Abstract
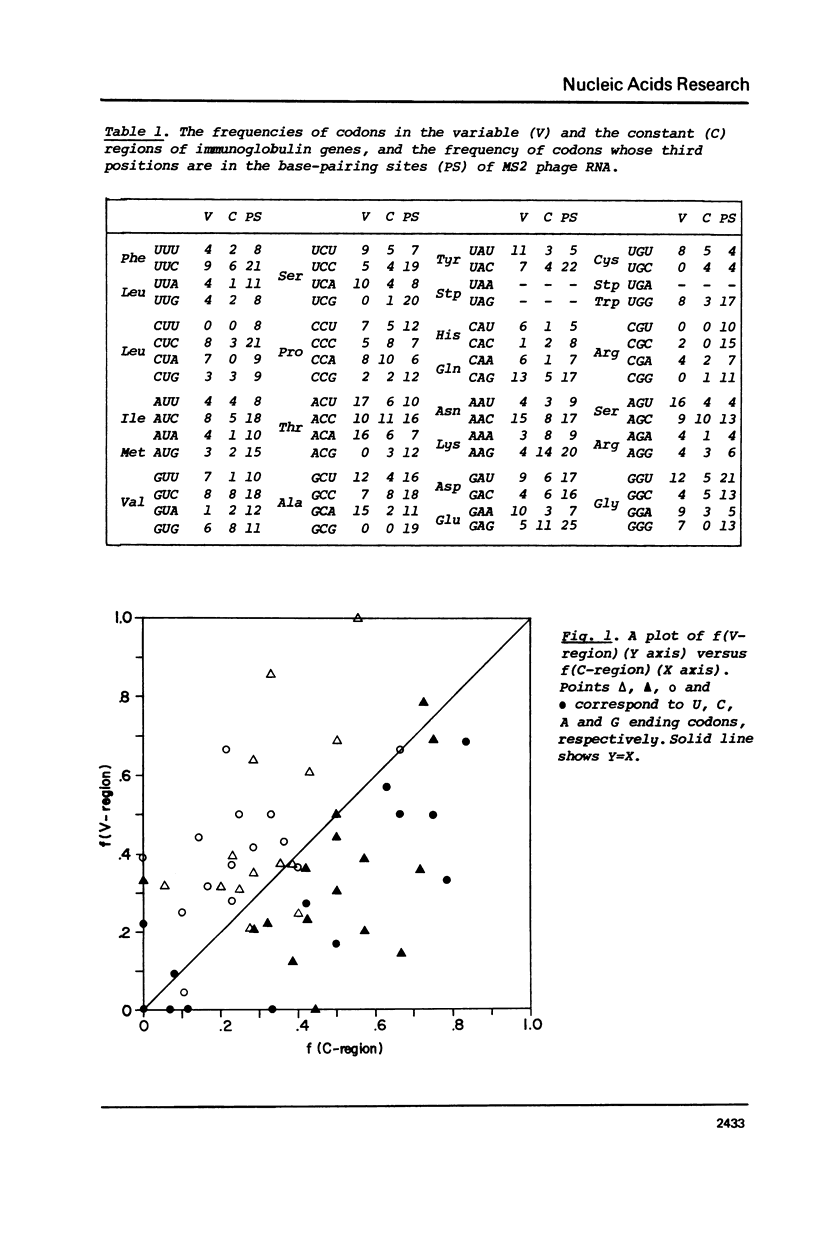
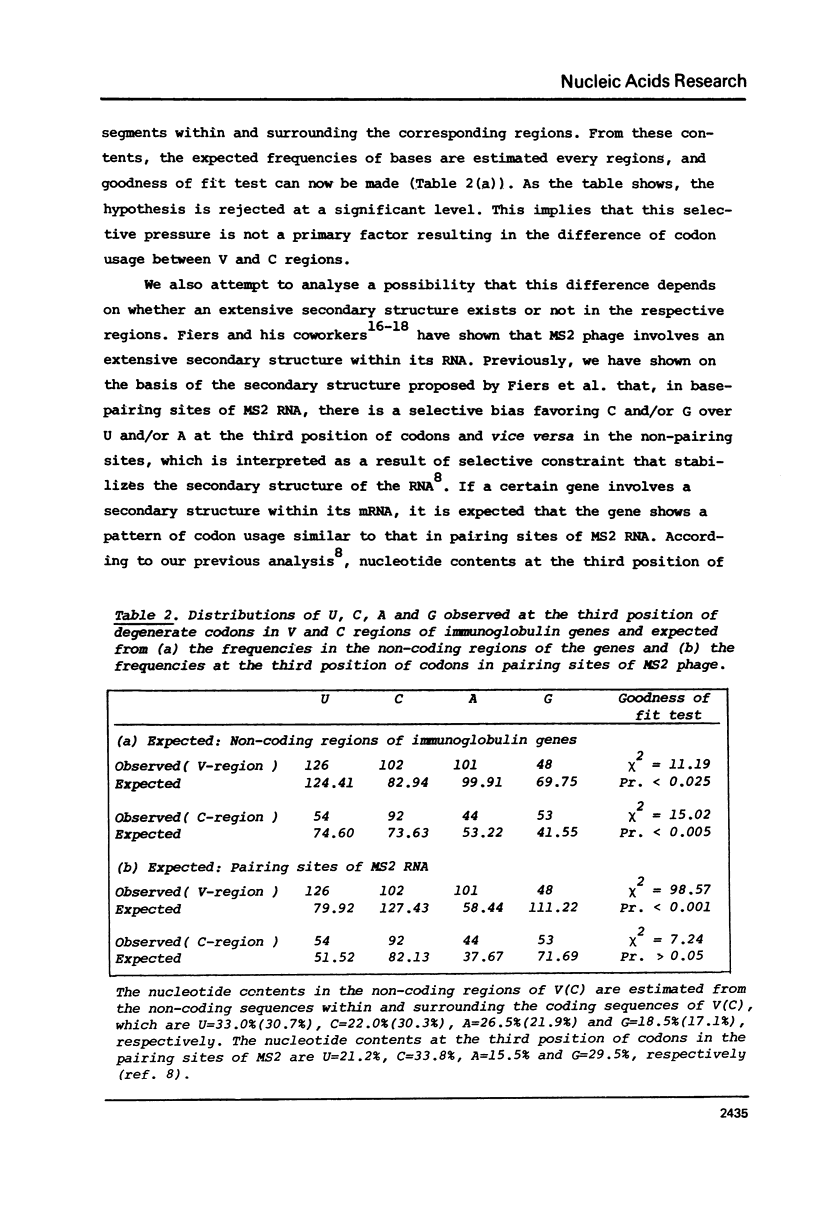
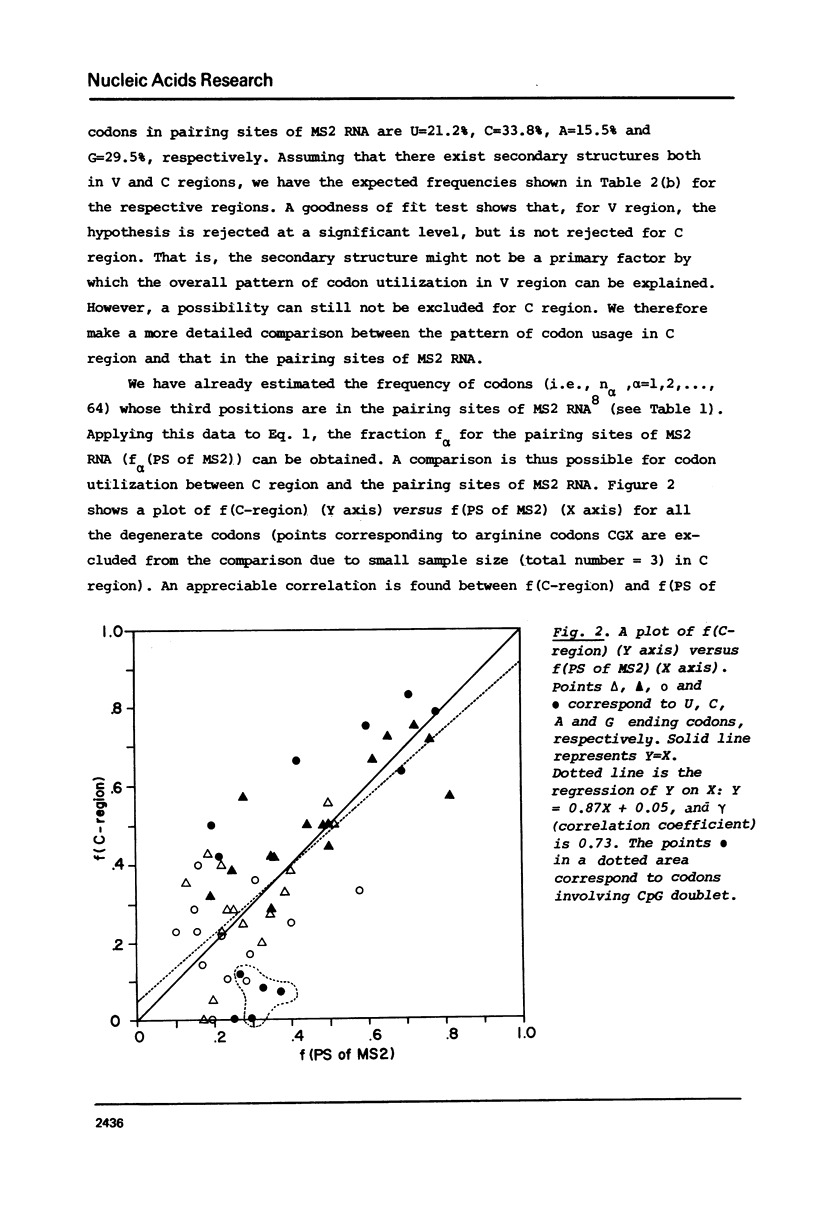
The pattern of codon utilization in the variable and constant regions of immunoglobulin genes are compared. It is shown that, in these regions, codon utilizations are quite distinct from one another: For most degenerate codons, there is a selective bias that prefers C and/or G ending codons to U and/or A ending codons in the constant region compared with the bias in the variable region. This would strongly suggest that, in immunoglobulin genes, the bias in code word usage is determined by other factors than those concerning with the translational mechanism such as tRNA availability and codon-anticodon interaction. A possibility is also suggested that this differance of code word usage between them is due to the existence of secondary structure in the constant region but not in the variable region.
Full text
PDF




Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Berger E. M. Pattern and chance in the use of the genetic code. J Mol Evol. 1978 Feb 21;10(4):319–323. doi: 10.1007/BF01734221. [DOI] [PubMed] [Google Scholar]
- Bernard O., Hozumi N., Tonegawa S. Sequences of mouse immunoglobulin light chain genes before and after somatic changes. Cell. 1978 Dec;15(4):1133–1144. doi: 10.1016/0092-8674(78)90041-7. [DOI] [PubMed] [Google Scholar]
