Abstract
Because we can observe oscillation within individual cells and in the tissue as a whole, the suprachiasmatic nucleus (SCN) presents a unique system in the mammalian brain for the analysis of individual cells and the networks of which they are a part. While dispersed cells of the SCN sustain circadian oscillations in isolation, they are unstable oscillators that require network interactions for robust cycling. Using cluster analysis to assess bioluminescence in acute brain slices from PERIOD2∷Luciferase (PER2∷LUC) knockin mice, and immunochemistry of SCN from animals harvested at various circadian times, we assessed the spatiotemporal activation patterns of PER2 to explore the emergence of a coherent oscillation at the tissue level. The results indicate that circadian oscillation is characterized by a stable daily cycle of PER2 expression involving orderly serial activation of specific SCN subregions, followed by a silent interval, with substantial symmetry between the left and right side of the SCN. The biological significance of the clusters identified in living slices was confirmed by co-expression of LUC and PER2 in fixed, immunochemically stained brain sections, with the spatiotemporal pattern of LUC expression resembling that revealed in the cluster analysis of bioluminescent slices. We conclude that the precise timing of PER2 expression within individual neurons is dependent on their location within the nucleus, and that small groups of neurons within the SCN give rise to distinctive and identifiable subregions. We propose that serial activation of these subregions is the basis of robustness and resilience of the daily rhythm of the SCN.
Keywords: circadian rhythms, luciferase, networks, PER2, SCN, synchronization
Introduction
Investigations of neuronal connectivity in the mammalian brain confront a fundamental problem of access. At present, specific cells can be studied when brain tissue is suitably sliced, allowing neurons to be individually identified and visualized. Available visualization techniques require the destruction of at least some of the network connections being studied. Here, we make a first attempt to address this problem by investigating spatiotemporal structure of a specific neural circuit using novel statistical methods applied to bioluminescent cells in imaged slices, supplemented by analysis of brain sections from animals euthanized at corresponding specific time points. While the paper focuses on the brain clock, the analytic techniques we develop are applicable to any spatially organized data that have a temporal dimension.
Several features of the SCN make it an attractive system in which to study cells and circuits. The functions and basic organization of the mammalian suprachiasmatic nucleus (SCN) and its individual cells are well characterized. This hypothalamic nucleus is comprised of a diverse population of ~20 000 neurons, and it is the locus of a master circadian clock. In one view, based on electrical activity of individual neurons (Welsh et al., 1995) and on partial lesions of the nucleus (Rusak, 1977; Davis & Gorski, 1988; Harrington et al., 1993), the cellular elements of the SCN appear to be equipotential and equifunctional, operating as an undifferentiated mass of oscillators, self-organized through weak, Huygens-type coupling (Strogatz & Stewart, 1993). Initially, this view was supported by evidence indicating that as long as 25% of the nucleus survived ablation, circadian behavior was sustained. In another view, the SCN cell population is functionally heterogeneous, with peptidergically similar cells clustered together within the nucleus (van den Pol & Tsujimoto, 1985; Moore & Silver, 1998; Abrahamson & Moore, 2001; Antle & Silver, 2005). Questions regarding the significance of these spatially organized cell groupings became salient with the discovery of local variations across the nucleus in phase and amplitude measured by electrical activity, clock gene and clock-controlled gene expression (Hamada et al., 2001, 2004; Jobst & Allen, 2002; Yan & Okamura, 2002; Quintero et al., 2003; Yamaguchi et al., 2003; Karatsoreos et al., 2004; Nakamura et al., 2005; Rohling et al., 2006). The emergent problem is to characterize this local variation and to explain its functional significance.
Many mechanisms mediating intercellular synchronization are known, including contacts through electrochemical synapses or electrical coupling, changes in glial activity, expression of neural cell adhesion molecule, polysialic acid, receptor expression and protein trafficking (Servière & Lavialle, 1996; Shen et al., 1997, 2001; Liu & Reppert, 2000; Harmar et al., 2002; Aton & Herzog, 2005; Brown et al., 2005, 2007; Maywood et al., 2007; LeSauter et al., 2009). A key step in unraveling SCN network architecture is to explore how its cellular elements give rise to the characteristic daily spatiotemporal changes at the tissue level. The molecular clockwork consists of oscillatory transcriptional and translational networks (Reppert & Weaver, 2002; Ukai & Ueda, 2010) and individual SCN neurons in dispersed cell cultures are capable of sustaining oscillations (Welsh et al., 1995; Webb et al., 2009). However, individual cells are weak, inducible, and probabilistic oscillators that rely on network interactions for stability (Liu et al., 2007; Webb et al., 2009). Clearly, the tissue is the issue.
Previous work has attempted to characterize spatiotemporal changes in rhythmic neuronal activation based on a small number of SCN cells, and/or using qualitative or a priori methods to capture the temporal domain (Yamaguchi et al., 2003; Davidson et al., 2009; Yan et al., 2007; Quintero et al., 2003). In the present paper we explore how gene expression in individual cells emerges to produce a coherent oscillation at the tissue level by examining the spatiotemporal pattern of bioluminescence in the SCN using brain slices from transgenic PERIOD2∷Luciferase (PER2∷LUC) knockin mice, and regional localization of PERIOD2 (PER2) in immunochemically stained tissue. An algorithmic method for identifying successive regions of activation was developed using statistical cluster analysis. Confounds attributable to scatter of light and noise were assessed. The statistical analysis indicates functional differentiation of subregions of SCN cells based on anatomical position in the nucleus, and provides supporting evidence based on immunochemical analysis that this regional differentiation occurs in vivo.
Materials and methods
Animals and housing
For studies of bioluminescence, SCN slices were obtained from circadian reporter mice expressing a fusion protein in which firefly luciferase (LUC) is fused in frame to the C-terminus of mPER2 (Yoo et al., 2004). The PER2∷LUC fusion protein is a highly faithful bioluminescent reporter of circadian clock function, reflecting both transcriptional and post-transcriptional regulation of the mPer2 locus. Some mice were of an alternate strain incorporating an SV40 polyadenylation site after the luciferase gene. SCN slices from this strain are ~30% brighter but otherwise indistinguishable from slices from the original strain lacking SV40. PER2∷LUC mice were bred as homozygotes on a mixed B6/129 genetic background, and maintained in light : dark (LD) 12 : 12 h throughout gestation and from birth until used for experiments.
For immunochemical studies of LUC expression, transgenic PER2∷LUC mice (N = 15), aged 5–6 weeks, were maintained in a 12 : 12 h LD cycle and C57BL/6 mice (Charles River Laboratories, Wilmington, MA, USA) housed in either LD (N = 16) or in constant darkness (DD; N = 19) were euthanized at different times of day. Animals were anesthetized with 200 mg/kg pentobarbital and then perfused intracardially using 50 mL of saline followed by 100 mL of 4% paraformaldehyde in 0.1 m phosphate buffer (PB), pH 7.3. The brains were postfixed in paraformaldehyde for 24 h at 4 °C, then cryoprotected with 20% sucrose in 0.1 m PB overnight. Coronal brain slices (50 μm) containing the SCN were cut on a cryostat. All animal studies were conducted in accordance with the regulations of the Committees on Animal Care and Use at University of California San Diego and Columbia University.
SCN slice culture
SCN slices were dissected and cultured in HEPES-buffered serum-free explant medium containing B-27 and luciferin, as previously described (Yamazaki, 2000). The HEPES-buffered, air-equilibrated medium was based on DMEM (12100-046; GIBCO/Invitrogen, Carlsbad, CA, USA), and supplemented with 10 mm HEPES, 1.2 g/L NaHCO3, 25 U/mL penicillin, 25 μg/mL streptomycin, 2% B-27 (GIBCO/Invitrogen) and 0.1 mm luciferin (L-8220; BioSynth International, Itasca, IL, USA), pH 7.4. SCN slices were cut by tissue chopper (Stoelting, Kiel, WI, USA) in the coronal plane to a thickness of 300–400 μm, trimmed to ~2 × 2 mm, and cultured on Millicell-CM membrane inserts (PICMORG50; Fisher Scientific, Pittsburg, PA, USA). Inserts were placed in 35-mm culture dishes containing ~1 mL of medium, and covered by 40 mm circular coverslips (40CIR1; Fisher Scientific) sealed in place with vacuum grease to prevent evaporation. SCN slices were prepared from early postnatal mice (4–7 days old), and then allowed to thin in culture for 1–7 weeks. During this time, slices were monitored in a LumiCycle luminometer (Actimetrics Inc., Wilmette, IL, USA), which was kept inside a standard tissue culture incubator at 36 °C. Medium was replaced every 1–2 weeks. Just before imaging, medium was changed to fresh medium identical to that used for LumiCycle recording except that it contained 1 mm luciferin.
Bioluminescence imaging
Imaging of PER2∷LUC bioluminescence was performed as previously described (Liu et al., 2007). Dishes were placed on the stage of an inverted microscope (IX71; Olympus, Center Valley, PA, USA) in a dark room. A heated lucite chamber around the microscope stage (Solent Scientific, Segensworth, UK) kept the cells at a constant 36 °C. Images were collected using an Olympus 10x UPlanApo objective and transmitted to a CCD camera (Spectral Instruments SI800, Tucson, AZ, USA) cooled to −90 °C. Some experiments used an Orca IIER camera (Hamamatsu, Bridgewater, NJ, USA) with signal/noise enhanced by 2 × 2 binning of pixels. A total of 16 unilateral SCNs were studied from eight separate slices. One slice (no. 6) was initially out of focus and the camera was adjusted, so that the first 2 days of imaging were lost. Images of 30–60 min exposure duration were collected continuously for 3–5 days. Integration of bioluminescence over the entire imaging field gave population patterns similar to those measured in the luminometer.
Image processing
Using MetaMorph (Molecular Devices, Sunnyvale, CA, USA), images were corrected for bias and hot pixels by background subtraction. Cosmic rays and other artifacts were removed by setting the highest and lowest 0.0001 quantiles of pixel values in the stack to the mean of their neighborhood (surrounding pixels in the previous, same and next frames) using Mathematica 7 (Wolfram Research, Champaign, IL, USA).
