Abstract
To evaluate the potential importance in autistic subjects of copy number variants (CNVs) that alter genes of relevance to bioenergetics, ionic metabolism, and synaptic function, we conducted a detailed microarray analysis of 69 autism probands and 35 parents, compared to 89 CEU HapMap controls. This revealed that the frequency CNVs of ≥ 100 kb and CNVs of ≥ 10 Kb were markedly increased in probands over parents and in probands and parents over controls. Evaluation of CNVs ≥ 1 Mb by chromosomal FISH confirmed the molecular identity of a subset of the CNVs, some of which were associated with chromosomal rearrangements. In a number of the cases, CNVs were found to alter the copy number of genes that are important in mitochondrial oxidative phosphorylation (OXPHOS), ion and especially calcium transport, and synaptic structure. Hence, autism might result from alterations in multiple bioenergetic and metabolic genes required for mental function.
Keywords: autism, mitochondria, OXPHOS, copy number variants (CNVs), calcium channels, synapses
Introduction
The term autism is applied to a triad of symptoms including communication deficits, impaired social interactions, and repetitive stereotypic behaviours [1]. Autism manifestations also occur in diseases with defined genetic etiology, e.g. Timothy syndrome, Angelman syndrome, Tuberous sclerosis, Fragile X mental retardation, Rett syndrome, etc. [2]. However, in the majority of cases the underlying pathogenic mechanisms that lead to this symptom complex are not defined.
There is growing evidence that genome dosage changes can contribute to the etiology of autism. These can result from cytogenetically detectable chromosomal changes to sub-microscopic changes detected through microarray analyses and comparative genomic hybridization. Among the earliest surveys of autism patients for sub-microscopic genomic alterations (deletions or duplications) it was reported that copy number variants (CNVs) were found in 10% of sporadic autism patients, 3% of multiplex families, and 1% of controls [3, 4].
While a specific nuclear DNA (nDNA) CNVs may lead to autism in some individuals it might manifest as cognitive impairment, attention deficit hyperactivity disorders, or psychiatric manifestations in others [5]. This suggests that neuropsychiatric disorders might have a common pathophysiological basis and that variation in the clinical manifestations might reflect interactions between the gene dosage affects caused by the CNV and other genetic, epigenetic, and/or environmental modifying factors.
It is still unclear if autism and other neuropsychiatric diseases are caused by one of a few relatively rare large (greater than 1 megabase) genome copy number dosage changes or whether smaller rare copy number variations in specific combinations also play roles in autism pathogenesis. Whatever the nature of the CNVs, it is clear that they must alter the copy of one or more genes in which gene dosage changes are sufficient to cause disease. Deletion, duplication, or mutations of such genes could then perturb cellular function sufficiently to cause neuronal symptoms. Mutation of an active copy of an imprinted gene could have such an effect or deletion of the normal copy of a gene whose homologue was mutated. Alternatively, the expressivity of one CNV might be influenced by the interaction with another CNV [6].
Given that the CNVs that have been found associated with autism are dispersed throughout the nDNA, then alteration of the copy number of anyone of a number of genes must be able to increase the risk of autism. This implies that the pathophysiology of autism might be the result of perturbation of a genetic network all of whose gene functions contribute to optimal neurological function. One such network could be the roughly one to two thousand nDNA genes plus the thousands of copies of the mitochondrial DNA (mtDNA) genes that are required to assemble the mitochondrion and the mitochondrial energy generating pathway, OXPHOS. In addition to generating most of the cellular energy, the mitochondrion regulates cellular oxidation-reduction states, reactive oxygen species production, cytosolic and mitochondrial calcium levels, and apoptosis via activation of the mitochondrial permeability transition pore (mtPTP). Since the brain has the highest mitochondrial energy demand, then partial defects in mitochondrial energy production or calcium homeostasis could be expected to preferentially affect synaptic function and result in neuropsychiatric disease [2, 7, 8].
Alterations in mitochondrial structure and function have been repeatedly observed in autistic patients [1, 9–15], and alterations in the mtDNA have been reported in some cases of autism spectrum disorder [16–18]. The activity of the mitochondrial inner membrane Calcium-regulated aspartate/glutamate carrier (AGC) gene, SLC25A12, and/or its expression have also been reported to be increased in autistic patients, at least in part due to elevated levels of calcium in the brains of autism patients [19–21].
Therefore, we reasoned that CNV alteration in anyone of a wide variety of bioenergetic, ion transport, of synaptic structural genes might predispose to autism. To test this hypothesis, we surveyed the CNVs in a set or carefully phenotyped autism patients and in a number of cases their parents and then asked if the CNVs encompassed genes that would be important in mitochondrial function, ion metabolism, and synaptic structure.
Methods
These studies were carried out at University of California Irvine with Institutional Review Board approvals (1996-616 and 2002–2608) and at the University of California San Diego with Institutional Review Board approvals (090243 and 0807260) for human subject research. Parents/guardians gave informed consent to participate in the study for probands and for themselves and other children. When appropriate, informed assent was obtained from probands.
Criteria used to establish autism diagnosis included ADOS (autism diagnostic observation schedule), ADI (autism diagnostic inventory), and in some cases CARS (Childhood Autism Rating Scale). Subjects underwent testing of cognitive abilities using Mullen or Stanford Binet tests depending on the proband age and abilities. Family history was obtained and geneticists with expertise in dysmorphology and neurologists examined probands. Clinical chemistry and metabolic analyses, including organic and amino acid profiles were obtained. In two autistic probands where blood chemistries indicated possible mitochondrial functional impairment, muscle biopsy was obtained for biochemical studies.
Blood was drawn from probands and parents and in some cases from siblings and was used to establish lymphoblastoid cell lines. DNA from the cells was used in microarray analyses.
The total number of autistic probands studied was 69, including 7 sets of Monozygotic twins and 4 affected sib pairs. We also studied 35 parents. For control data we utilized data information on DNA of lymphoblastoid cell lines of Hap Map CEU individuals (control population of northern and western European ancestry).