- Brack C., Hirama M., Lenhard-Schuller R., Tonegawa S. A complete immunoglobulin gene is created by somatic recombination. Cell. 1978 Sep;15(1):1–14. doi: 10.1016/0092-8674(78)90078-8. [DOI] [PubMed] [Google Scholar]
- Fiers W., Contreras R., Duerinck F., Haegeman G., Iserentant D., Merregaert J., Min Jou W., Molemans F., Raeymaekers A., Van den Berghe A. Complete nucleotide sequence of bacteriophage MS2 RNA: primary and secondary structure of the replicase gene. Nature. 1976 Apr 8;260(5551):500–507. doi: 10.1038/260500a0. [DOI] [PubMed] [Google Scholar]
- Fiers W., Contreras R., Duerinck F., Haegmean G., Merregaert J., Jou W. M., Raeymakers A., Volckaert G., Ysebaert M., Van de Kerckhove J. A-protein gene of bacteriophage MS2. Nature. 1975 Jul 24;256(5515):273–278. doi: 10.1038/256273a0. [DOI] [PubMed] [Google Scholar]
- Fiers W., Grosjean H. On codon usage. Nature. 1979 Jan 25;277(5694):328–328. doi: 10.1038/277328a0. [DOI] [PubMed] [Google Scholar]
- Fitch W. M. Is there selection against wobble in codon-anticodon pairing? Science. 1976 Dec 10;194(4270):1173–1174. doi: 10.1126/science.996548. [DOI] [PubMed] [Google Scholar]
- Fitch W. M. The large extent of putative secondary nucleic acid structure in random nucleotide sequences or amino acid derived messenger-RNA. J Mol Evol. 1974;3(4):279–291. doi: 10.1007/BF01796043. [DOI] [PubMed] [Google Scholar]