Optical noise
Noise includes signal noise (‘shot noise’), read noise and thermal noise from the camera. Noise of the recording system was estimated from the SD of 10 × 50 pixel patches in parts of the movie frame that did not contain brain tissue. The ratio of noise to oscillation size across slices ranged from 2.6 to 4%. This value has essentially no circadian oscillation, indicating that noise in this system is very low and does not play a role in the following analysis.
Analysis of bioluminescence
Raw data
The raw data are stacks of image files with brightness represented by integers. For the cluster analysis, each stack was analyzed in two ways, first with the bilateral SCN and then separately, for each unilateral SCN, in two new stacks containing the left and right nucleus. The stacks for each slice were adjusted to be close to the same size, and to include corresponding areas of brain tissue.
Visualization of LUC in serial frames of the image stacks
To visualize changes in bioluminescence over time, raw image stacks were made into individual image sequences using ImageJ (NIH: http://rsb.info.nih.gov/ij/), and the average brightness of the full frame was taken for each image to create a time series. Peaks and troughs in the time series were computed by taking local maxima and minima over a moving 15-h window. Using the first trough as time zero, frames at 3-h intervals were pseudocolored in Volocity (Improvision Inc, Lexington, MA, USA). The rainbow color scale was calibrated to the brightest slice at the top and to extra-SCN areas at the bottom, making the non-SCN tissue the same color for each frame.
Cluster analysis algorithm
The initial analysis of images used an unsupervised learning algorithm, termed ‘k-means cluster analysis’ (Tibshirani et al., 2001), which can be applied to any set of observations when a quantitative index of the ‘similarity’ of the observations can be specified. The cluster analysis algorithm operates without any prior information input by the researcher as to the probable structure of the clusters. The algorithm takes the set of observations and the similarity function as inputs, and creates ‘clusters’ that have a high statistical degree of similarity within each cluster and large differences among clusters.
The following standard clustering algorithm was used – given an arbitrary set of observations [x1, …, xn], and a similarity function (or distance function) d[xi, xj] with the properties d[.,.] ≥ 0, d[x, x] = 0 (each object is similar to itself), several (k) observations are randomly chosen as prototypes for clusters. Candidate clusters are formed by assigning each remaining observation to the group whose prototype is most similar to it. For each candidate cluster, a mean reference is computed. The distance from each member of the cluster to the reference (i.e. dispersion) is also computed.
An observation is taken at random, and switched from one cluster to another and the dispersion is recomputed. If the mean dispersion is less than it was previously, the clusters are updated. If not, the algorithm tries again with a different object. This step is repeated until every possible move from one cluster to another increases the mean dispersion, ensuring minimum within-cluster dispersion.
In the threshold analysis the Gap method of choosing k clusters was used with a tolerance parameter. Tibshirani et al. (2001) describe the computation of the Gap[k] statistic from the minimal dispersions for k clusters, and recommend the choice of the smallest k that satisfies Gap[k] > Gap[k + 1] − T * sk+1 where sk+1 is an estimate of the SD of the Gap[k + 1] statistic, and T is the tolerance. Thus, so long as the Gap is below the desired statistical threshold, the algorithm increases the number of clusters and repeats the optimization. Once the desired statistical threshold has been reached, the algorithm outputs the completed cluster analysis in the form of a list of the clusters (subsets) of the original objects.
A primary advantage of the k-means clustering analysis using the gap method is that it is unsupervised. Thus, the experimenter does not provide any assumptions about the underlying structure of the data. As such, a unique insight gained by using the cluster analysis is that a recognizable ‘map’ and ‘rhythm’ of distinct SCN subregions is specified solely by the spatiotemporal distribution of bioluminescent signal.
Implementation of cluster analysis of bioluminescence in SCN slices
The cluster analysis was computed in Mathematica 7 and applied to the image stacks (reference code is available at: http://www.columbia.edu/cu/psychology/silver/cluster/). Each image in the stack is first compressed into superpixels by taking the average value of 5 × 5 blocks of pixels (as a point of reference, one pixel equal to ~1.32 μm). The total level of noise at the superpixel level was small due to spatial averaging and does not contribute to the remaining analyses. For each of these bins, a time series of brightness values is generated from the stack. The distance (degree of dis-similarity) between the time series for each superpixel and all other superpixels is computed using a distance function (see below for definitions; the default distance function is cosine distance). Thus the distance between any superpixel and itself is 0 and superpixels that have similar time patterns of brightness are regarded as similar by the clustering algorithm.
We did the cluster analyses using three different distance functions (correlation, Bray–Curtis, and cosine distance). (Note that correlation distance is the cosine distance with the vectors normalized by subtraction of their mean values). The distance between the time series of superpixel u and superpixel v is defined as follows:
Correlation distance:
Bray–Curtis distance:
Cosine distance:
We present all the results using the cosine distance analysis because it emphasizes phase differences in the time series, a key aspect of circadian rhythms, while retaining sensitivity to the difference in amplitudes among superpixels. Next, using the cosine distance function, three different cluster analyses were performed on each slice, as follows. First, an unconstrained gap method clustering (termed ‘Gap’) was computed on the bilateral SCNs with a tolerance of 6, meaning that there had to be a difference of at least six SDs between the gap statistic for k clusters and k + 1 clusters (see sk+1 above). Second, in order to facilitate visualization of the time series analysis, another clustering on the bilateral SCNs was computed with k constrained to 5 (termed ‘5Clusters’). Third, to reveal right–left similarities and/or differences in the SCN, a ‘unilateral’ clustering analysis (Unil Clus) was computed separately for the left and right side of the nucleus with k set to the same number of clusters previously found on the bilateral SCN slice using the ‘Gap’ method. The ‘Gap’ method and ‘5Clusters’ analyses were also performed using Bray–Curtis and correlation distance to compare the spatiotemporal patterns of bioluminescence with the different statistical analyses (results are shown for a representative slice; see Supporting Information Data S1, Fig S1).
Quantitative analysis of the clusters
Once the clustering operations were computed, the following analyses were completed on each slice – maps, time series of cluster bioluminescence, and time series of the SD of bioluminescence. Maps indicate the location of each cluster in each slice using a color-coded spatial map of superpixels, where each color denotes the cluster uncovered in the analysis. The time series of cluster bioluminescence shows the amplitude and phase of each cluster over time. The time series of the SD of bioluminescence shows the dispersion within each cluster over time. The time series of cluster bioluminescence and the SD of bioluminescence time series within each cluster are color-coded to the spatial map. To facilitate visualization of the time series for cluster brightness and SD, the 5Cluster analysis is shown on a single cycle. While facilitating visualization, this analysis forces the algorithm to merge parts of various clusters identified in the unconstrained analysis.
Assessment of scatter of light
An unwelcome feature of measuring bioluminescence is that oscillations are detected in regions that are known to lack rhythmicity, such as parts of the frame lacking neural tissue, the ventricle and areas below the optic tracts. For this reason, we next assessed the scatter of light and/or physical properties of the detection system that give rise to noise.
To be useful in analyzing the internal organization of the SCN, the statistical clustering method must distinguish SCN subregions independent of scatter of light from bright areas of the tissue. Importantly, scatter results in apparent brightness fluctuations in areas of the image that cannot emit bioluminescence. We assess this effect of scatter by measuring the bioluminescent signal below the optic chiasm because this region has no brain tissue (available for three slices). We manually selected regions of interest (ROIs) outside the tissue. Mean brightness was calculated in these ROIs in three ways: (i) as a function of distance from the tissue; (ii) through time; and (iii) from the left to the right edge of the frame (Fig. 1A).
Fig. 1.
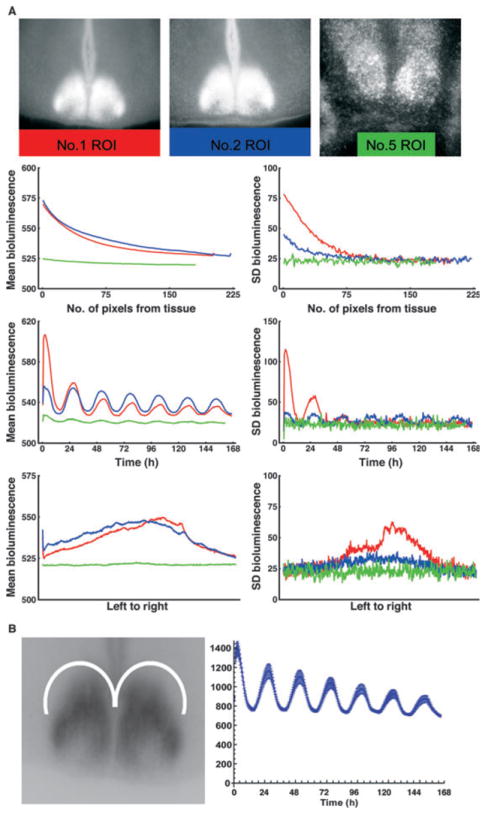
Analysis of scatter of light. (A) Scatter in non-tissue ROIs. (Top row) Time-averaged frame of bioluminescence showing non-tissue ROI, delineated by a colored rectangle for each case (color code: red, slice 1; blue, slice 2; green, slice 5). (Second row) Mean bioluminescence and SD bioluminescence, averaged over distance from tissue. Zero distance corresponds to the topmost line of pixels in the ROI, which are closest to the tissue. (Third row) Mean bioluminescence and SD bioluminescence of the ROI averaged at each frame. (Bottom row) Mean bioluminescence and SD bioluminescence averaged from leftmost row of pixels to rightmost row of pixels in each ROI. Note that circadian oscillation is detected despite the absence of tissue to produce LUC. (B) Scatter of light in extra-SCN (ROI). The left panel shows the crescent-shaped ROI used to measure time series of oscillation (± SD) of bioluminescence in a region outside the SCN in each slice used of this study. Right panel shows the ROI mean bioluminescence for slice 1.