Microarray analyses were performed using the Affymetrix 6.0 SNP arrays. The SNP (single nucleotide polymorphism) genotypes and CNVs were determined using the Genotyping Console version 3.0.2 (Affymetrix Inc., 2008). Cell intensity files (CEL files) were created which contain the images of the scanned probe set, including copy number and allele-specific SNP probes. The software determines the number of copies present by comparing the signal intensity of the probes and comparing them to the Hap Map reference data. The copy number variants can then be visually appreciated using the Genotyping Console. Two Quality Control (QC) measures are used in the Genotyping Console to insure the data is sufficient for analysis. The first QC step is the Contrast QC measurement. The second QC measure is the Median of the Absolute values of all Pairwise Differences (MAPD). The MAPD measure analyzes the variability of log2 ratios of adjacent probes. All samples used in the analyses presented here met QC standards on both measures.
Data were analyzed using the Affymetrix genotyping console and two sets of search parameters were used. In the first set we searched for CNVs that included 20 markers, and extended over segments that were greater than 100 kb in length, with no more than 60% overlap with known polymorphic variants designated in the Database for Genomic variants (abbreviated as 20m100k60o). In a second search we documented genomic changes detected by 20 markers within segments larger than 10 kb and with no overlap with known copy number variants (abbreviated as 20m10k0o). Microarray data used in these analyses passed Affymetrix quality control measures, i.e. MAPD <0.35 and contrast QC>1.7.
Cell lines from 14 of the 15 autistic probands who were found to harbor CNVs larger than 1 megabase were examined by metaphase chromosomes fluorescence in situ hybridization (FISH).
Two of the patients, one with a few (2) CNVs and another with a large number (13) of CNVs, were then analyzed for systemic mitochondrial OXPHOS dysfunction. A muscle biopsy was collected and the electron transport chain enzymes assayed [22, 23].
Results
Frequency of copy number variants detected in microarray analyses
If mutations in anyone of a number of genes involved in an integrated pathway could cause autism, we hypothesized that individuals with more CNVs would be more likely to manifest autism. To determine if autism probands harbor more CNVs than their parents or population controls, we used Affymetrix 6.0 microarray analysis to identify CNVs in lymphoblastoid cell lines derived from the autism probands and their parents and we compared these to those reported in the lymphoblastoid cell lines derived from 89 CEU Hap Map subjects that had been analyzed using the same microarray platform. The number and frequency of variants detected using criteria 20m100k60o and criteria 20m10k0o are presented in Supplemental Tables 1A and 1B, respectively, with gains and losses listed separately. Because estimations of the overall frequency of CNVs may be influenced by cases with large numbers of variants, we graphed the frequency of patients versus the numbers of CNVs per individual in categories: 0 CNVs, 1 CNV, 2–5 CNVs, 6–10 CNVs, etc.
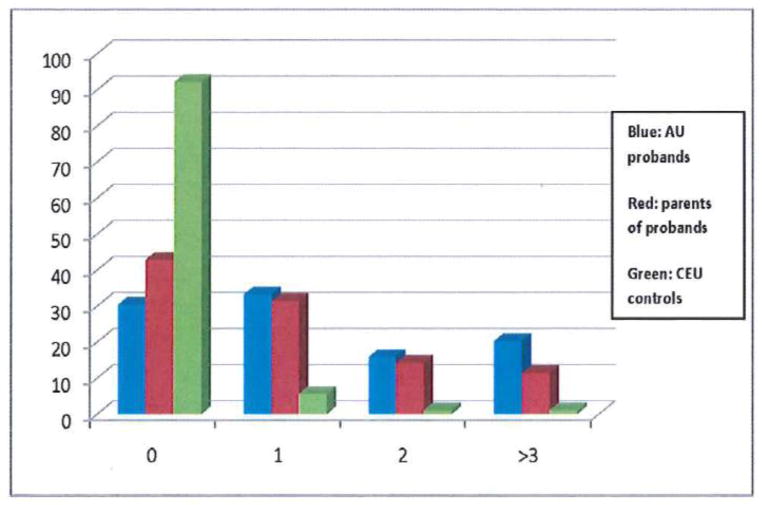
The numbers of CNVs per subject using search parameters 20m100k60o for probands, parents, and CEU controls show highly significant differences (p < 0.00005) in CNV frequency between autistic probands and CEU controls and between parents of autistic subjects and CEU controls (Figure 1, Supplemental Table 1a). Furthermore, CEU control samples were more likely to have no large CNVs detected and only 2% of the CEU control samples had more than one large CNV. This compares to 36% of the proband samples and 25% of the parental samples. The mean number of CNVs per subject in the category 20m100k60o is 1.6 for probands, 0.94 for parents, and 0.16 for CEU controls.
Figure 1. The Relative Frequency of CNVs in Probands, Parents, and CEF Controls by Standard Criteria.
Criteria for inclusion in this analysis are CNVs that encompass ≥ 20 SNP markers, are ≥ 100 kilobases in length, and cannot overlap more than 60% with a previously identified variant (20m100k60o). The frequency of CNVs is much greater in the patients than in parents, and CNV frequency is greater in parents than in controls (Kruskal-Wallis non-parametric test, p < 0.00005).
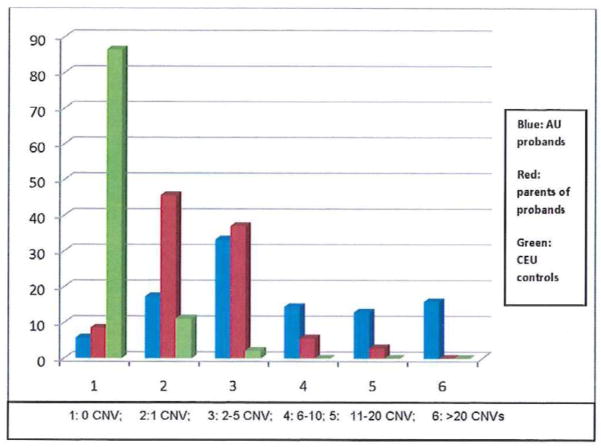
Highly significant differences in frequency of CNVs in autistic probands and their parents were also observed in the category of variants detected using the 20m10k0o criteria (p = 0.0001). The mean number of CNVs per subject in this category is 9.0 for proband, 2.5 for parents of autistic probands, and 0.14 for CEU controls (Figure 2, supplemental Table 2).
Figure 2. The Relative Frequency of CNVs in Probands, Parents, and CEF Controls by the Most Exacting Criteria.
Criteria for inclusion in this analysis are CNVs that encompass ≥ 20 SNP markers, are ≥ 10 kilobases in length, but do not overlap at all with any previously identified variant (20m100k0o). The frequency of CNVs is much greater in the patients than in parents, and CNV frequency is greater in parents than in controls (Kruskal-Wallis non-parametric test, p < 0.00005).
