Summary
The deubiquitinating module (DUBm) of the SAGA coactivator contains the Ubp8 isopeptidase, Sgf11, Sus1 and Sgf73, which form a highly interconnected complex. While Ubp8 contains a canonical USP catalytic domain, it is only active when in complex with the other DUBm subunits. The Sgf11 zinc finger (Sgf11-ZnF) binds near the Ubp8 active site and is essential for full activity, suggesting that the Sgf11-ZnF helps maintain the active conformation of Ubp8. We report structural and solution studies showing that deletion of the Sgf11-ZnF destabilizes incorporation of Ubp8 within the DUBm, giving rise to domain swapping with a second complex and misaligning active site residues. Activating mutations in Ubp8 that partially restore activity in the absence of the Sgf11-ZnF promote the monomeric form of the DUBm. Our data suggest an unexpected role for Sgf11 in compensating for the absence of structural features that maintain the active conformation of Ubp8.
Introduction
The SAGA (Spt-Ada-Gcn5-Acetyltransferase) transcriptional co-activator is a 1.8 MDa, 21-protein complex that regulates inducible yeast genes by performing multiple functions including acetylating core histones, recruiting the RNA polymerase II preinitiation complex, and deubiquitinating histone H2B (reviewed in (Baker & Grant, 2007; Koutelou, Hirsch, & Dent, 2010; Rodríguez-Navarro, 2009)). At genes activated by SAGA, the E2 ubiquitin conjugating enzyme, Rad6, and the E3 ubiquitin ligase, Bre1, mono-ubiquitinate histone H2B at Lys123 (Hwang et al., 2003). This modification promotes recruitment of Set1, which methylates histone H3 at Lys4 and triggers the recruitment of the SAGA complex (Daniel et al., 2004). SAGA acetylates histone H3 and deubiquitinates histone H2B (H2B-Ub), which is required for downstream events including phosphorylation of the C-terminal domain (CTD) of RNA polymerase II by Ctk1 and methylation of H3K36 by Set2 (Weake and Workman, 2010; Wyce et al., 2007). Mutations and deletions that abrogate the deubiquitinating activity of SAGA give rise to defects in both transcription activation and elongation (Daniel et al., 2004; Wyce et al., 2007; Henry et al., 2003), pointing to a key role for deubiquitination in the transcription of inducible genes. The deubiquitinating activity of SAGA resides in a distinct sub-complex called the deubiquitinating module (DUBm), which consists of four proteins that are conserved across eukaryotes: Ubp8, Sgf11, Sus1 and Sgf73, of which residues 1–96 are sufficient for in vitro DUBm activity (Kohler et al., 2008). The enzymatic subunit, Ubp8, contains a C-terminal catalytic domain belonging to the Ubiquitin Specific Protease (USP) family of deubiquitinating enzymes (DUBs) (Reyes-Turcu and Wilkinson, 2010; Henry et al., 2003) and an N-terminal ZnF-UBP domain (Henry et al., 2003; Ingvarsdottir et al., 2005; Reyes-Turcu et al., 2006) that is required for incorporation into the SAGA complex (Ingvarsdottir et al., 2005). USP enzymes are papain-like cysteine proteases that contain a conserved Cys-His catalytic dyad as well as conserved oxyanion hole residues that are required for a conserved general mechanism of deubiquitination (Zhang et al., 2011; Komander et al., 2009; Hu et al., 2002). Although Ubp8 contains a canonical USP domain, this subunit is inactive unless it is in complex with the other three DUBm subunits (Kohler et al., 2008; Lee et al., 2009). Relatively small deletions in Sgf11 and Sgf73 disrupt DUBm activity (Kohler et al., 2008, 2010), pointing to the importance that these proteins play in contributing to enzymatic activity.