Light also scatters within neural tissue, slightly differently for each slice. To evaluate the scatter of light in each slice, crescent-shaped ROIs were chosen in brain tissue dorsal to each SCN (Fig. 1B). For each slice the ROI time series of mean pixel brightness and SD of pixel brightness were computed using the same methods as for the clusters, to establish a floor level for regional oscillation. The SDs for the ROIs are pixel-based, and so are divided by 5 to conform to the 5 × 5 superpixels used in the cluster analysis. In each slice, SCN regions that oscillate below the value established by this ROI time series (with SD error bars) lack measurable regional oscillations that can be discerned from scatter.
The patterns revealed by this analysis are consistent with the hypothesis that the apparent brightness oscillations we observe in regions lacking brain tissue and in extra-SCN regions are artifacts of scattered light. While this is distracting, it does not have implications for the validity of the cluster analysis in identifying subregions of the SCN tissue areas with different time profiles of brightness, and higher amplitude than scattered light.
Perfusion, fixation and immunochemistry
To determine how clusters found in the spatial–temporal pattern of expression of LUC in real-time bioluminescence imaging of SCN slices in vitro correspond to those seen in immunochemically stained sections, we evaluated brain sections from animals euthanized at specific times of day. To identify the precise spatial localization of LUC or PER2 expression within the SCN, each section was also labeled for vasopressin (VP). To determine whether LUC and PER2 occur in the same cells, double label staining was performed.
Fixed sections were washed with 0.1 m PB containing 0.1% Triton-X-100. Sections were blocked (1 h) with normal donkey serum (NDS; 1 : 50 in PB + 0.3% Triton) followed by incubation in the primary antibodies in PB + 0.3% Triton. After incubation of primary for 48 h at 4 °C, sections were washed in PB + 0.1% Triton then stained with the appropriate secondary antibody conjugated to Cy2 or Cy3 fluorescent chromogens made in donkey (Jackson ImmunoResearch, West Grove, PA, USA; 1 : 200 in PB + 0.3% Triton) for 2 h. Sections were mounted, dehydrated and coverslipped with Krystalon (EM Science, Gibbstown, NJ, USA).
The following antibodies were used. (i) Polyclonal anti-PER2 made in rabbit against human PER2 (1 : 2000; Alpha Diagnostic International, San Antonio, TX, USA; Catalog number PER21-A; affinity pure). The polyclonal PER2 antibody is collected from rabbit immunized with a 23-amino-acid peptide sequence (gene accession no. 054943) of mouse PER2 conjugated to keyhole limpet hemocyanin; epitope location ~ C-terminus. Staining was eliminated by preincubation of the diluted antiserum (1 : 1000) with 2 μg/mL of the protein. No staining was detected in PER1-PER2 mutant mice (results from our lab). (ii) Polyclonal anti-VP made in guinea pig against synthetic VP (Peninsula Laboratories Inc., San Carlos, CA, USA; Catalog number T-5048). The polyclonal AVP antibody is collected from guinea pigs immunized with a synthetic peptide as the immunogen. Staining was eliminated by preincubation of the diluted antiserum (1 : 5000) with 2 μg/mL of the protein. No staining was detected in Brattleboro rats (results from our lab). (iii) Polyclonal anti-luciferase made in rabbit (catalog – ab21176) or in goat (catalog – ab498) against firefly luciferase (1 : 1000; Abcam, Cambridge, MA, USA). The polyclonal LUC antibody is collected from rabbit or goat immunized with the full-length native protein (purified) [Firefly (Photinus pyralis)]. No staining was detected in wild-type animals (results from our lab). The secondary antibodies (Jackson Immuno-Research) used were Cy2 donkey antirabbit for single label, and Cy2 donkey antirabbit and Cy3 donkey antiguinea pig or Cy2 donkey antigoat and Cy3 donkey antirabbit for double labels.
Analysis of immunostained sections
Sections were examined on a Nikon Eclipse E800 microscope (Nikon, Tokyo, Japan) using a Plan-Apo 20 × lens numerical aperture 0.75 (Nikon). Images were captured using QCapture software (version 2.95.0; Quantitative Imaging, Surrey, BC, Canada) connected to a Q-Imaging Retiga EXi, fast 1394 camera (Quantitative Imaging). In addition, confocal imaging was performed as described previously (Karatsoreos et al., 2004) using a Zeiss Axiovert 200 m fluorescence microscope (Carl Zeiss, Thornwood, NY, USA) with a Zeiss LSM 510-META scanning confocal attachment. Sections were examined with lasers using excitation wavelength of 488 nm for Cy2 and 543 nm for Cy3.
Visualization and quantification of the immunochemical data
For 3-D visualization of LUC expression, confocal images (scaling: X, 0.44 μm; Y, 0.44 μm; Z, 1.5 μm) where taken with excitation wavelength of 488 nm on alternate sections of the SCN. Image stacks from each alternate brain slice from rostral to caudal were composed of 9, 10, 10, 10, 8 and 7 serial confocal images. Stacks were imported into Volocity to form a 3-D image of the SCN. The 3-D image was rotated and snapshots taken of the frontal, medial and caudal aspects. These images were also pseudocolored in Volocity as above. The image of the frontal view was imported into NIH Image, converted to grayscale and adjusted with the threshold function. For visualization of sections immunostained for LUC, PER2 and VP, grayscale images were taken with the Q-Imaging Retiga camera, were imported into Adobe Photoshop 7.0 (Adobe Systems Inc., San Jose, CA, USA) color-inverted and contrast-normalized using the ‘Image-Adjustment-Levels’ dialog box. In order to enhance visualization of the relative brightness of staining in various SCN regions, grayscale images of LUC- and PER2-stained sections were pseudocolored using QCapture Pro by using the ‘process’, ‘pseudocolor’ commands and choosing the blue-to-red color spread, the upper limit of 255 and 5 divisions. For quantification of the time course of PER2 and LUC expression the levels were measured by relative optical density (ROD) on three sections from rostral, mid and caudal SCN identified in sections double-labeled for VP. Specifically, the VP region was outlined in Photoshop. The outline was overlaid on the LUC- or PER2-labeled section and ‘Mean Gray Value’ was measured in ImageJ. This value was subtracted from Mean Gray Value of non-SCN tissue of the same section to obtain the ROD. Mean RODs were calculated and plotted using Excel (Microsoft Corp., Redmond, WA, USA).
Results
In order to understand how the preparation of coronal brain slices might influence network architecture measured by bioluminescence, we examined confocal image stacks of sections from the rostral to caudal extent of the SCN at zeitgeber time (ZT) 13 (approximate LUC peak expression time), rotated horizontally so as to permit visualization of regional variation in LUC expression. To facilitate visualization, the LUC expression data are pseudocolored and coded for intensity (Fig. 2). The results point to noteworthy regional heterogeneity in the rostrocaudal axis. There is a crescent-shaped region of high LUC expression in the mid-region of the nucleus. This region is better visualized in the inset. A roughly oval area in the ventrolateral SCN below the crescent has scattered LUC-positive cells but generally low expression levels. As seen in the medial view of the SCN, the region with reduced LUC expression (delineated by the black oval) lies in the caudal aspect of the nucleus. This result suggests that different network architectures are likely to be detected depending on which components of the nucleus (rostral, mid or caudal) are included when the SCN slice is prepared. That is, a prepared slice containing the rostral to mid SCN will result in a different network architecture than a section containing the caudal region.
Fig. 2.
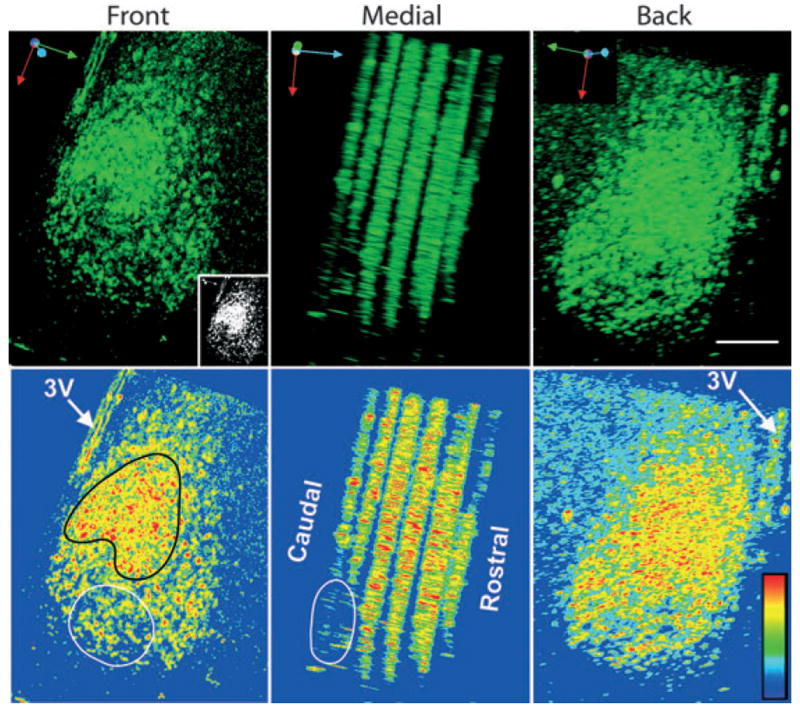
Three-dimensional image of a unilateral SCN at ZT13, immunostained for LUC. Confocal image stacks of alternate sections from the rostral to caudal extent of the nucleus are seen from the front, back and medial aspects. The six stacks from rostral to caudal are composed of 10, 10, 10, 10, 8 and 7 confocal 5-μm optical slices. An SCN measures ~700 μm in its rostrocaudal extent. Here the alternate sections and the rostral- and caudal-most sections are not included. The black spaces between slices in this panel are the tops and bottoms of the scanned optical slices. The top row shows LUC-ir while the bottom row shows the same images, pseudocolored to enhance visualization of intensity of label and to match the pseudocoloring used in Figs 3 and 4. The image is rotated horizontally to expose regional variation in LUC expression. The orientation arrows in the upper left corner of each image indicate rotational axis (red, dorsal-to-ventral; green, medial-to-lateral; blue, caudal-to-rostral). The highest expression of LUC occurs in a crescent-shaped region (outlined in black) similar to that in slices 1 and 2 in Figs 3 and 4. The inset (black–white) enables visualization of relative expression levels of LUC (prepared using the threshold function in NIH image). The ovals (white outline) show regions of low LUC expression lying ventrocaudally in the nucleus. The rostrocaudal differences highlight regional variation in LUC expression. Pseudocolor rainbow scale (blue, low and red, high expression) is shown on lower left side of the last panel. Scale bar, 100 μm.
