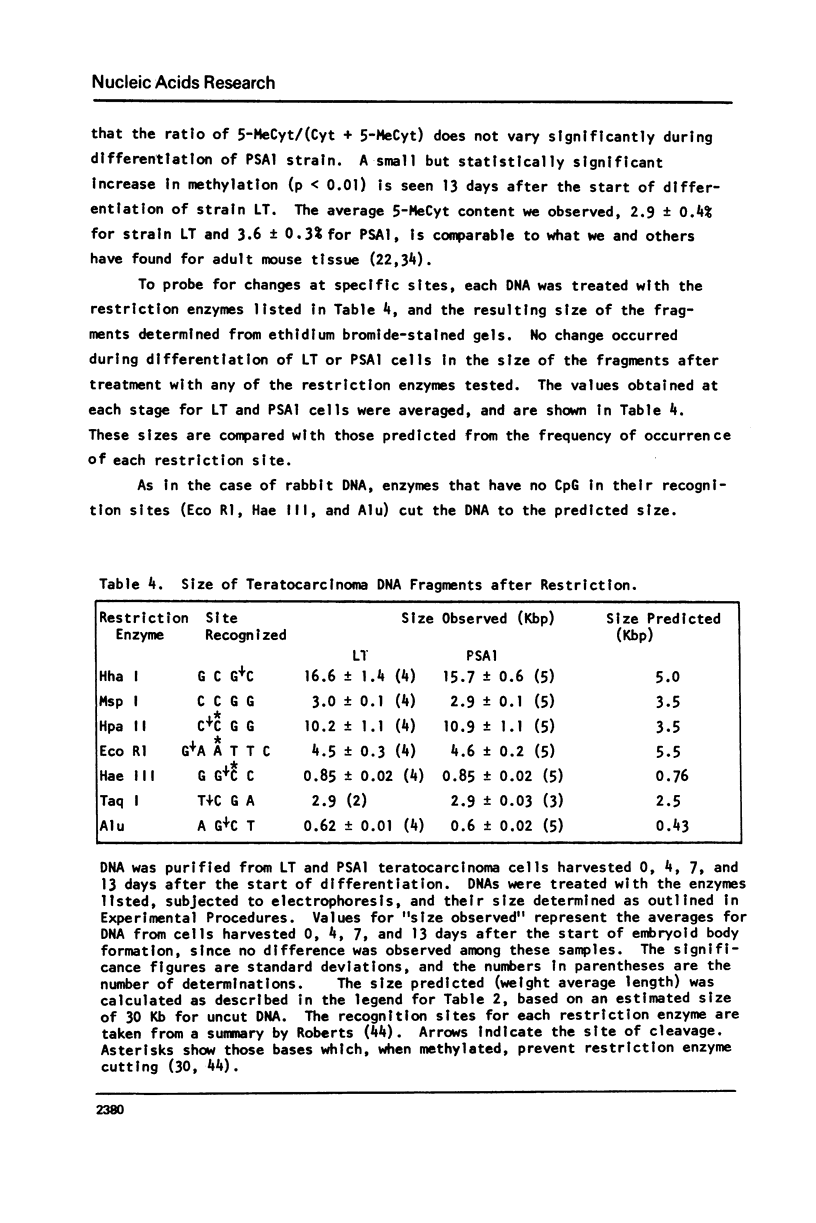
Abstract
The distribution and amount of 5-methylcytosine (5-MeCyt) in DNA was measured for early embryos of mouse strain CF1 (2 to 4 cell stage to blastocyst) and mouse teratocarcinoma cells. In each case, the pattern of methylation was examined by use of the restriction enzymes Hha I and HPA II HPA II, which cut DNA at the sites 5'GCGC and 5'CCGG respectively, when the cytosines at these sites are not methylated. Mouse embryo DNA was found to have the same level of methylation as adult mouse tissues, and no changes in methylation were seen during differentiation of the teratocarcinoma cells. The ratio of 5-MeCyt/Cyt in DNA was measured by high performance liquid chromatography for the differentiating teratocarcinoma cells and for several adult mouse and rabbit tissues. The variation between tissues or between teratocarcinoma cells at different stages of differentiation was less than 10 percent. These results are discussed in view of proposals that 5-MeCyt plays a role in differentiation.