We also examined the frequency of inherited CNVs in trios where samples were available from both parents and their autistic child. Using the search criteria 20m10k0o we determined that 16 of the 145 CNVs (11%) observed in probands were inherited from a parent.
Genes involved in megabase sized multigenic CNVs
If CNVs associated with autism generate the phenotype because they alter the genes of an integrated bioenergetic, ion metabolism, and synaptic structural gene network, then we hypothesized that autism associated CNVs should encompasses one of more genes important for these functions. Accordingly, we performed a detailed analysis of CNVs larger than 1 megabase (mb) in 15 autistic probands. In 14 of these cases we also analyzed the metaphases using FISH in an attempt to verify the molecular defect.
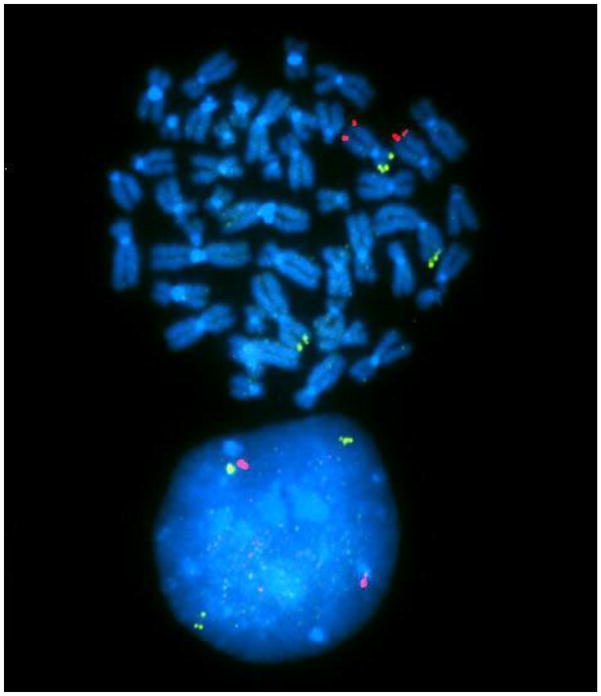
In one sib pair affected with autism and developmental delay we identified an approximately 4-megabase deletion on chromosome 12p and a similar sized duplication on chromosome 2p. Previously, these siblings had been reported as having normal karyotypes on the basis of clinical cytogenetic studies. By FISH analysis we found that the siblings carried a 2p:12p translocation. This translocation led to replacement of the terminal region of chromosome 12 (12p) with material from 2p (Figure 3). This translocation resulted in the deletion from one homologue of chromosome 12 of 27 genes including the calcium ion channel genes CACNA1C and CACNA2D4. The duplication of 2p25 material led to increased copy number of 12 genes, including SNTG2 (Table 1). These siblings showed marked growth retardation with height at the 10th percentile and weight at the 5th percentile.
Figure 3. FISH Analysis of 2p:12p Translocation Resulting in Deletion of CACNA1C and CACNA2D4.
Green = chromosome 2p probe which marks the translocated region and hence results in three copies, Red = chromosome 12q probe which marks the bottom of chromosome 12, this region is not rearranged. Translocation of the tip of the short arm of chromosome 2 (2p) onto the tip of the short arm of chromosome 12 (12p), replaces a section of the short arm of chromosome 2 (2p) with the transferred end of the short arm of chromosome 12 (12p). Loss of one of the two chromosomal copies of 12p removes 27 genes including CACNA1C and CACNA2D4. Generation of an extra copy of the 2p duplicates 12 genes including SNTG2.
Table 1.
CNV larger than 1 megabase in autistic probands detected by Affymetrix 6.0 SNP arrays and FISH
| Case ID | Chrom.region. | Prox.bp | Distal bp | Change | Genes |
|---|---|---|---|---|---|
| AU28–202 | 15q24.1–q24.2 | 70,750kb | 73,860kb | deletion | 37 |
| AU52–201 | 4q32.1–q34.2 | 157,975kb | 176,835kb | deletion | 32 |
| AU09–202 | 15q11–q13.3 | 18,200kb | 30,700kb | duplication | 29 |
| AU64–201 | 15q11–q13.3 | 18,200kb | 32,000kb | duplication | 29 |
| 29-AH | 15q13.2–q13.3 | 28,700kb | 30,320kb | deletion | 7 |
| AU210202 | 15q11.2–q13.1 | 21,192K | 26,500k | duplication | 12 |
| AU24–201 | 15pter-q11 | 1k | 8000kb | Extra chrom. | 3 |
| AU78–202 | 2q37.2–q37.3 | 235,663kb | 243697kb | deletion | 31 |
| AU50–201 | 13q13.3–q14.11 | 33,949kb | 43,438kb | deletion | 39 |
| 69 AF | Xp11.23-p11.22 | 47,961kb | 52,723kb | duplication | 66 |
| AU55–202 | 2pter-p25.2 | 2kb | 4,981kb | duplication | 12 |
| AU55–204 | 2pter-p25.2 | 2kb | 4,981kb | duplication | 12 |
| AU55–202 | 12pter-p13.2 | 6kb | 4.006kb | deletion | 25 |
| AU55–204 | 12pter-p13.2 | 6kb | 4.006kb | deletion | 25 |
| AU69–201 | 7p22.2-p21.3 | 10.338kb | 11.732kb | deletion | 3 |
| AU69–202 | 7p22.2-p21.3 | 10.338kb | 11.732kb | deletion | 3 |
| AU69–202 | 2pter-2cent. | 1kb | 80MB | duplication* | Many |
| AU69–202 | 7pter-7p22.3 | 160kb | 4,400kb | deletion | 9 |
| AU81–201 | 1q21.1 | 144,570kb | 147,300kb | duplication+ | 11 |
The chromosome regions involved in the 1 megabase CNV and the numbers of genes encompassed are listed in Table 1. In Table 2 we summarize genes of particular interest in the regions that showed CNVs and note genes involved in mitochondrial functions and ion metabolism.