- Grantham R. Viral, prokaryote and eukaryote genes contrasted by mRNA sequence indexes. FEBS Lett. 1978 Nov 1;95(1):1–11. doi: 10.1016/0014-5793(78)80041-6. [DOI] [PubMed] [Google Scholar]
- Grosjean H., Sankoff D., Jou W. M., Fiers W., Cedergren R. J. Bacteriophage MS2 RNA: a correlation between the stability of the codon: anticodon interaction and the choice of code words. J Mol Evol. 1978 Dec 29;12(2):113–119. doi: 10.1007/BF01733262. [DOI] [PubMed] [Google Scholar]
- Hamlyn P. H., Browniee G. G., Cheng C. C., Gait M. J., Milstein C. Complete sequence of constant and 3' noncoding regions of an immunoglobulin mRNA using the dideoxynucleotide method of RNA sequencing. Cell. 1978 Nov;15(3):1067–1075. doi: 10.1016/0092-8674(78)90290-8. [DOI] [PubMed] [Google Scholar]
- Hensgens L. A., Grivell L. A., Borst P., Bos J. L. Nucleotide sequence of the mitochondrial structural gene for subunit 9 of yeast ATPase complex. Proc Natl Acad Sci U S A. 1979 Apr;76(4):1663–1667. doi: 10.1073/pnas.76.4.1663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jukes T. H. Codons and nearest-neighbor nucleotide pairs in mammalian messenger RNA. J Mol Evol. 1978 Jun 20;11(2):121–127. doi: 10.1007/BF01733888. [DOI] [PubMed] [Google Scholar]