Full text
PDF











Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Adams R. L. Delayed methylation of DNA in developing sea urchin embryos. Nat New Biol. 1973 Jul 4;244(131):27–29. doi: 10.1038/newbio244027a0. [DOI] [PubMed] [Google Scholar]
- Bird A. P., Southern E. M. Use of restriction enzymes to study eukaryotic DNA methylation: I. The methylation pattern in ribosomal DNA from Xenopus laevis. J Mol Biol. 1978 Jan 5;118(1):27–47. doi: 10.1016/0022-2836(78)90242-5. [DOI] [PubMed] [Google Scholar]
- Blin N., Stafford D. W. A general method for isolation of high molecular weight DNA from eukaryotes. Nucleic Acids Res. 1976 Sep;3(9):2303–2308. doi: 10.1093/nar/3.9.2303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolivar F., Betlach M. C., Heyneker H. L., Shine J., Rodriguez R. L., Boyer H. W. Origin of replication of pBR345 plasmid DNA. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5265–5269. doi: 10.1073/pnas.74.12.5265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burton W. G., Grabowy C. T., Sager R. Role of methylation in the modification and restriction of chloroplast DNA in Chlamydomonas. Proc Natl Acad Sci U S A. 1979 Mar;76(3):1390–1394. doi: 10.1073/pnas.76.3.1390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cedar H., Solage A., Glaser G., Razin A. Direct detection of methylated cytosine in DNA by use of the restriction enzyme MspI. Nucleic Acids Res. 1979;6(6):2125–2132. doi: 10.1093/nar/6.6.2125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donahue R. P. Maturation of the mouse oocyte in vitro. I. Sequence and timing of nuclear progression. J Exp Zool. 1968 Oct;169(2):237–249. doi: 10.1002/jez.1401690210. [DOI] [PubMed] [Google Scholar]
- Gautier F., Bünemann H., Grotjahn L. Analysis of calf-thymus satellite DNA: evidence for specific methylation of cytosine in C-G sequences. Eur J Biochem. 1977 Oct 17;80(1):175–183. doi: 10.1111/j.1432-1033.1977.tb11869.x. [DOI] [PubMed] [Google Scholar]
- Grippo P., Iaccarino M., Parisi E., Scarano E. Methylation of DNA in developing sea urchin embryos. J Mol Biol. 1968 Sep 14;36(2):195–208. doi: 10.1016/0022-2836(68)90375-6. [DOI] [PubMed] [Google Scholar]
- Helling R. B., Goodman H. M., Boyer H. W. Analysis of endonuclease R-EcoRI fragments of DNA from lambdoid bacteriophages and other viruses by agarose-gel electrophoresis. J Virol. 1974 Nov;14(5):1235–1244. doi: 10.1128/jvi.14.5.1235-1244.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holliday R., Pugh J. E. DNA modification mechanisms and gene activity during development. Science. 1975 Jan 24;187(4173):226–232. [PubMed] [Google Scholar]
- Kallos J., Fasy T. M., Hollander V. P., Bick M. D. Estrogen receptor has enhanced affinity for bromodeoxyuridine-substituted DNA. Proc Natl Acad Sci U S A. 1978 Oct;75(10):4896–4900. doi: 10.1073/pnas.75.10.4896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalousek F., Morris N. R. The purification and properties of deoxyribonucleic acid methylase from rat spleen. J Biol Chem. 1969 Mar 10;244(5):1157–1163. [PubMed] [Google Scholar]
- Laskey R. A., Mills A. D. Quantitative film detection of 3H and 14C in polyacrylamide gels by fluorography. Eur J Biochem. 1975 Aug 15;56(2):335–341. doi: 10.1111/j.1432-1033.1975.tb02238.x. [DOI] [PubMed] [Google Scholar]
- Lin S. Y., Riggs A. D. Lac operator analogues: bromodeoxyuridine substitution in the lac operator affects the rate of dissociation of the lac repressor. Proc Natl Acad Sci U S A. 1972 Sep;69(9):2574–2576. doi: 10.1073/pnas.69.9.2574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin S., Lin D., Riggs A. D. Histones bind more tightly to bromodeoxyuridine-substituted DNA than to normal DNA. Nucleic Acids Res. 1976 Sep;3(9):2183–2191. doi: 10.1093/nar/3.9.2183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luthardt F. W., Donahue R. P. DNA synthesis in developing two-cell mouse embryos. Dev Biol. 1975 May;44(1):210–216. doi: 10.1016/0012-1606(75)90389-9. [DOI] [PubMed] [Google Scholar]
- Mann M. B., Smith H. O. Specificity of Hpa II and Hae III DNA methylases. Nucleic Acids Res. 1977 Dec;4(12):4211–4221. doi: 10.1093/nar/4.12.4211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin G. R., Epstein C. J., Travis B., Tucker G., Yatziv S., Martin D. W., Jr, Clift S., Cohen S. X-chromosome inactivation during differentiation of female teratocarcinoma stem cells in vitro. Nature. 1978 Jan 26;271(5643):329–333. doi: 10.1038/271329a0. [DOI] [PubMed] [Google Scholar]
- Martin G. R., Wiley L. M., Damjanov I. The development of cystic embryoid bodies in vitro from clonal teratocarcinoma stem cells. Dev Biol. 1977 Dec;61(2):230–244. doi: 10.1016/0012-1606(77)90294-9. [DOI] [PubMed] [Google Scholar]