Spatiotemporal changes in SCN bioluminescence
Changes in bioluminescence of the bilateral SCN are presented in pseudocolored single frames captured at 3-h intervals throughout the circadian cycle (Fig. 3). Visual inspection of these images points to identifiable regions, with a distinct spatial–temporal topography of LUC expression at each phase of oscillation. A second characterization of changes in LUC over time is captured in frames showing the pattern of activation at times of peak and trough amplitude, and the halfway points from trough to peak and peak to trough (Fig. 4). The rightmost column in Fig. 4 quantifies the overall amplitude of rhythms in each slice.
Fig. 3.
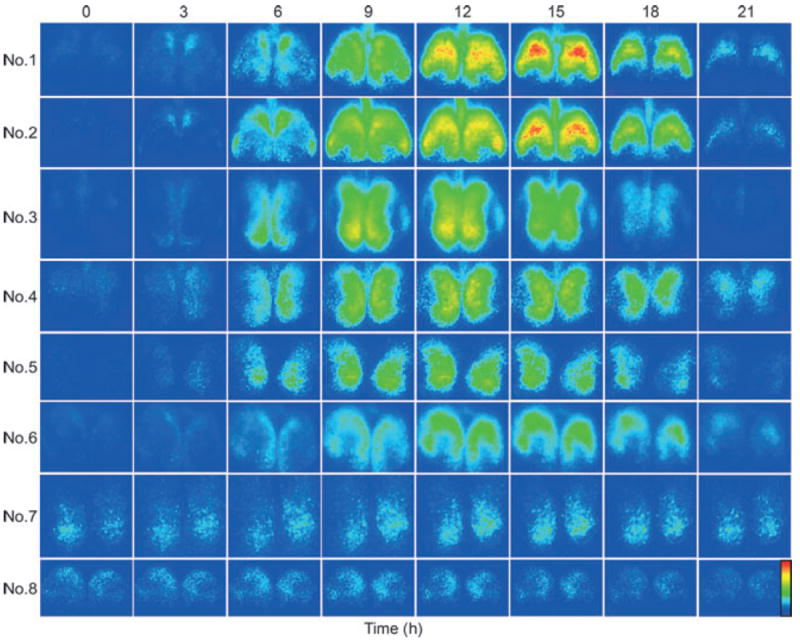
Spatiotemporal pattern in PER2∷LUC bioluminescence in SCN slices at 3-h intervals. Time zero was defined as the trough of the first cycle (see Materials and Methods for details). The pseudocolored images are normalized to the brightest slice. Rainbow scale (blue, low and red, high expression) is shown on right side of the last panel.
Fig. 4.
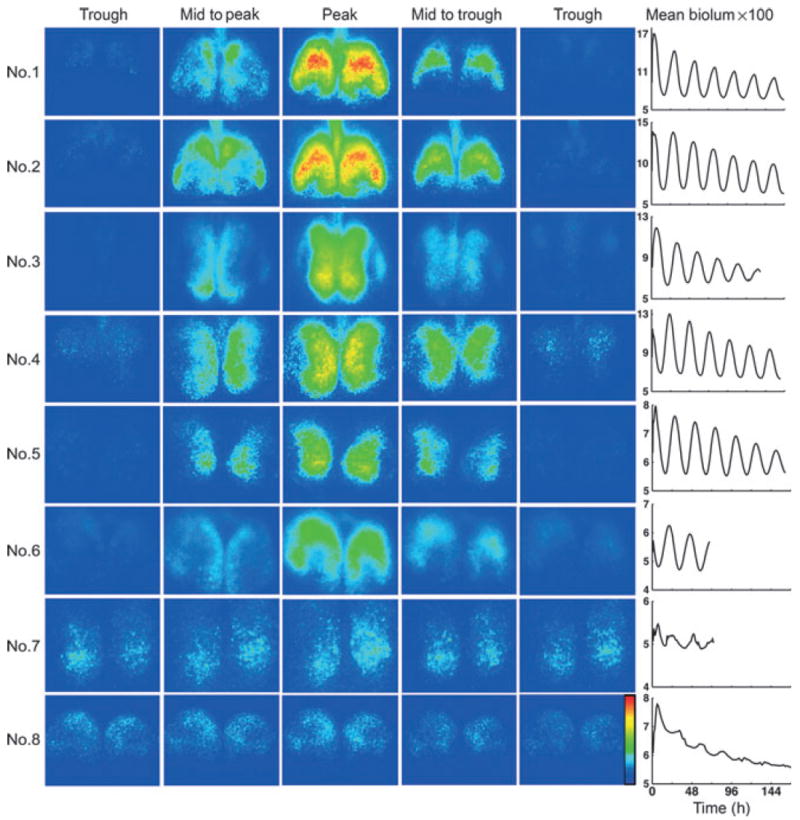
Spatiotemporal pattern in PER2∷LUC bioluminescence (same SCN slices as in Fig. 3). The frames are selected to show the time of peak and trough expression, as well as half-way to peak and trough for each slice. The circadian rhythm for each slice, computed on the whole-frame mean time series, is shown in the right column. Rainbow scale is shown on right side of the last panel.
In slices 1–6, the results point to a regularly recurring pattern of changes, with peak LUC expression at ~1200–1500 h and a distinct interval during which time LUC expression in the SCN is not detected. Importantly, some areas within the SCN tissue, delineated by GRP-containing cells in mouse SCN, do not express detectable rhythmicity in clock genes at any phase of the cycle, as has previously been described (Karatsoreos 2004).
Analysis of the overall brightness for slices 1–6 indicates that the peak expression of LUC occurs ~12 h after the trough (Table 1). In contrast, slices 7 and 8 present a somewhat different picture – here there is no phase of the cycle in which LUC is entirely absent (Figs 3 and 4). Furthermore the oscillation is irregular (Fig. 3, right column), the left side of slice 7 cycles outside the circadian range, and the time of peak expression for slice 8 is at hour 0800 while all the other slices peak close to mid-cycle (Table 1).
Table 1.
Analysis of first cycle peaks and troughs
| Brain slice | Weeks in culture | Period peak to peak (h)
|
Interval (h) from trough to peak (1st cycle)
|
||||
|---|---|---|---|---|---|---|---|
| Left | Right | Bilateral | Left | Right | Bilateral | ||
| 1 | 1 | 26 | 25.5 | 25.5 | 12.5 | 13.5 | 13 |
| 2 | 4 | 25.5 | 25.5 | 25.5 | 12.5 | 11.5 | 12.5 |
| 3 | 2 | 26 | 26 | 26 | 11 | 11.5 | 11 |
| 4 | 3 | 25.5 | 25 | 25.5 | 11 | 12 | 11 |
| 5 | 2 | 24.5 | 24 | 24.5 | 11 | 11.5 | 11.5 |
| 6 | 3 | 25 | 25 | 25 | 12.5 | 13 | 13 |
| 7 | 7 | 32 | 26 | 26 | 9 | 9 | 9 |
| 8 | 1 | 24 | 24 | 24 | 8 | 8 | 8 |
Cluster analysis
As noted in Materials and Methods, the results of the cosine analysis using the Gap method clustering are presented for each SCN as a map, time series for brightness, and time series for SD of the brightness. Based on the Gap algorithm, there are 2–7 clusters with signal exceeding the level of light scatter. In some cases, the large number of clusters makes it difficult to visualize the time series results. To facilitate visualization and comparisons among slices, the 5Cluster data for map and for both time series are shown for a single cycle. The results are presented in the following sequence – analyses of slices 1 and 2 are shown in Figs 5 and 6, analyses of slices 7 and 8 are shown in Figs 7 and 8, and the results for all the remaining slices (3–6) are shown in Supporting Information Figs S2–S5.
Fig. 5.
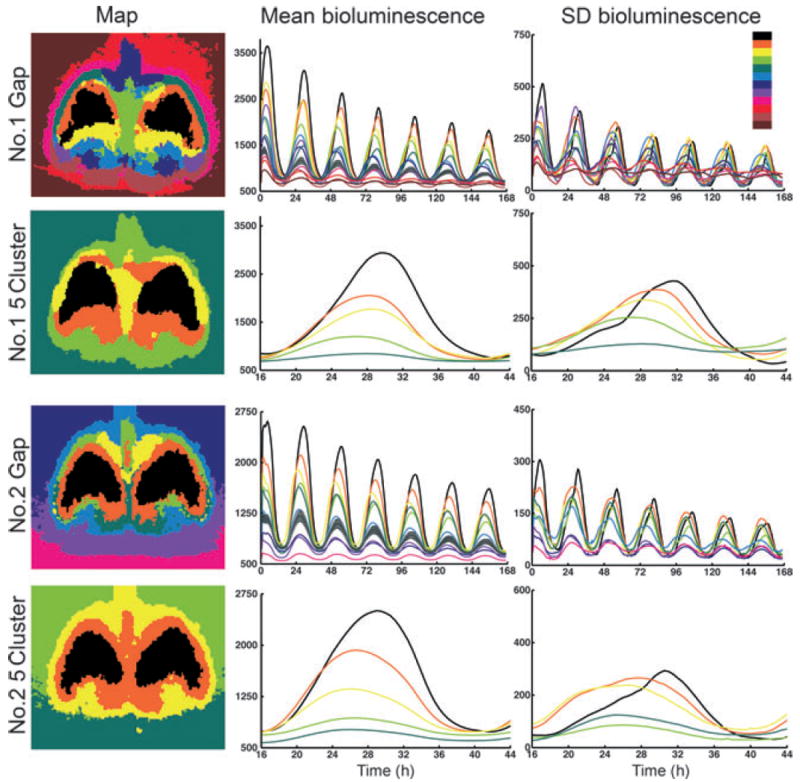
Cluster analysis of bilateral SCN for slices 1 and 2. The color code in the upper right shows the order of cluster coloring from highest (black) to lowest (dark brown) mean brightness. (Left column) Maps of bilateral cluster analysis. The top two maps are the SCNs in slice 1 while the bottom two maps are the SCNs in slice 2. The top two and bottom two maps show the Gap and 5Cluster analysis of slices 1 and 2, respectively. (Middle column) Time series of the mean cluster bioluminescence corresponding to the map found directly to the left. The Gap method time series show the full duration of the experiment. The 5Cluster analysis emphasizes detail for a single cycle by plotting from hours 16 to 44. The grey line denotes the mean ± SD value for scatter of light. Clusters with oscillation amplitude below this line have rhythms that cannot be discerned from light scatter in extra-SCN tissue. (Right column) Time series of the SD of bioluminescence of the clusters shown to the left. Again, the Gap method analysis is plotted for the complete experiment duration whereas the 5Cluster analysis is plotted from hours 16 to 44.