- Min Jou W., Haegeman G., Ysebaert M., Fiers W. Nucleotide sequence of the gene coding for the bacteriophage MS2 coat protein. Nature. 1972 May 12;237(5350):82–88. doi: 10.1038/237082a0. [DOI] [PubMed] [Google Scholar]
- Post L. E., Strycharz G. D., Nomura M., Lewis H., Dennis P. P. Nucleotide sequence of the ribosomal protein gene cluster adjacent to the gene for RNA polymerase subunit beta in Escherichia coli. Proc Natl Acad Sci U S A. 1979 Apr;76(4):1697–1701. doi: 10.1073/pnas.76.4.1697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogers J., Clarke P., Salser W. Sequence analysis of cloned cDNA encoding part of an immunoglobulin heavy chain. Nucleic Acids Res. 1979 Jul 25;6(10):3305–3321. doi: 10.1093/nar/6.10.3305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakano H., Rogers J. H., Hüppi K., Brack C., Traunecker A., Maki R., Wall R., Tonegawa S. Domains and the hinge region of an immunoglobulin heavy chain are encoded in separate DNA segments. Nature. 1979 Feb 22;277(5698):627–633. doi: 10.1038/277627a0. [DOI] [PubMed] [Google Scholar]
- Seidman J. G., Leder A., Edgell M. H., Polsky F., Tilghman S. M., Tiemeier D. C., Leder P. Multiple related immunoglobulin variable-region genes identified by cloning and sequence analysis. Proc Natl Acad Sci U S A. 1978 Aug;75(8):3881–3885. doi: 10.1073/pnas.75.8.3881. [DOI] [PMC free article] [PubMed] [Google Scholar]