The structure of the DUBm revealed an unexpected arrangement of Ubp8, Sgf11, Sus1, Sgf73, in which each protein contacts the other three to form a highly interconnected complex (Samara et al., 2010; Kohler et al., 2010). The DUBm proteins are organized into two lobes around the globular domains of Ubp8 (Figure 1A). The ZnF-UBP (Samara et al., 2010), or assembly lobe (Kohler et al., 2010) of the complex is centered around the long N-terminal helix of Sgf11, which has one face that binds to the ZnF-UBP domain of Ubp8, while the other face binds Sus1 (Samara et al., 2010; Kohler et al., 2010). An extended ordered linker joins the Sgf11 N-terminal helix to its C-terminal zinc finger domain (Sgf11-ZnF), which binds to the Ubp8 USP domain immediately adjacent to the catalytic site (Figures 1A and 1B). The Sgf11-ZnF makes extensive contacts with a loop (referred to as loop L2) that contains active site residues, in addition to connecting the USP domain of Ubp8 to its ZnF-UBP domain. Sgf731–96, which has a low degree of secondary structure and contains a C-terminal zinc finger, is partially buried between the two lobes of the DUB module and makes extensive contacts with both domains of Ubp8 (Figure 1A). The zinc fingers of Sgf11 and Sgf73 are critical for DUB module activity: deletion of the Sgf11-ZnF drastically reduces enzymatic activity, while deletion of the Sgf73 zinc finger causes Sgf73 to dissociate from the DUB module, thus abrogating activity (Kohler et al., 2010, 2008). The dependence of Ubp8 deubiquitinating activity on the presence of additional subunits is not unique: USP1, USP12 and USP46 all require the regulatory protein UAF1 for activity (Kee et al., 2010; Cohn et al., 2009, 2007), and the H2A DUB, BAP1, requires ASX for activity (Scheuermann et al., 2010). The mechanism by which these DUBs are regulated by additional subunits is not known.
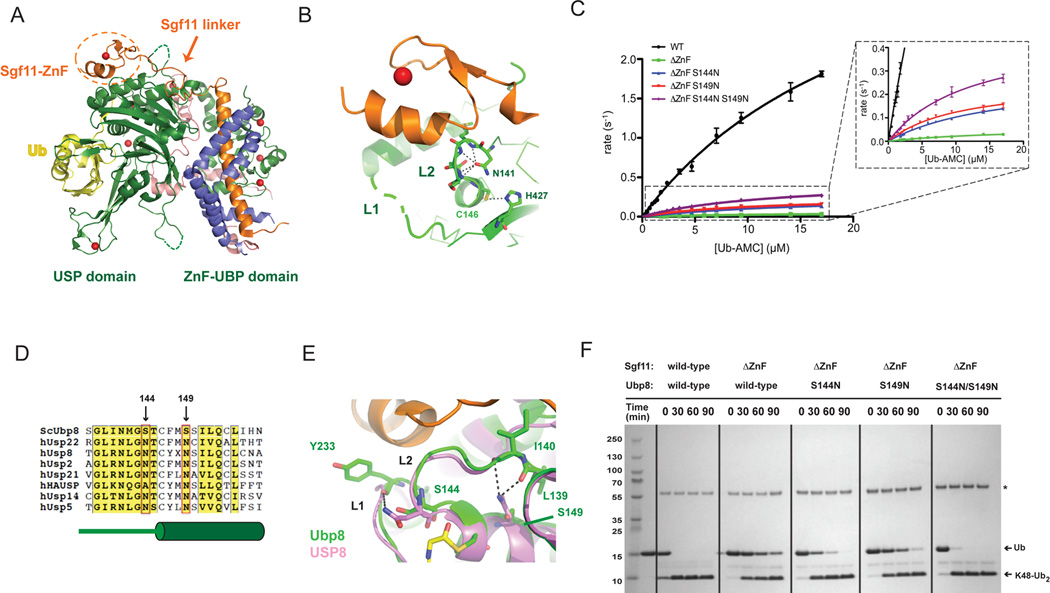
Figure 1. Gain-of-function mutations in Ubp8 that compensate for deletion of the Sgf11 zinc finger.