Fig. 6.
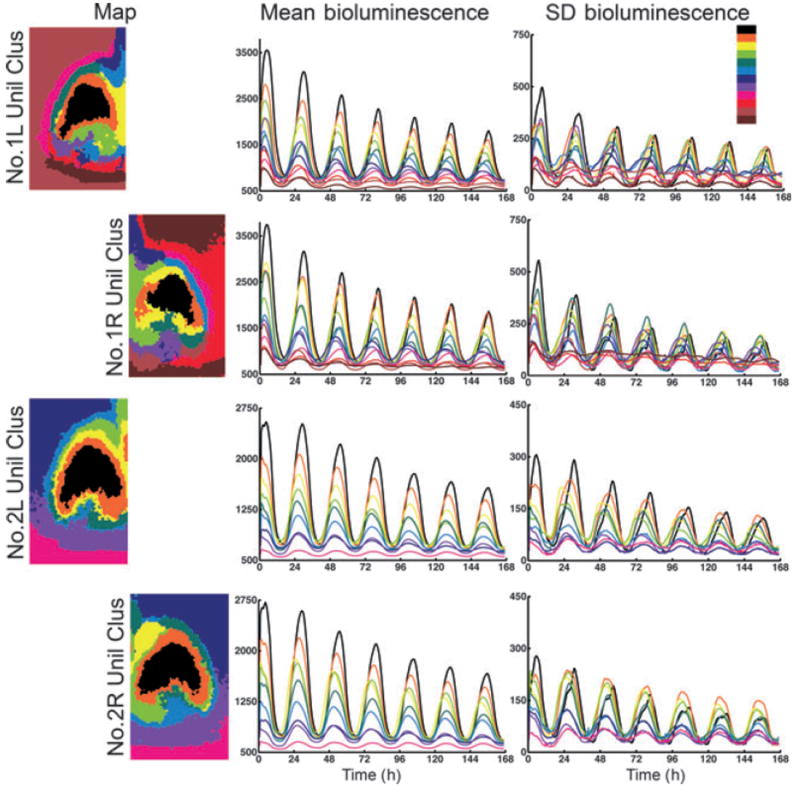
Cluster analysis of unilateral SCN. The color code is as in Fig. 5. (Left column) Maps of the left (L) and right (R) SCN with unilateral cluster analysis (Unil Clus), with the number of clusters constrained to that revealed in the bilateral Gap analysis of each SCN. The top two maps are the left and right SCNs in slice 1, while the bottom two maps are the left and right SCNs in slice 2. Note the right dorsal medial region found in slice 2 (yellow) does not have a corresponding contralateral cluster. (Middle column) Time series of the mean cluster bioluminescence corresponding to the map found directly to the left. (Right column) Time series of SD bioluminescence of the same clusters found to the left.
Fig. 7.
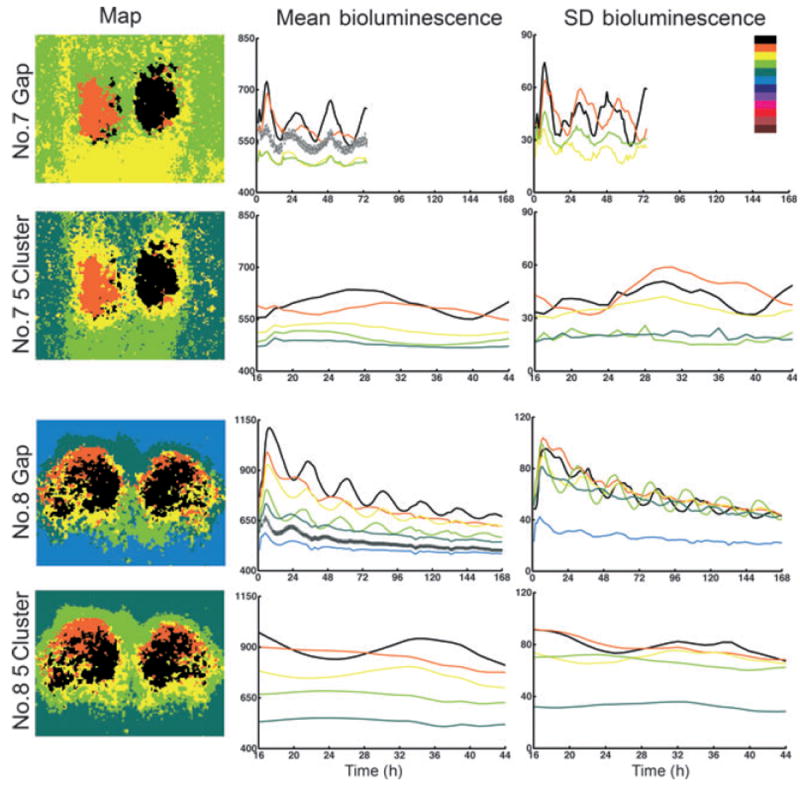
Cluster analysis of bilateral SCN in slices 7 and 8. The color code and legend are as in Fig. 5.
Fig. 8.
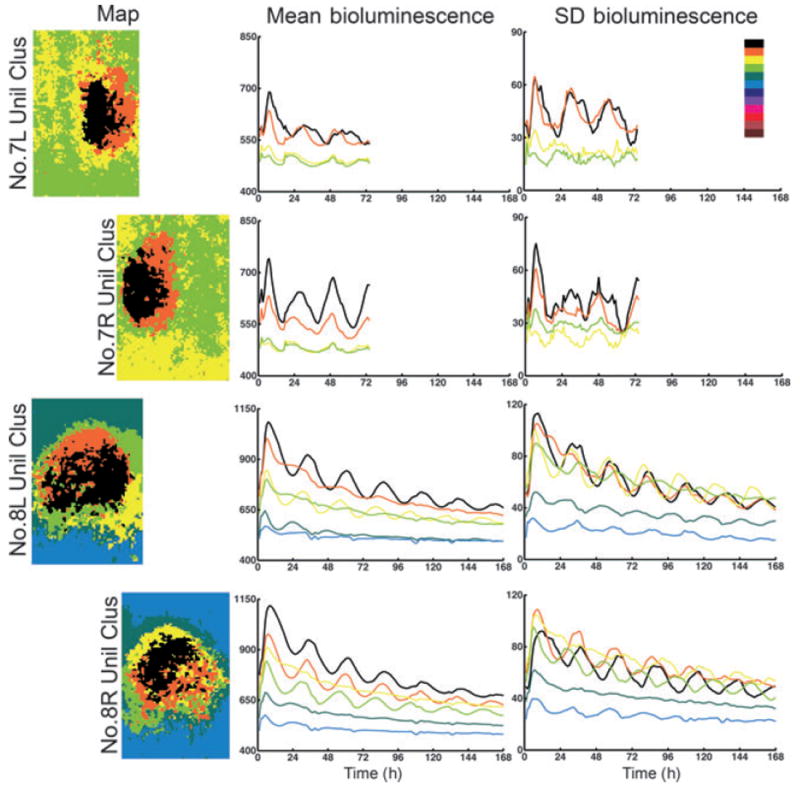
Cluster analysis of unilateral SCN. The color code is as in Fig. 5; analysis of map, bioluminescence time series and SD of bioluminescence time series as in Fig. 6.
Analysis of bilateral slices 1 and 2
As seen in the maps of color-coded subregions of the image, the cluster analysis (using cosine distance to minimize the effects of scatter) successfully distinguishes the SCN from surrounding brain tissue, and from areas of the image lacking brain tissue (Fig. 5, left column). A major result of the cluster analysis is the discovery of consistent features in the maps across slices, especially obvious when the number of clusters is constrained, seen in the 5Cluster analysis. The brightness time series results show oscillations of each cluster (middle panels, Fig. 5) and indicate that the clusters maintain a fixed phase relationship to one another as well as the same relative amplitude over serial cycles.
For all slices, the onset of bioluminescence of the various clusters appears more synchronized than does the offset. The SD of the brightness varies rhythmically over the circadian cycle (right column, Fig. 5). For each cluster, SD is correlated with the overall amplitude, indicating that as the cluster gets brighter its constituent pixels spread out in their values. For example, in slices 1 and 2 the highest amplitude (black) cluster has maximum SD late in the cycle, as other clusters turn off, while the yellow cluster has maximum SD nearer the onset. Note that there is a region within the SCN in both slices that appears to have very low amplitude LUC expression at all times; this low amplitude rhythm is probably attributable, at least in part, to scatter of light from other regions rather than to bioluminescence originating in neurons within the region.
Analysis of unilateral slices 1 and 2
The comparison of the right and left SCN, analyzed separately, is given in Fig. 6. The maps showing spatial distribution of clusters within each nucleus indicates substantial bilateral symmetry, indicated by similar localization of peak clusters (black and orange) and surrounding rings of clusters (yellow and both greens). A salient feature of slice 2 is a distinct cluster in the dorsomedial region (yellow) in the right SCN. The brightness time series indicates that the right dorsomedial region (yellow) in slice 2 mentioned above has an early onset. The rise time of bioluminescence in each cycle is more coherent than the fall, as seen clearly in the cosine distance, correlation, and Bray–Curtis cluster analysis of slice 2 (Fig. 5, Supporting Information Data S1 and Fig. S1).