Table 2.
| A. Genes Affected by CNVs ≥ 1Mb studied by microarrays and FISH | ||||
|---|---|---|---|---|
| Proband ID | Chromosome region | Position and extent In Kb | Number of genes | Genes of Interest |
| 28–202 | 15q24.1–15q24.2 | 73,420–75,932 del. | 45 | PPCDC*, COX5A*, SIN3A, SEMA7A, CYP11A1*, CYP1A1, CYP1A2 |
| 64–201 | 15q11–15q13.3 | 2,000–32,940 dup. | >80 | NDN, SNURPN, SNORD-HBI, UBE3A, ATP10A, GABRA5,^ GABRB3^, GABRG3^ HERC2, OCA2, APBA2, CHRNA7^, SCG5 |
| 09–201 | 15q11–15q13.3 | 2,000–32,940 dup. | >80 | NDN, SNURPN, SNORD-HBI, UBE3A, ATP10A, GABRA5^, GABRB3^, GABRG3^, HERC2, OCA2, APBA2, SCG5 |
| 210-202 | 15q11.2–15q13.1 | 21,200–28,700 dup. | >80 | NDN, SNURPN, SNORD-HBI, UBE3A, ATP10A, GABRA5^, GABRB3^, GABRG3^, HERC2 |
| AH | 15q13.3 | 30,670–32,460 del. | 14 | ARHGAP11B, TRPM1, KFL13, CHRNA7^ |
| 069AF | Xp11.23-p11.21 | 47,961–52,723 dup | 14 | CLCN5, AKAP4, DGKK+, SHROOM4 |
| 50–201 | 13q13.2–13q14.11 | 35,400–43,903 del | 44 | NBEA, SLC25A15*, MTRF1+, DGKH, ENOX1, AKAP11 |
| 52–201 | 4q32.1–4q32.2 | 157,977–176,836 del | 61 | ETFDH*, CBR4* CLCN3, GLRB^ GPM6^ |
| 78–202 | 2q37.2–q37.3 | 236,307–243819 del | 78 | NDUFA10*, D2HGDH*, CENTG2, HDACA4, UBE2F, AGXT |
| B. CNVs detected first by microarrays and then confirmed by FISH | ||||
|---|---|---|---|---|
| Proband ID | Chromosome region | Extent in KB | Number of genes | Genes of Interest |
| 55–202, 55–204 | 2pter-p25.3 dup | 0–4,498 dup. | 12 | SNTG2, PXDN, TPO+ |
| 55-02, 55–204 | 12pter-p25.2 | 0–3,930 del. | 27 | CACNA1C^, CACNA2D4^ SLC6A12, SLC6A13, RAD52, ERC1, FKBP4 |
| CNV in both members of monozygotic twin pair microarray only | ||||
|---|---|---|---|---|
| 69–201 69–202 |
7p21.3 | 10,340K–11,720K | 2 | NDUFA4*, PHF4 |
Mitochondrial location or function;
metabolic function,
Ion channel related
Mitochondrial Functions
15q24.1–15q24.2
PPCDC: Phosphopantothenoylcysteine decarboxylase involved in the biosynthesis Coenzyme A.
COX5: Cytochrome c oxidase subunit Va, a subunit of OXPHOS complex IV.
CYP11A: Mitochondrial inner membrane enzyme that catalyzes the conversion of cholesterol to pregnenolone.
13q13.2–14.11
SLC25A15: Member of the mitochondrial carrier family, transports ornithine across the inner mitochondrial membrane from the cytosol to the mitochondrial matrix.
MTRF1: Mitochondrial protein with similarity to the peptide chain release factors (RFs)
4q32.1–q32.2
ETFDH: Electron-transferring-flavoprotein dehydrogenase reduces ubiquinone in the mitochondrial membrane using electrons derived from fatty acid oxidation within the matrix borne by the mitochondrial electron-transfer flavoprotein.
CBR4: Carbonyl reductase 4 mitochondrial location NAD(P)H dehydrogenase quinone activity.
2q37.2–q37.3
NDUFA10: A component of the NADH dehydrogenase (ubiquinone) oxidoreductase (OXPHOS complex I).
D2HGDH: D-2hydroxyglutarate dehydrogenase, a mitochondrial enzyme belonging to the FAD-binding oxidoreductase/transferase type 4 family
7p21.3
NDUFA4: A component of the NADH dehydrogenase (ubiquinone) oxidoreductase (OXPHOS complex I).
Ion channel functions
15q11–q13.3
ATP10A: A member of the family of P-type cation transport ATPases
CHRNA7: A subunit of the nicotinic acetylcholine receptors (nAChRs), members of a superfamily of ligand-gated ion channels that mediate fast signal transmission at synapses.
GABRA5, GABRB3, GABRG3: Distinct subunits of a multisubunit chloride channels that serves as the receptors for gamma-aminobutyric acid.
TRPM1: A calcium permeable cation channel4q32.1–q32.2
CLCN3: A member of the voltage-gated chloride intracellular channel family.
GPM6: A neuronal membrane glycoprotein with calcium channel activity.
12pter-p25.2
CACNA1C: An alpha-1 subunit of a voltage-dependent calcium channel.
CACNA2D4: a member of the alpha-2/delta subunit family, a protein in the voltage-dependent calcium channel complex.
Multi-gene duplications of 15q11.1–15q11.3 were found in three cases and included the SNRPN and UBE3A genes (see Table 1). In two cases the duplications also involved the GABA receptor genes GABRB3, GABRA5 and GABRG3. Our studies on parents in these cases revealed that the abnormalities arose de novo in the affected children. Studies of polymorphic markers revealed that the largest duplications (cases AU09–202 and AU64–201) arose on maternal chromosomes, the duplication in case AU210-202 arose on a paternal chromosome. Analysis of autistic manifestations and degree of cognitive impairment in the three cases with 15q duplications revealed that the phenotype was much milder in AU210-202, where the interstitial duplication of 15q11.2–q13.1 arose on a paternal chromosome.
Our microarray studies confirmed the presence of a 15q13.3 deletion in a case of autism that had prior clinical cytogenetic studies. We also studied the parents of this proband and determined that the 15q13.3 deletion occurred de novo in the patient. In this patient biochemical evaluation revealed slight increases in long chain organic acids and increased acylcarnitine during one hospitalization for evaluation of a seizure disorder. On carnitine supplementation organic acid levels were normal and free carnitine and acylcarnitine blood levels were in the normal range.