(A) Structure of the wild-type DUBm. The four subunits are Ubp8 (green), Sgf11 (orange), Sus1 (blue), Sgf73 (salmon); zinc atoms are depicted as red spheres. The USP and ZnF-UBP domains of Ubp8 are indicated below. The Sgf11-ZnF (in dotted circle) binds to the USP domain of Ubp8 and is connected to its helical region by an extended linker.
(B) The Sgf11-ZnF (orange) binds near the Ubp8 (green) active site, contacting loops L1 and L2. The catalytic residues, C146 and H427, and the oxyanion hole residue, N131 are depicted as sticks. Loop L1 (dashed gray line) is disordered in the absence of ubiquitin.
(C) Rate of Ub-AMC cleavage as a function of substrate concentration by wild-type DUBm and DUBm lacking the Sgf11-ZnF containing either wild-type or Ubp8 with substitutions S144N, S149N, or S144N/S149.
(D) Sequence alignment of USP DUBs indicated residues 144 and 149 of Ubp8. Diagram in green below sequence indicates secondary structure in Ubp8.
(E) Superposition of Ubp8 (green) with hUSP8 (pink) showing hydrogen bonding between N149 in hUSP8 with loop L2 backbone residues. Residue S149 in Ubp8 does not contact loop L2.
(F) Cleavage of K48-diubiquitin by wild-type and mutant DUBm. Gel stained with Coomassie.
See also Figure S2.
A potential model for how Sgf11-ZnF activates Ubp8 was suggested by structural studies of HAUSP/Usp7 (Hu et al., 2002; Faesen et al., 2011) and OTUB1 (Edelmann et al., 2009; Wiener et al., 2012). In those deubiquitinating enzymes, the active site residues are misaligned in the absence of ubiquitin and become correctly aligned when the ubiquitin substrate is bound (Edelmann et al., 2009; Wiener et al., 2012; Hu et al., 2002). Unlike HAUSP and OTUB1, the active site residues of Ubp8 are aligned for catalysis in both the absence and presence of ubiquitin (Samara et al., 2010; Kohler et al., 2010). One way in which the other DUBm subunits could activate Ubp8 would be by maintaining the correct conformation of the Ubp8 active site. The Sgf11-ZnF makes extensive hydrogen bonding interactions with loop L2 in Ubp8, which contains the catalytic cysteine, C146, and the oxyanion hole residue, N141, (Figure 1B). Sgf11 also contacts active site loop L1 (Figure 1B), which contains residues (aa 235–237) that interact with the C-terminal tail of ubiquitin but is disordered in the absence of bound ubiquitin (Samara et al., 2010). These important interactions, coupled with the observation that deletion of the Sgf11-ZnF significantly reduces DUBm activity (Kohler et al., 2010), raised the possibility that the Sgf11-ZnF may directly affect enzymatic activity by helping to orient the active site residues in the proper conformation for catalysis.
We report here structural and solution studies aimed at determining the role that Sgf11 plays in activating Ubp8 within the DUBm. Based on comparisons with USP enzymes that do not require partner proteins for activity, we introduced gain-of-function mutations into Ubp8 that were designed to stabilize the structure of the active site and thus overcome the deleterious effect of deleting the Sgf11-ZnF. We were able to identify two substitutions in Ubp8 that partially restored DUBm activity in the absence of the Sgf11-ZnF. However, crystal structures of the DUBm lacking the Sgf11-ZnF and containing either wild-type or mutant Ubp8 revealed an unanticipated role for the Sgf11-ZnF in maintaining the structural integrity of the DUB module, which in turn is important for Ubp8 activation. We find that deleting the Sgf11-ZnF destabilizes the compact folding of the DUBm through its interactions with Ubp8 loop L2, which joins the USP and ZnF-UBP domains. In the absence of contact with the Sgf11-ZnF, the loop adopts an extended conformation that permits the Ubp8 ZnF-UBP domain to peel away from the remainder of the DUBm and participate in domain-swapping interactions with another DUBm. In the resulting domain-swapped DUBm dimer, some of the active site residues in Ubp8 are misaligned. Analytical ultracentrifugation studies show that one of the mutations that restore DUBm activity in the absence of Sgf11 stabilizes the monomeric, compact conformation of Ubp8, which in turn ensures the correct configuration of the active site. The second activating mutation only partially shifts the equilibrium towards the monomer and may play an additional role in organizing the active site. These results suggest an unanticipated mechanism for maintaining Ubp8 in an active state, whereby Sgf11 promotes the proper organization of the DUBm and compensates for the intrinsic flexibility of a key portion of Ubp8.