Analysis of slices 3–6
The data for the slices 3–6 are shown in Supporting Information Figs S2–S5. The maps indicate that the shapes of the clusters are somewhat different than those of slices 1 and 2, in that the brightest region appears to lie more medially (a pattern previously reported: Quintero et al., 2003; Yamaguchi et al., 2003). This is probably due to differences in plane of section, or in precise extent of the SCN harvested during slice preparation – the importance of slice architecture has previously been shown for rhythm-generating circuits of the thalamus (Gloveli et al., 2005).
Analysis of slices 7 and 8
The bilateral SCN map for slices 7 and 8 reveals 4 and 6 clusters, respectively (Fig. 7). In slice 7 the power in the circadian range is very low, and it appears that the right side is rhythmic while the left is not (black vs. orange line, 5Cluster analysis; Fig. 7). The clusters in slice 8 are especially poorly organized with intermingled superpixels. The brightness time series shows that, in this slice, the highest three clusters have similar low amplitudes. Analysis of each SCN individually (Fig. 8) indicates that the left SCN has a single oscillating cluster while the right SCN has better organization with a larger contiguous cluster that oscillates more strongly than does the corresponding region on the other side. In both the right and left SCN, there is a ventromedial cluster (light green) that slightly leads the highest amplitude ‘black’ one.
Spatiotemporal changes in LUC and PER2 expression in vivo
Differences in the half-life of LUC and PER2 (Collaco & Geusz, 2003) may make spatial distribution of the former protein easier to detect than the latter. We next sought to understand whether the clusters seen in the slices could also be detected in brain sections harvested at various circadian times and therefore we examined PER2 and LUC expression in vivo. There were no significant differences in RODs from animals maintained in LD or in DD and the data from the two groups were combined. Figure 9A and B shows the time of expression and spatial localization of LUC and PER2 in relation to VP in the mid-SCN region, in double-labeled sections. The pseudocolored image interposed between immunostained sections facilitates comparison with corresponding time points of slices in Figs 3 and 4. The results indicate that the spatial localization of immunochemically identified LUC and PER2 map well to each other (Fig. 9A and B) and to the patterns seen in the bioluminescent signals in the slice preparation (Figs 3 and 4). LUC expression was first detected at ~ZT4 in the dorsomedial to dorsal SCN. This corresponds closely to the pattern of bioluminescence seen at ZT3 in slices 1 and 2 (Fig. 3). LUC expression was subsequently detected at ZT13–16 through the rest of the shell region, corresponding closely to the pattern of bioluminescence seen at ZT12-15 in slices 1 and 2 (Fig. 3), with highest expression also forming a crescent in the medial SCN. Importantly, in both preparations, LUC was not expressed at any time of the cycle in the ventral SCN region. Similar to LUC, PER2 expression was increased at ZT6 in the dorsomedial region, and then spread through most of the shell region by ZT16-18. The highest expression of PER2 was localized similarly to LUC. As for LUC, a ventrolateral region expresses very little PER2. Quantification of the time-course of PER and LUC indicates that, in vivo, both proteins peaked at ~1200–1600 h and reached a trough at ~2300–0400 h (Fig. 9C). In a section taken from an animal euthanized at about the time of peak expression, confocal and light microscopic analyses confirm that PER2 and LUC were colocalized (Supporting Information Fig. S6).
Fig. 9.
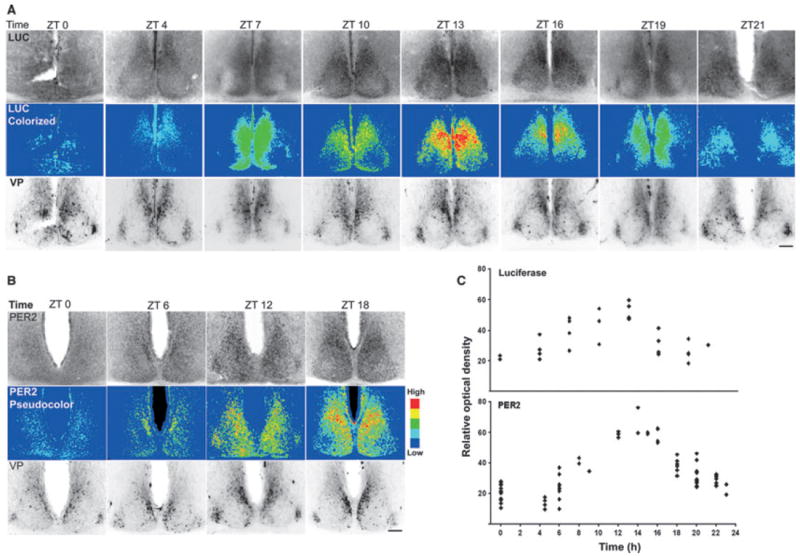
(A) Time-course of LUC expression of PER2:LUC transgenic mice in vivo. First row shows microphotographs of the SCN immunostained for LUC. Middle row shows the same images as in top row, pseudocolored to facilitate comparison of LUC expression in Figs 2 and 3. Bottom row shows the same sections double-labeled with VP to delineate the mid-SCN, ensuring that similar regions were examined across animals. (B) Time-course of PER2 expression in C57BL/6 mice in vivo. As in A, the first row shows PER2 expression, middle row shows the same images pseudocolored and bottom row shows VP staining. Overall, the immunochemical staining corresponds well with bioluminescence seen in slices 1 and 2 (Figs 3 and 4). (C) Graph showing ROD of LUC and PER2-ir in each unilateral SCN throughout the cycle. Scale bars, 100 μm.
Discussion
The results reveal that the characteristic changes in clock gene expression of autonomous cellular oscillators within the SCN depend on a cell’s spatial location within the nucleus. This is consistent with the hypothesis of orderly connectivity among cells, and inconsistent with the hypothesis that the SCN is comprised of independent autonomous oscillators synchronized by weak coupling. The statistical cluster analysis used here finds robust differentiation of the nucleus into distinct subsystems. An area centered in the dorsomedial SCN has a low-amplitude oscillation and is the start point of the daily cycle. A second region in the central SCN has a high-amplitude oscillation. A third area lacks detectable rhythmic LUC beyond that attributable to scatter of light. The clusters are very similar on the right and left sides of the nucleus. Finally, there appears to be a slow expansion and regression of bioluminescent signal centered on the region of the highest-amplitude signal. Whether this is a consequence of scatter of light or the results of a gradient or tidal pattern (Yan et al., 2007) in the expression pattern of clock genes remains to be determined. Supporting evidence from anatomical studies of animals euthanized at specific times indicates that this regional differentiation faithfully reflects changes that recur on a daily basis within the animal rather than being an artifact of bioluminescent imaging. Taken together, the results indicate that an individual oscillator’s behavior depends on the cell’s position and its connections within the SCN neuronal network.
Scatter and noise of bioluminescent signals
In the cluster analysis, noise is not a serious problem because of spatial averaging. On the other hand, distinguishing between oscillation in a region of tissue due to the scatter of light and the bioluminescence due to local LUC expression is an important problem in interpreting these results. Part of the bioluminescent signal measured at any location in the slice is contaminated by scatter – light produced at a given location which has passed through the tissue and medium and is emitted at another location. Assigning the bioluminescent signal strictly to its source is difficult because neural tissue scatters photons in a broad and unpredictable manner (Polimeni et al., 2005). Scatter of light leads to ‘ghost’ oscillations in non-oscillating parts of the SCN and in non-tissue parts of the images. While scatter limits detectable spatial resolution of the cluster analysis, the results indicate that it is nevertheless possible to assess the relative contributions of endogenous bioluminescence and scatter through clustering. This is demonstrated in the fact that the cluster analysis reliably distinguishes subregions across multiple slices, even though the slices sample slightly different portions of the SCN network architecture.
Oscillatory properties of SCN tissue vs. cells
Individual SCN neurons are competent circadian oscillators (Welsh et al., 1995; Webb et al., 2009), and it is important to understand how the SCN’s connectivity contributes to its ability to express circadian oscillations. Isolated individual SCN neurons are weak, inducible and probabilistic oscillators (Webb et al., 2009). Furthermore, PER2∷LUC imaging in individual dispersed cells, combined with immunostaining for VP and vasoactive intestinal polypeptide, does not reveal an association between circadian phenotype and peptidergic expression.
This stands in sharp contrast to the present results showing that within an SCN slice (in contrast to dissociated cells), the pattern of oscillation of each of the major clusters (and thus of the composite of cells within each cluster) is robust and stable across serial cycles. Furthermore, cells within some clusters do not express any detectable rhythms, either in the statistical cluster analyses or in slices stained for LUC or clock genes (Hamada et al., 2001; Karatsoreos et al., 2004). These results highlight the conclusion that the connectivity of SCN individual neurons is a major contributor to the stable circadian oscillation of the brain’s master clock. Importantly, while weak coupling of individual oscillators can lead to organized activity in circadian oscillators (Abraham et al., 2010; Granada et al., 2009), such coupling cannot account for the stable pattern of spatial and temporal changes seen in the SCN in the present study.
Distinct function of specific clusters
A significant constraint in understanding SCN networks is the lack of knowledge about the connections among its neurons, especially those among subregions bearing specific peptidergic phenotypes. The cluster analysis suggests the backbone of a possible local circuit organization that must now be explored with other methods, both biological and statistical. That said, we do nevertheless have some information with regard to the distinct zones identified in the present cluster analysis. In studies of the spatial arrangement of oscillator cells that examined the canonical clock genes Per1 and Per2, and the clock-controlled gene VP, the most dorsomedial cells were the first in each circadian cycle to express both Per1 and VP mRNA (Hamada et al., 2004), or Per1-Luc and PER2∷LUC (Yamaguchi et al., 2003). Here, gene expression then spread very slowly through much of the nucleus for the next 12 h before receding to baseline levels. The present work confirms a slowly expanding and receding zone of PER2 expression, with a subsequent return to baseline. The occurrence of a cluster that lacks circadian oscillation above the level attributable to scatter determines a third zone, and is confirmed in previous findings (Hamada et al., 2001; Karatsoreos et al., 2004). Accordingly, we modeled the SCN as bearing intrinsic oscillators and nonrhythmic gate cells, in which nonrhythmic neurons contribute to daily oscillation by gating input to the SCN while receiving feedback from rhythmic cells (Antle et al., 2003, 2007).