We carried out microarray studies to more accurately define chromosome deletion breakpoints and impacted genes in four patients with autism where cytogenetic and clinical data where previously reported. Deletions in these patients involved 15q24.1 (AU28–202), 4q32.1–q34.2 (AU52–201), 2q37.2–q37.3 (AU78–202) and 13q13.3–q14.11 (AU50–201). In autism case AU28–202, a 15q deletion, was found to remove the promyelocytic leukemic (PML) locus and surrounding genes [24]. This region also encompassed other important mitochondria genes (see Table 2). Deletion of 15q24.1–15q24.2 removed the mitochondrial genes COX5A and PPCDC. COX5A is a subunit of complex IV and PPCDC encodes phosphopantothenoyl-cysteine decarboxylase which is involved in the generation of the acyl carrier molecule, coenzyme A. Coenzyme A, in turn, is involved in the synthesis of Coenzyme Q, and both are central to mitochondrial function. Consistent with these associations, physiological studies of this patient revealed elevated alanine levels which are commonly seen in patients with mitochondrial dysfunction.
Female monozygotic twins, AU69–201 and AU69–202, who met ADOS criteria for autism and with developmental delay, were evaluated at age 10 years. These twins were both found to harbor duplication on chromosome 7p that was 1.4 mb in size. We further mapped the duplication to 7p21.3–p22.2, extending from 10,338K to 11,732K. This region encompassed the NDUFA4 gene that encodes a subunit of mitochondrial OXPHOS complex I as well as PHF4 involved in epigenetic chromatin regulation and THSD7A that encodes a sub-unit of thrombospondin.
Two cases were examined for mitochondrial OXPHOS defects in skeletal muscle mitochondria, case 1 with two CNVs (a low number) and case 2 with 13 CNVs (a high number) (Table 3). Case 1 shows a partial reduction in complex IV which might reflect a heterozygous complex IV gene deletion. However, this level of complex IV reduction would not be considered clinically significant in the diagnosis of patients with multisystem disease. Case 2 had a 44% reduction in complex I as well as a striking increased activity of both citrate synthase and the ratio of complexes II/III (308% increase). While the partial reduction of complex I would not be considered significant in a patient with multisystem disease, the up-regulation of citrate synthase and complex II are seen in severe mitochondrial diseases cases and have been interpreted as a compensatory response to a chronic OXPHOS defect. Therefore, at least some autism patients have significant mitochondrial defects, though at present we cannot determine if there is a correlation between the number of CNVs and the severity of mitochondrial dysfunction.
Table 3.
Relative Skeletal Muscle Mitochondrial Electron Transport Chain Activities in Autistic Probands with high and low CNV Level.
| Case | CNV # | CS | CI | CI/CS | CII/CIII | CII/CIII/CS | CIV | CIV/CS |
|---|---|---|---|---|---|---|---|---|
| 1 | 2 | 930 | 89.4 | 96.1 | 155 | 167 | 0.68 | 0.73 |
| 2 | 13 | 1746 | 62.8 | 36.0 | 571 | 327 | 1.06 | 0.61 |
CS = citrate synthase activity: CI, CII, CIII, and CIV–OXPHOS complexes I–IV. Specific activities presented as nmole/min/mg protein
Discussion and Significance
Our study has demonstrated that there is a direct correlation between the autism phenotype and the number of CNVs, with the number of CNVs in the parents of autistic children being intermediate between the probands and the controls. Hence, an increase in the number of CNVs increases the probability of developing autism.
Analysis of the genes encompassed by well defined CNVs confirmed that CNVs frequently altered genes involved in mitochondrial, ion channel, and synaptic function. Since both mitochondrial function and ion transport are important for synaptic function, alterations in any of these genes could affect synaptic function and create a neuropsychiatric phenotype [2].
While some rearrangements were seen more than once, others were seen only once in our sample. The most common CNV occurred at 15q11–q13.3. This region encompasses the UBE3A gene, deficiency of which we have shown in a mouse mutant specifically alters the hippocampal mitochondrial morphology and brain complex II/III specific activity [25]. Other CNVs altered the copy number of genes known to be important in mitochondrial function including NDUFA4, NDUFA10, PPCDC, COX5, CYP11A, SLC25A15, MTRF, ETFDH, and CBR4 (Table 2).
There is now evidence that recurrent 15q24 microdeletion events occur in individuals in various populations and that this microdeletion is associated with intellectual disability, speech problems, and frequently with autism. A study of 15 cases with 15q24.1–15q24.2 deletions also led to the conclusion that several genes in this region likely played roles in determining intellectual disability [26]. Since this region encompasses the COX5A and PPCDC genes, this recurrent microdeletion is consistent with our hypothesis that a subset of autism CNVs do seem to coincide with important nuclear encoded mitochondrial expressed gene loci [27].
Other CNVs were found to alter ion transport genes. A 2p:12p chromosomal translocation was found in two autistic siblings. It leads to deletion of 12p that removes one copy each of the CACNA1C and CACNA2D4 calcium channel genes. Mutations in the CACNA1C gene, generally at codon G406R, cause Timothy syndrome [28]. The CACNA1C protein is a T-type voltage-gated Cav1.2 channel and its mutation in Timothy syndrome results in Long QT syndrome and autism. Cav1.2 shows its highest expression in the hippocampus, amygdala, and putamen [29]. The CACNA2D4 gene has a more limited range of tissue expression than CACNA1C, and expressed primarily in eye and retina, in the pituitary, and adrenal gland.
An independent study reported findings on a two children who manifested developmental delay and impaired social interactions. They were found to have an interstitial deletion on 12p13.33 that impacted eight genes including CACNA1C, CACNA2D4 and ERC1 [30]. A survey of the CACNA1H gene, a close paralog of the Timothy syndrome Ca++ channel, in 461 autism DNAs revealed six new mutations. Functional studies of two of these mutant proteins, R212C and R902W, revealed extended Ca++ influx into the cytosol [31]. There is growing evidence for the importance of CACNA1C in brain function and evidence that specific allelic variants and genotypes of CACNA1C occur with higher frequency in patients with mental illness including bipolar disease, recurrent major depression and schizophrenia [32, 33]. Also CACNA1C genotype can influence verbal fluency in healthy individuals [34].