- McGhee J. D., Ginder G. D. Specific DNA methylation sites in the vicinity of the chicken beta-globin genes. Nature. 1979 Aug 2;280(5721):419–420. doi: 10.1038/280419a0. [DOI] [PubMed] [Google Scholar]
- Pollock J. M., Jr, Swihart M., Taylor J. H. Methylation of DNA in early development: 5-methyl cytosine content of DNA in sea urchin sperm and embryos. Nucleic Acids Res. 1978 Dec;5(12):4855–4861. doi: 10.1093/nar/5.12.4855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riggs A. D. X inactivation, differentiation, and DNA methylation. Cytogenet Cell Genet. 1975;14(1):9–25. doi: 10.1159/000130315. [DOI] [PubMed] [Google Scholar]
- Roberts R. J. Restriction endonucleases. CRC Crit Rev Biochem. 1976 Nov;4(2):123–164. doi: 10.3109/10409237609105456. [DOI] [PubMed] [Google Scholar]
- Roizes G. A possible structure for calf satellite DNA I. Nucleic Acids Res. 1976 Oct;3(10):2677–2696. doi: 10.1093/nar/3.10.2677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- SWARTZ M. N., TRAUTNER T. A., KORNBERG A. Enzymatic synthesis of deoxyribonucleic acid. XI. Further studies on nearest neighbor base sequences in deoxyribonucleic acids. J Biol Chem. 1962 Jun;237:1961–1967. [PubMed] [Google Scholar]
- Sager R., Kitchin R. Selective silencing of eukaryotic DNA. Science. 1975 Aug 8;189(4201):426–433. [PubMed] [Google Scholar]
- Sheid B., Srinivasan P. R., Borek E. Deoxyribonucleic acid methylase of mammalian tissues. Biochemistry. 1968 Jan;7(1):280–285. doi: 10.1021/bi00841a034. [DOI] [PubMed] [Google Scholar]
- Singer J., Roberts-Ems J., Riggs A. D. Methylation of mouse liver DNA studied by means of the restriction enzymes msp I and hpa II. Science. 1979 Mar 9;203(4384):1019–1021. doi: 10.1126/science.424726. [DOI] [PubMed] [Google Scholar]
- Singer J., Stellwagen R. H., Roberts-Ems J., Riggs A. D. 5-Methylcytosine content of rat hepatoma DNA substituted with bromodeoxyuridine. J Biol Chem. 1977 Aug 10;252(15):5509–5513. [PubMed] [Google Scholar]
- Sista H. S., Loder R. T., Caruthers M. H. Studies on gene control regions X. The effect of specific adenine-thymine transversions on the lac repressor-lac operator interaction. Nucleic Acids Res. 1979 Jun 11;6(7):2583–2599. doi: 10.1093/nar/6.7.2583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sneider T. W., Potter V. R. Methylation of mammalian DNA: studies on Novikoff hepatoma cells in tissue culture. J Mol Biol. 1969 Jun 14;42(2):271–284. doi: 10.1016/0022-2836(69)90043-6. [DOI] [PubMed] [Google Scholar]
- Sneider T. W., Teague W. M., Rogachevsky L. M. S-adenosylmethionine: DNA-cytosine 5-methyltransferase from a Novikoff rat hepatoma cell line. Nucleic Acids Res. 1975 Oct;2(10):1685–1700. doi: 10.1093/nar/2.10.1685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tosi L., Granieri A., Scarano E. Enzymatic DNA modifications in isolated nuclei from developing sea urchin embryos. Exp Cell Res. 1972 May;72(1):257–264. doi: 10.1016/0014-4827(72)90588-5. [DOI] [PubMed] [Google Scholar]
- Turnbull J. F., Adams R. L. DNA methylase: purification from ascites cells and the effect of various DNA substrates on its activity. Nucleic Acids Res. 1976 Mar;3(3):677–695. doi: 10.1093/nar/3.3.677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanyushin B. F., Mazin A. L., Vasilyev V. K., Belozersky A. N. The content of 5-methylcytosine in animal DNA: the species and tissue specificity. Biochim Biophys Acta. 1973 Mar 28;299(3):397–403. doi: 10.1016/0005-2787(73)90264-5. [DOI] [PubMed] [Google Scholar]
- Vovis G. F., Horiuchi K., Zinder N. D. Kinetics of methylation of DNA by a restriction endonuclease from Escherichia coli B. Proc Natl Acad Sci U S A. 1974 Oct;71(10):3810–3813. doi: 10.1073/pnas.71.10.3810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waalwijk C., Flavell R. A. DNA methylation at a CCGG sequence in the large intron of the rabbit beta-globin gene: tissue-specific variations. Nucleic Acids Res. 1978 Dec;5(12):4631–4634. doi: 10.1093/nar/5.12.4631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waalwijk C., Flavell R. A. MspI, an isoschizomer of hpaII which cleaves both unmethylated and methylated hpaII sites. Nucleic Acids Res. 1978 Sep;5(9):3231–3236. doi: 10.1093/nar/5.9.3231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan R., Meselson M. A specific complex between a restriction endonuclease and its DNA substrate. Proc Natl Acad Sci U S A. 1970 Feb;65(2):357–362. doi: 10.1073/pnas.65.2.357. [DOI] [PMC free article] [PubMed] [Google Scholar]