While studies of dispersed cells bearing either vasoactive intestinal polypeptide or VP do not provide evidence for a specialized class of non-oscillatory cells (Webb et al., 2009), the cluster analysis of slices reveals that, when their connections are maintained, cells lacking detectable oscillation are spatially localized within the nucleus. This result does not mean that there are no rhythmic cells at all within this region, as a few scattered cells can be seen expressing PER2 or LUC. The result does reveal, however, that scatter of light, measurable in areas lacking brain tissue and in non-SCN regions of the brain, is the major source of the low-amplitude oscillation detected within that zone and in non-SCN brain regions. Finally the cluster analysis reveals an extremely robust synchrony between the right and left SCN. This could be a product of neural (Moore et al., 2002), glial (Servière & Lavialle, 1996) or humoral (Silver et al., 1990; Kramer et al., 2001; Cheng et al., 2002; Kraves & Weitz, 2006; Li et al., 2009) connections between the nuclei.
Inter-slice variability
Though not previously studied, the present results demonstrate significant inter-slice variations that are probably always present in studies using slice preparations. Elucidating these variations is a major strength of quantitative statistical analysis protocols. This is most dramatically seen in the differences between slices 1–6 (bearing stable circadian rhythms) and 7 and 8 (lacking stable circadian rhythms). In this preparation, brain slices were prepared in coronal sections and then trimmed to remove non-SCN regions. In blocking the tissue, slightly different parts of the SCN and nearby extra-SCN brain regions are necessarily included in the slice. Differences in SCN circuits included in the slice are probably major sources of variability in the precise pattern of LUC activation detected, as previously demonstrated in thalamic slice preparations (Gloveli et al., 2005).
One can reasonably ask whether the low-amplitude oscillations such as those seen in slices 7 and 8 are biologically significant. While such oscillations are unlikely within the SCN of the living normal animal, our evidence indicates that they do occur in vitro in SCN brain slices harvested from wild-type animals. This is consistent with evidence that the quality of oscillation of individual cells within a slice may be influenced by the precise composition of the SCN network in which it is encased (Yamaguchi et al., 2003), and that the behavior of individual cells in slices with high-amplitude, highly organized oscillation may differ from those seen in weakly oscillating slices.
Expression of PER2 in vivo
The daily expression of PER2∷LUC or PER2 protein in animals maintained in an LD cycle or in DD is consistent with the expression seen in the brain slice, with a dorsomedial cluster expressing LUC or PER2 first, then the expression extending to most of the SCN (second zone); finally, a third zone expresses few LUC or PER2-ir cells. This confirms that the changes seen in the brain slice accurately reflect daily changes in the animal.
Conclusion
The results of this analysis point to two important and related lines of research on the SCN, one centered on the anatomical differentiation of the SCN tissue and the other on our understanding of its function. We have previously suggested a view of the SCN as a ‘gated’ oscillator system in which circadian oscillation is organized by interaction of the core and shell parts of the tissue. The present research carries this program further in delineating the spatial organization of oscillating cells in the shell. The picture we have at this point raises the question of exactly what we mean in describing the neurons of the SCN as ‘cell-autonomous oscillators’. In the living tissue individual cells undergo an oscillation in expression of genes and proteins as they participate in complex interactions across the circadian cycle of the tissue as a whole SCN. In this sense they are clearly ‘oscillating cells’, but their temporal behavior depends critically on their location in the SCN tissue, most probably as a reflection of intercellular communication. SCN cells are also capable of sustaining oscillatory patterns in vitro when isolated from their connections, and in this sense are also ‘oscillator cells’. But the present results suggest that the evolution of the SCN as a structure has exploited the periodicity of these cells primarily to establish the characteristic time period of the composite oscillation rather than depending on the individual cells as independent timekeepers. In sum, the tissue is the issue.
Supplementary Material
Fig. S1. Analysis of clusters to compare alternate similarity functions using slice #2.
Fig. S2. Bilateral and unilateral cluster analysis for slice #3.
Fig. S3. Bilateral and unilateral cluster analysis for slice #4.
Fig. S4. Bilateral and unilateral cluster analysis for slice #5.
Fig. S5. Bilateral and unilateral cluster analysis for slice #6.
Fig. S6. Photomicrographs show a coronal SCN section double-labeled for LUC (left) and PER2 (middle) and the overlay (right).
Data S1. Supplemental results.
Acknowledgments
Supported by NIH grants RO1 NS37919 and MH075045 (R.S.); R01 MH082945 (D.K.W.) and a V.A. Career Development Award to D.K.W. We thank Alexis Hatzis and Tabby Khan for assistance in data analysis and immunochemistry, and Jennifer Evans for preparing some slices for immunochemistry.
Abbreviations
- DD
constant darkness
- LD
light : dark
- LUC
luciferase
- PB
phosphate buffer
- PER2
PERIOD2
- PER2∷LUC
PERIOD2∷Luciferase
- ROD
relative optical density
- ROI
regions of interest
- SCN
suprachiasmatic nucleus
- VP
vasopressin
- ZT
zeitgeber time
Footnotes
Supporting Information
Additional supporting information can be found in the online version of this article:
Please note: As a service to our authors and readers, this journal provides supporting information supplied by the authors. Such materials are peer-reviewed and may be re-organized for online delivery, but are not copy-edited or typeset by Wiley-Blackwell. Technical support issues arising from supporting information (other than missing files) should be addressed to the authors.
References
- Abraham U, Granada AE, Westermark PO, Heine M, Kramer A, Herzel H. Coupling governs entrainment range of circadian clocks. Mol Syst Biol. 2010;6:438. doi: 10.1038/msb.2010.92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abrahamson EE, Moore RY. Suprachiasmatic nucleus in the mouse: retinal innervation, intrinsic organization and efferent projections. Brain Res. 2001;916:172–191. doi: 10.1016/s0006-8993(01)02890-6. [DOI] [PubMed] [Google Scholar]
- Antle MC, Silver R. Orchestrating time: arrangements of the brain circadian clock. Trends Neurosci. 2005;28:145–151. doi: 10.1016/j.tins.2005.01.003. [DOI] [PubMed] [Google Scholar]
- Antle MC, Foley DK, Foley NC, Silver R. Gates and oscillators: a network model of the brain clock. J Biol Rhythms. 2003;18:339–350. doi: 10.1177/0748730403253840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antle MC, Foley NC, Foley DK, Silver R. Gates and oscillators II: zeitgebers and the network model of the brain clock. J Biol Rhythms. 2007;22:14–25. doi: 10.1177/0748730406296319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aton SJ, Herzog ED. Come together, right…now: synchronization of rhythms in a mammalian circadian clock. Neuron. 2005;48:531–534. doi: 10.1016/j.neuron.2005.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown TM, Hughes AT, Piggins HD. Gastrin-releasing peptide promotes suprachiasmatic nuclei cellular rhythmicity in the absence of vasoactive intestinal polypeptide-VPAC2 receptor signaling. J Neurosci. 2005;25:11155–11164. doi: 10.1523/JNEUROSCI.3821-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown TM, Colwell CS, Waschek JA, Piggins HD. Disrupted neuronal activity rhythms in the suprachiasmatic nuclei of vasoactive intestinal polypeptide-deficient mice. J Neurophysiol. 2007;97:2553–2558. doi: 10.1152/jn.01206.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng MY, Bullock CM, Li C, Lee AG, Bermak JC, Belluzzi J, Weaver DR, Leslie FM, Zhou QY. Prokineticin 2 transmits the behavioural circadian rhythm of the suprachiasmatic nucleus. Nature. 2002;417:405–410. doi: 10.1038/417405a. [DOI] [PubMed] [Google Scholar]
- Collaco AM, Geusz ME. Monitoring immediate-early gene expression through firefly luciferase imaging of HRS/J hairless mice. BMC Physiol. 2003;3:8. doi: 10.1186/1472-6793-3-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davidson AJ, Yamazaki S, Arble DM, Menaker M, Block GD. Resetting of central and peripheral circadian oscillators in aged rats. Neurobiol Aging. 2008;29:471–477. doi: 10.1016/j.neurobiolaging.2006.10.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis FC, Gorski RA. Development of hamster circadian rhythms: role of the maternal suprachiasmatic nucleus. J Comp Physiol. 1988;162:601–610. doi: 10.1007/BF01342635. [DOI] [PubMed] [Google Scholar]
- Gloveli T, Dugladze T, Rotstein HG, Traub RD, Monyer H, Heinemann U, Whittington MA, Kopell NJ. Orthogonal arrangement of rhythm-generating microcircuits in the hippocampus. Proc Natl Acad Sci USA. 2005;102:13295–13300. doi: 10.1073/pnas.0506259102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Granada A, Hennig RM, Ronacher B, Kramer A, Herzel H. Phase response curves elucidating the dynamics of coupled oscillators. Methods Enzymol. 2009;454:1–27. doi: 10.1016/S0076-6879(08)03801-9. [DOI] [PubMed] [Google Scholar]