Calcium levels have been found to be elevated in the brains of autism patients and this appears to activate the mitochondrial AGC which is central to the cytosol-mitochondrial NADH shuttle system [19]. Cytosolic calcium levels are regulated by mitochondrial calcium uptake, driven by the mitochondrial inner membrane potential (ΔP). This involves the mitochondrial Ca++ uniporter [35, 36] which interfaces with the type 3 inositol triphosphate receptor (IP3R) receptors in the endoplasmic reticulum (ER) located in the mitochondrial associated membranes (MAMs). The promyelocytic leukemia (PML) protein whose gene at 15q is deleted in AU28–202 is associated with this complex [37]. Excessive mitochondrial uptake of calcium can activate the mitochondrial permeability transition pore (mtPTP) causing cell death [38]. Other CNVs also encompassed transport genes including GPM6, TRPM1, (GABRA5, GABRB3, GABRG3), CLCN3, CHRNA7 and ATP10A (Table 2).
One patient report links mitochondrial dysfunction with ion channel disruption. This patient carried a de novo translocation between chromosomes 1 and 15, t (1; 15) (p36.11: q24.2) which interrupted the CLIC4 intracellular chloride channel 4 gene on 1p36.11 and the PPCDC gene on 15q24 which was split between exons 1 and 3. In addition, the translocation decreased the expression of the SCAMP5 gene close to the 15 translocation breakpoint [39].
Other CNVs appear to impact synaptic structure and function. The duplication of 2p25 material added an additional copy of the SNTG2 gene (Figure 3). Previously, we reported that patient 50–201 with a 13q deletion was hemizygosity for a number of genes including the very large neurobeachin (NBEA) gene [40]. Disruption of the NBEA gene has been reported in another autism patient [41] and knockout of NBEA in mice led to a reduced number of dendritic spine synapses in cultured neuronal cells [42].
Conclusion
Our study confirmed the correlation between increased numbers of CNVs and risk of developing autism and revealed that some patients with CNVs can also manifest mitochondrial dysfunction. By carefully mapping the CNVs we have observed that CNVs in autistic patients commonly encompass genes important in mitochondrial function, ion transport, and synaptic structure and function. Since the nucleus contains between one and two thousand mitochondrial genes plus numerous ion transport genes, it is conceivable that the removal of any one of a large number of different genes necessary to maintain this integrated bioenergetic, signaling, and synaptic structural gene network could adversely affect synaptic function and thus result in autism. If this speculation is correct, it might explain why it has been so difficult to identify individual structural genes that cause neuropsychiatric disorders.
Supplementary Material
Highlights.
CNVs are significantly increased in autism.
Some autism patients have systemic OXPHOS defects.
CNVs often encompass mitochondrial, ion channel, and synaptic genes.
Acknowledgments
The work was supported by Autism Speaks Foundation grant 5668, Simons Foundation Grant #205844, and NIH grants NS070298, AG24373, and NS21328 awarded to DCW.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Contributor Information
Moyra Smith, Email: dmsmith@uci.edu.
Pamela L. Flodman, Email: pflodman@uci.edu.
John J. Gargus, Email: jjgargus@uci.edu.
Mariella T Simon, Email: simonm@uci.edu.
Kimberley Verrell, Email: kverrell@uci.edu.
Richard Haas, Email: rhaas@ucsd.edu.
Gail E. Reiner, Email: gereiner@ucsd.edu.
Robert Naviaux, Email: rnaviaux@ucsd.edu.
Katherine Osann, Email: kosann@uci.edu.
M. Anne Spence, Email: maspence@uci.edu.
Douglas C. Wallace, Email: wallaced1@email.chop.edu.
References
- 1.Filipek PA. Medical Aspects of Autism. In: Volkmar FR, Klin A, Paul R, Cohen DJ, editors. Handbook of Autism and Pervasive Developmental Disorders. Chapter 520 John Wiley and Sons; New York: 2005. pp. 534–578. [Google Scholar]
- 2.Wallace DC, Fan W. Energetics, epigenetics, mitochondrial genetics. Mitochondrion. 2010;10:12–31. doi: 10.1016/j.mito.2009.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sebat J, Lakshmi B, Malhotra D, Troge J, Lese-Martin C, Walsh T, Yamrom B, Yoon S, Krasnitz A, Kendall J, Leotta A, Pai D, Zhang R, Lee YH, Hicks J, Spence SJ, Lee AT, Puura K, Lehtimaki T, Ledbetter D, Gregersen PK, Bregman J, Sutcliffe JS, Jobanputra V, Chung W, Warburton D, King MC, Skuse D, Geschwind DH, Gilliam TC, Ye K, Wigler M. Strong association of de novo copy number mutations with autism. Science. 2007;316:445–449. doi: 10.1126/science.1138659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zhao X, Leotta A, Kustanovich V, Lajonchere C, Geschwind DH, Law K, Law P, Qiu S, Lord C, Sebat J, Ye K, Wigler M. A unified genetic theory for sporadic and inherited autism. Proc Natl Acad Sci USA. 2007;104:12831–12836. doi: 10.1073/pnas.0705803104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Guilmatre A, Dubourg C, Mosca AL, Legallic S, Goldenberg A, Drouin-Garraud V, Layet V, Rosier A, Briault S, Bonnet-Brilhault F, Laumonnier F, Odent S, Le Vacon G, Joly-Helas G, David V, Bendavid C, Pinoit JM, Henry C, Impallomeni C, Germano E, Tortorella G, Di Rosa G, Barthelemy C, Andres C, Faivre L, Frebourg T, Saugier Veber P, Campion D. Recurrent rearrangements in synaptic and neurodevelopmental genes and shared biologic pathways in schizophrenia, autism, and mental retardation. Arch Gen Psychiatry. 2009;66:947–956. doi: 10.1001/archgenpsychiatry.2009.80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Veltman JA, Brunner HG. Understanding variable expressivity in microdeletion syndromes. Nat Genet. 2010;42:192–103. doi: 10.1038/ng0310-192. [DOI] [PubMed] [Google Scholar]