- Hamada T, LeSauter J, Venuti JM, Silver R. Expression of Period genes: rhythmic and nonrhythmic compartments of the suprachiasmatic nucleus pacemaker. J Neurosci. 2001;21:7742–7750. doi: 10.1523/JNEUROSCI.21-19-07742.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamada T, Antle MC, Silver R. Temporal and spatial expression patterns of canonical clock genes and clock-controlled genes in the suprachiasmatic nucleus. Eur J Neurosci. 2004;19:1741–1748. doi: 10.1111/j.1460-9568.2004.03275.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harmar AJ, Marston HM, Shen S, Spratt C, West KM, Sheward WJ, Morrison CF, Dorin JR, Piggins HD, Reubi JC, Kelly JS, Maywood ES, Hastings MH. The VPAC(2) receptor is essential for circadian function in the mouse suprachiasmatic nuclei. Cell. 2002;109:497–508. doi: 10.1016/s0092-8674(02)00736-5. [DOI] [PubMed] [Google Scholar]
- Harrington ME, Rahmani T, Lee CA. Effects of damage to SCN neurons and efferent pathways on circadian activity rhythms of hamsters. Brain Res Bull. 1993;30:655–669. doi: 10.1016/0361-9230(93)90097-u. [DOI] [PubMed] [Google Scholar]
- Jobst EE, Allen CN. Calbindin neurons in the hamster suprachiasmatic nucleus do not exhibit a circadian variation in spontaneous firing rate. Eur J Neurosci. 2002;16:2469–2474. doi: 10.1046/j.1460-9568.2002.02309.x. [DOI] [PubMed] [Google Scholar]
- Karatsoreos IN, Yan L, LeSauter J, Silver R. Phenotype matters: identification of light-responsive cells in the mouse suprachiasmatic nucleus. J Neurosci. 2004;24:68–75. doi: 10.1523/JNEUROSCI.1666-03.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kramer A, Yang FC, Snodgrass P, Li X, Scammell TE, Davis FC, Weitz CJ. Regulation of daily locomotor activity and sleep by hypothalamic EGF receptor signaling. Science. 2001;294:2511–2515. doi: 10.1126/science.1067716. [DOI] [PubMed] [Google Scholar]
- Kraves S, Weitz CJ. A role for cardiotrophin-like cytokine in the circadian control of mammalian locomotor activity. Nat Neurosci. 2006;9:212–219. doi: 10.1038/nn1633. [DOI] [PubMed] [Google Scholar]
- LeSauter J, Bhuiyan T, Shimazoe T, Silver R. Circadian trafficking of calbindin-ir in fibers of SCN neurons. J Biol Rhythms. 2009;24:488–496. doi: 10.1177/0748730409350876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li JD, Burton KJ, Zhang C, Hu SB, Zhou QY. Vasopressin receptor V1a regulates circadian rhythms of locomotor activity and expression of clock-controlled genes in the suprachiasmatic nuclei. Am J Physiol Regul Integr Comp Physiol. 2009;296:R824–R830. doi: 10.1152/ajpregu.90463.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu C, Reppert SM. GABA synchronizes clock cells within the suprachiasmatic circadian clock. Neuron. 2000;25:123–128. doi: 10.1016/s0896-6273(00)80876-4. [DOI] [PubMed] [Google Scholar]
- Liu AC, Welsh DK, Ko CH, Tran HG, Zhang EE, Priest AA, Buhr ED, Singer O, Meeker K, Verma IM, Doyle FJ, III, Takahashi JS, Kay SA. Intercellular coupling confers robustness against mutations in the SCN circadian clock network. Cell. 2007;129:605–616. doi: 10.1016/j.cell.2007.02.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maywood ES, O’ Neill JS, Chesham JE, Hastings MH. Minireview: the circadian clockwork of the suprachiasmatic nuclei – analysis of a cellular oscillator that drives endocrine rhythms. Endocrinology. 2007;148:5624–5634. doi: 10.1210/en.2007-0660. [DOI] [PubMed] [Google Scholar]
- Moore RY, Silver R. Suprachiasmatic nucleus organization. Chronobiol Int. 1998;15:475–487. doi: 10.3109/07420529808998703. [DOI] [PubMed] [Google Scholar]
- Moore RY, Speh JC, Leak RK. Suprachiasmatic nucleus organization. Cell Tissue Res. 2002;309:89–98. doi: 10.1007/s00441-002-0575-2. [DOI] [PubMed] [Google Scholar]
- Nakamura W, Yamazaki S, Takasu NN, Mishima K, Block GD. Differential response of Period 1 expression within the suprachiasmatic nucleus. J Neurosci. 2005;25:5481–5487. doi: 10.1523/JNEUROSCI.0889-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van den Pol AN, Tsujimoto KL. Neurotransmitters of the hypothalamic suprachiasmatic nucleus: immunocytochemical analysis of 25 neuronal antigens. Neuroscience. 1985;15:1049–1086. doi: 10.1016/0306-4522(85)90254-4. [DOI] [PubMed] [Google Scholar]
- Polimeni JR, Granquist-Fraser D, Wood RJ, Schwartz EL. Physical limits to spatial resolution of optical recording: clarifying the spatial structure of cortical hypercolumns. Proc Natl Acad Sci U S A. 2005;102:4158–4163. doi: 10.1073/pnas.0500291102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quintero JE, Kuhlman SJ, McMahon DG. The biological clock nucleus: a multiphasic oscillator network regulated by light. J Neurosci. 2003;23:8070–8076. doi: 10.1523/JNEUROSCI.23-22-08070.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reppert SM, Weaver DR. Coordination of circadian timing in mammals. Nature. 2002;418:935–941. doi: 10.1038/nature00965. [DOI] [PubMed] [Google Scholar]
- Rohling J, Meijer JH, VanderLeest HT, Admiraal J. Phase differences between SCN neurons and their role in photoperiodic encoding; a simulation of ensemble patterns using recorded single unit electrical activity patterns. J Physiol Paris. 2006;100:261–270. doi: 10.1016/j.jphysparis.2007.05.005. [DOI] [PubMed] [Google Scholar]
- Rusak B. The role of the suprachiasmatic nucleus in the generation of circadian rhythms in the golden hamster, Mesocricetus auratus. J Comp Physiol. 1977;118:145–164. [Google Scholar]
- Servière J, Lavialle M. Astrocytes in the mammalian circadian clock: putative roles. In: Buijs RM, Kalsbeek A, Romijin CMA, Pennartz CMA, Mirmiran M, editors. Hypothalamic Integration of Circadian Rhythms. Elsevier; Amsterdam: 1996. pp. 57–73. [DOI] [PubMed] [Google Scholar]
- Shen H, Watanabe M, Tomasiewicz H, Rutishauser U, Magnuson T, Glass JD. Role of neural cell adhesion molecule and polysialic acid in mouse circadian clock function. J Neurosci. 1997;17:5221–5229. doi: 10.1523/JNEUROSCI.17-13-05221.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen H, Watanabe M, Tomasiewicz H, Glass JD. Genetic deletions of NCAM and PSA impair circadian function in the mouse. Physiol Behav. 2001;73:185–193. doi: 10.1016/s0031-9384(01)00468-1. [DOI] [PubMed] [Google Scholar]
- Silver R, Lehman MN, Gibson M, Gladstone WR, Bittman EL. Dispersed cell suspensions of fetal SCN restore circadian rhythmicity in SCN-lesioned adult hamsters. Brain Res. 1990;525:45–58. doi: 10.1016/0006-8993(90)91319-c. [DOI] [PubMed] [Google Scholar]
- Strogatz SH, Stewart I. Coupled oscillators and biological synchronization. Sci Am. 1993;269:102–109. doi: 10.1038/scientificamerican1293-102. [DOI] [PubMed] [Google Scholar]
- Tibshirani R, Walther G, Hastie T. Estimating the number of clusters in a data set via the gap statistic. J R Statist Soc B. 2001;63:411–423. [Google Scholar]
- Ukai H, Ueda HR. Systems biology of mammalian circadian clocks. Annu Rev Physiol. 2010;72:579–603. doi: 10.1146/annurev-physiol-073109-130051. [DOI] [PubMed] [Google Scholar]
- Webb AB, Angelo N, Huettner JE, Herzog ED. Intrinsic, nondeterministic circadian rhythm generation in identified mammalian neurons. Proc Natl Acad Sci USA. 2009;106:16493–16498. doi: 10.1073/pnas.0902768106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welsh DK, Logothetis DE, Meister M, Reppert SM. Individual neurons dissociated from rat suprachiasmatic nucleus express independently phased circadian firing rhythms. Neuron. 1995;14:697–706. doi: 10.1016/0896-6273(95)90214-7. [DOI] [PubMed] [Google Scholar]
- Yamaguchi S, Isejima H, Matsuo T, Okura R, Yagita K, Kobayashi M, Okamura H. Synchronization of cellular clocks in the suprachiasmatic nucleus. Science. 2003;302:1408–1412. doi: 10.1126/science.1089287. [DOI] [PubMed] [Google Scholar]
- Yamazaki S, Numano R, Abe M, Hida A, Takahashi R, Ueda M, Block GD, Sakaki Y, Menaker M, Tei H. Resetting central and peripheral circadian oscillators in transgenic rats. Science. 2000;288:682–685. doi: 10.1126/science.288.5466.682. [DOI] [PubMed] [Google Scholar]
- Yan L, Okamura H. Gradients in the circadian expression of Per1 and Per2 genes in the rat suprachiasmatic nucleus. Eur J Neurosci. 2002;15:1153–1162. doi: 10.1046/j.1460-9568.2002.01955.x. [DOI] [PubMed] [Google Scholar]
- Yan L, Karatsoreos I, Lesauter J, Welsh DK, Kay SA, Foley DK, Silver R. Exploring spatiotemporal organization of SCN circuits. Cold Spring Harb Symp Quant Biol. 2007;72:527–541. doi: 10.1101/sqb.2007.72.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoo SH, Yamazaki S, Lowrey PL, Shimomura K, Ko CH, Buhr ED, Siepka SM, Hong HK, Oh WJ, Yoo OJ, Menaker M, Takahashi JS. PERIOD2∷LUCIFERASE real-time reporting of circadian dynamics reveals persistent circadian oscillations in mouse peripheral tissues. Proc Natl Acad Sci U S A. 2004;101:5339–5346. doi: 10.1073/pnas.0308709101. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1. Analysis of clusters to compare alternate similarity functions using slice #2.
Fig. S2. Bilateral and unilateral cluster analysis for slice #3.
Fig. S3. Bilateral and unilateral cluster analysis for slice #4.
Fig. S4. Bilateral and unilateral cluster analysis for slice #5.
Fig. S5. Bilateral and unilateral cluster analysis for slice #6.
Fig. S6. Photomicrographs show a coronal SCN section double-labeled for LUC (left) and PER2 (middle) and the overlay (right).
Data S1. Supplemental results.