- 7.Wallace DC. A mitochondrial paradigm of metabolic and degenerative diseases, aging, and cancer: a dawn for evolutionary medicine. Annu Rev Genet. 2005;39:359–407. doi: 10.1146/annurev.genet.39.110304.095751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wallace DC, Fan W, Procaccio V. Mitochondrial energetics and therapeutics. Annu Rev Path. 2010;5:297–348. doi: 10.1146/annurev.pathol.4.110807.092314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gargus JJ, Imtiaz F. Mitochondrial energy-deficient endophenotype in autism. Am J Biochem Biotechnol. 2008;4:198–207. [Google Scholar]
- 10.Lombard J. Autism: a mitochondrial disorder? Med Hypotheses. 1998;50:497–500. doi: 10.1016/s0306-9877(98)90270-5. [DOI] [PubMed] [Google Scholar]
- 11.Haas RH, Townsend J, Courchesne E, Lincoln AJ, Schreibman L, Yeung-Courchesne R. Neurologic abnormalities in infantile autism. J Child Neurol. 1996;11:84–92. doi: 10.1177/088307389601100204. [DOI] [PubMed] [Google Scholar]
- 12.Correia C, Coutinho AM, Diogo L, Grazina M, Marques C, Miguel T, Ataide A, Almeida J, Borges L, Oliveira C, Oliveira G, Vicente AM. Brief report: high frequency of biochemical markers for mitochondrial dysfunction in autism: no association with the mitochondrial aspartate/glutamate carrier SLC25A12 gene. J Autism Dev Disord. 2006;36:1137–1140. doi: 10.1007/s10803-006-0138-6. [DOI] [PubMed] [Google Scholar]
- 13.Oliveira G, Ataide A, Marques C, Miguel TS, Coutinho AM, Mota-Vieira L, Goncalves E, Lopes NM, Rodrigues V, Carmona da Mota H, Vicente AM. Epidemiology of autism spectrum disorder in Portugal: prevalence, clinical characterization, and medical conditions. Dev Med Child Neurol. 2007;49:726–733. doi: 10.1111/j.1469-8749.2007.00726.x. [DOI] [PubMed] [Google Scholar]
- 14.Poling JS, Frye RE, Shoffner J, Zimmerman AW. Developmental regression and mitochondrial dysfunction in a child with autism. J Child Neurol. 2006;21:170–172. doi: 10.2310/7010.2006.00032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Holtzman D. Autistic spectrum disorders and mitochondrial encephalopathies. Acta Paediatr. 2008;97:859–860. doi: 10.1111/j.1651-2227.2008.00883.x. [DOI] [PubMed] [Google Scholar]
- 16.Graf WD, Marin-Garcia J, Gao HG, Pizzo S, Naviaux RK, Markusic D, Barshop BA, Courchesne E, Haas RH. Autism associated with the mitochondrial DNA G8363A transfer RNA(Lys) mutation. J Child Neurol. 2000;15:357–361. doi: 10.1177/088307380001500601. [DOI] [PubMed] [Google Scholar]
- 17.Fillano JJ, Goldenthal MJ, Rhodes CH, Marin-Garcia J. Mitochondrial dysfunction in patients with hypotonia, epilepsy, autism, and developmental delay: HEADD syndrome. J Child Neurol. 2002;17:435–439. doi: 10.1177/088307380201700607. [DOI] [PubMed] [Google Scholar]
- 18.Pons R, Andreu AL, Checcarelli N, Vila MR, Engelstad K, Sue CM, Shungu D, Haggerty R, de Vivo DC, DiMauro S. Mitochondrial DNA abnormalities and autistic spectrum disorders. Journal of Pediatrics. 2004;144:81–85. doi: 10.1016/j.jpeds.2003.10.023. [DOI] [PubMed] [Google Scholar]
- 19.Palmieri L, Papaleo V, Porcelli V, Scarcia P, Gaita L, Sacco R, Hager J, Rousseau F, Curatolo P, Manzi B, Militerni R, Bravaccio C, Trillo S, Schneider C, Melmed R, Elia M, Lenti C, Saccani M, Pascucci T, Puglisi-Allegra S, Reichelt KL, Persico AM. Altered calcium homeostasis in autism-spectrum disorders: evidence from biochemical and genetic studies of the mitochondrial aspartate/glutamate carrier AGC1. Mol Psychiatry. 2010;15:38–52. doi: 10.1038/mp.2008.63. [DOI] [PubMed] [Google Scholar]
- 20.Ramoz N, Reichert JG, Smith CJ, Silverman JM, Bespalova IN, Davis KL, Buxbaum JD. Linkage and association of the mitochondrial aspartate/glutamate carrier SLC25A12 gene with autism. Am J Psychiatry. 2004;161:662–669. doi: 10.1176/appi.ajp.161.4.662. [DOI] [PubMed] [Google Scholar]
- 21.Lepagnol-Bestel AM, Maussion G, Boda B, Cardona A, Iwayama Y, Delezoide AL, Moalic JM, Muller D, Dean B, Yoshikawa T, Gorwood P, Buxbaum JD, Ramoz N, Simonneau M. SLC25A12 expression is associated with neurite outgrowth and is upregulated in the prefrontal cortex of autistic subjects. Mol Psychiatry. 2008;13:385–397. doi: 10.1038/sj.mp.4002120. [DOI] [PubMed] [Google Scholar]
- 22.Haas RH, Parikh S, Falk MJ, Saneto RP, Wolf NI, Darin N, Wong L-J, Cohen BH, Naviaux RK. The in-depth evaluation of suspected mitochondrial disease. Mol Genet Metab. 2008;94:16–37. doi: 10.1016/j.ymgme.2007.11.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Trounce IA, Kim YL, Jun AS, Wallace DC. Assessment of mitochondrial oxidative phosphorylation in patient muscle biopsies, lymphoblasts, and transmitochondrial cell lines. Methods Enzymol. 1996;264:484–509. doi: 10.1016/s0076-6879(96)64044-0. [DOI] [PubMed] [Google Scholar]
- 24.Smith M, Filipek PA, Wu C, Bocian M, Hakim S, Modahl C, Spence MA. Analysis of a 1-megabase deletion in 15q22-q23 in an autistic patient: identification of candidate genes for autism and of homologous DNA segments in 15q22–q23 and 15q11–q13. Am J Med Genet. 2000;96:765–770. doi: 10.1002/1096-8628(20001204)96:6<765::aid-ajmg13>3.0.co;2-l. [DOI] [PubMed] [Google Scholar]
- 25.Su H, Fan W, Coskun PE, Vesa J, Gold JA, Jiang YH, Potluri P, Procaccio V, Acab A, Weiss JH, Wallace DC, Kimonis VE. Mitochondrial dysfunction in CA1 hippocampal neurons of the UBE3A deficient mouse model for Angelman syndrome. Neurosci Lett. 2011;487:129–133. doi: 10.1016/j.neulet.2009.06.079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mefford HC, Rosenfeld JA, Shur N, Slavotinek AM, Cox VA, Hennekam RC, Firth HV, Willatt L, Wheeler P, Morrow EM, Cook J, Sullivan R, Oh A, McDonald MT, Zonana J, Keller K, Hannibal MC, Ball S, Kussmann J, Gorski J, Zelewski S, Banks V, Smith W, Smith R, Paull L, Rosenbaum KN, Amor DJ, Silva J, Lamb A, Eichler EE. Further clinical and molecular delineation of the 15q24 microdeletion syndrome. J Med Genet. 2012;49:110–118. doi: 10.1136/jmedgenet-2011-100499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Smith M, Spence MA, Flodman P. Nuclear and mitochondrial genome defects in autisms. Annals of the New York Academy of Science. 2008;1151:102–132. doi: 10.1111/j.1749-6632.2008.03571.x. [DOI] [PubMed] [Google Scholar]
- 28.Splawski I, Timothy KW, Decher N, Kumar P, Sachse FB, Beggs AH, Sanguinetti MC, Keating MT. Severe arrhythmia disorder caused by cardiac L-type calcium channel mutations. Proc Natl Acad Sci USA. 2005;102:8089–8096. doi: 10.1073/pnas.0502506102. discussion 8086–8088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gargus JJ. Genetic calcium signaling abnormalities in the central nervous system: seizures, migraine, and autism. Ann N Y Acad Sci. 2009;1151:133–156. doi: 10.1111/j.1749-6632.2008.03572.x. [DOI] [PubMed] [Google Scholar]
- 30.Abdelmoity AT, Hall JJ, Bittel DC, Yu S. 1.39 Mb inherited interstitial deletion in 12p13.33 associated with developmental delay. Eur J Med Genet. 2011;54:198–203. doi: 10.1016/j.ejmg.2010.11.010. [DOI] [PubMed] [Google Scholar]
- 31.Splawski I, Yoo DS, Stotz SC, Cherry A, Clapham DE, Keating MT. CACNA1H mutations in autism spectrum disorders. J Biol Chem. 2006;281:22085–22091. doi: 10.1074/jbc.M603316200. [DOI] [PubMed] [Google Scholar]
- 32.Krug A, Nieratschker V, Markov V, Krach S, Jansen A, Zerres K, Eggermann T, Stocker T, Shah NJ, Treutlein J, Muhleisen TW, Kircher T. Effect of CACNA1C rs1006737 on neural correlates of verbal fluency in healthy individuals. Neuroimage. 2010;49:1831–1836. doi: 10.1016/j.neuroimage.2009.09.028. [DOI] [PubMed] [Google Scholar]
- 33.Green EK, Grozeva D, Jones I, Jones L, Kirov G, Caesar S, Gordon-Smith K, Fraser C, Forty L, Russell E, Hamshere ML, Moskvina V, Nikolov I, Farmer A, McGuffin P, Holmans PA, Owen MJ, O’Donovan MC, Craddock N. The bipolar disorder risk allele at CACNA1C also confers risk of recurrent major depression and of schizophrenia. Mol Psychiatry. 2010;15:1016–1022. doi: 10.1038/mp.2009.49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Thimm M, Kircher T, Kellermann T, Markov V, Krach S, Jansen A, Zerres K, Eggermann T, Stocker T, Shah NJ, Nothen MM, Rietschel M, Witt SH, Mathiak K, Krug A. Effects of a CACNA1C genotype on attention networks in healthy individuals. Psychol Med. 2011;41:1551–1561. doi: 10.1017/S0033291710002217. [DOI] [PubMed] [Google Scholar]
- 35.Baughman JM, Perocchi F, Girgis HS, Plovanich M, Belcher-Timme CA, Sancak Y, Bao XR, Strittmatter L, Goldberger O, Bogorad RL, Koteliansky V, Mootha VK. Integrative genomics identifies MCU as an essential component of the mitochondrial calcium uniporter. Nature. 2011;476:341–345. doi: 10.1038/nature10234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.De Stefani D, Raffaello A, Teardo E, Szabo I, Rizzuto R. A forty-kilodalton protein of the inner membrane is the mitochondrial calcium uniporter. Nature. 2011;476:336–340. doi: 10.1038/nature10230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pinton P, Giorgi C, Pandolfi PP. The role of PML in the control of apoptotic cell fate: a new key player at ER-mitochondria sites. Cell Death Differ. 2011;18:1450–1456. doi: 10.1038/cdd.2011.31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gunter TE, Sheu SS. Characteristics and possible functions of mitochondrial Ca(2+) transport mechanisms. Biochim Biophys Acta. 2009;1787:1291–1308. doi: 10.1016/j.bbabio.2008.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Castermans D, Volders K, Crepel A, Backx L, De Vos R, Freson K, Meulemans S, Vermeesch JR, Schrander-Stumpel CT, De Rijk P, Del-Favero J, Van Geet C, Van De Ven WJ, Steyaert JG, Devriendt K, Creemers JW. SCAMP5, NBEA and AMISYN: three candidate genes for autism involved in secretion of large dense-core vesicles. Hum Mol Genet. 2010;19:1368–1378. doi: 10.1093/hmg/ddq013. [DOI] [PubMed] [Google Scholar]
- 40.Smith M, Escamilla JR, Filipek P, Bocian ME, Modahl C, Flodman P, Spence MA. Molecular genetic delineation of 2q37.3 deletion in autism and osteodystrophy: report of a case and of new markers for deletion screening by PCR. Cytogenet Cell Genet. 2001;94:15–22. doi: 10.1159/000048775. [DOI] [PubMed] [Google Scholar]
- 41.Castermans D, Wilquet V, Parthoens E, Huysmans C, Steyaert J, Swinnen L, Fryns JP, Van de Ven W, Devriendt K. The neurobeachin gene is disrupted by a translocation in a patient with idiopathic autism. J Med Genet. 2003;40:352–356. doi: 10.1136/jmg.40.5.352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Niesmann K, Breuer D, Brockhaus J, Born G, Wolff I, Reissner C, Kilimann MW, Rohlmann A, Missler M. Dendritic spine formation and synaptic function require neurobeachin. Nat Commun. 2011;2:557. doi: 10.1038/ncomms1565. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.