Results
Gain of function mutations in Ubp8 partially compensate for deletion of the Sgf11 zinc finger
Deletion of the Sgf11-ZnF significantly reduces DUBm activity (Figure 1C (Kohler et al., 2010)), with greater than 50-fold reduction in kcat/KM (Table 1). Since the Sgf11-ZnF contacts Ubp8 loop L1, which contacts the substrate ubiquitin, and loop L2, which contains catalytic residues (Figure 1B), we speculated that the decrease in Ubp8 enzymatic activity was due to the loss of stabilizing contacts between loops L1 and L2 and the Sgf11-ZnF. Using structural and sequence alignments with other USP enzymes that do not depend upon partner proteins for activity, we searched for residues that are conserved in the active site loops of other USP DUBs (Avvakumov et al., 2006; Hu et al., 2002, 2005; Renatus et al., 2006; Ye et al., 2011), but not in Ubp8, and could potentially account for the dependence of Ubp8 activity on Sgf11. The alignments revealed two serine residues in Ubp8, S144 and S149 that are typically asparagine in other USP DUBs (Figure 1D). Ubp8 residue S144, which is part of active site loop L2, is found in place of a conserved asparagine residue in hUSP8 that donates a hydrogen bond to the backbone carbonyl of Y864 on loop L1 of hUSP8 (Avvakumov et al., 2006). A similar interaction is found in other USP DUBs such as hUSP2, hUSP21, and hUSP14 (Hu et al., 2005; Renatus et al., 2006; Ye et al., 2011). Model building with the structure of the SAGA DUBm showed that substituting an asparagine for S144 in Ubp8 could give rise to an analogous hydrogen bond with the backbone carbonyl of the conserved tyrosine in Ubp8, Y233 (Figure 1E). Similarly, replacement of S149 in Ubp8 with an asparagine could potentially donate two hydrogen bonds to the backbone carbonyl groups of residues L139 and I140 in loop L2, which could stabilize loop L2 in the absence of the Sgf11-ZnF and thus potentially maintain enzymatic activity of the DUBm (Figure 1E).
Table 1.
Kinetic constants for wild-type and mutant DUBm cleavage of Ub-AMC See also Figure S1.
| DUB module mutant |
kcat (sec−1) | KM (µM) |
kcat/ KM (M−1 sec−1) |
Critical χ2 |
|---|---|---|---|---|
| WT | 4.4 ± 0.3 | 24 ± 2 | (1.8 ± 0.2) × 105 | 3.1 × 10−2 |
| S144N | 6.5 ± 0.3 | 28 ± 2 | (2.3 ± 0.2) × 105 | 2.0 × 10−2 |
| S149N | 3.1 ± 0.2 | 20 ± 2 | (1.6 ± 0.2) × 105 | 2.2 × 10−2 |
| ΔZnF | 0.063 ± 0.01 | 19 ± 1 | (3.3 ± 0.4) × 103 | 3.1 × 10−5 |
| ΔZnF S144N | 0.28 ± 0.02 | 17 ± 2 | (1.6 ± 0.2) × 104 | 4.5 × 10−4 |
| ΔZnF S149N | 0.26 ± 0.02 | 11 ± 1 | (2.4 ± 0.3) × 104 | 5.9 × 10−4 |
| ΔZnF S144N S149N | 0.47 ± 0.02 | 13 ± 1 | (3.6 ± 0.3) × 104 | 1.1 × 10−3 |
To test whether the presence of serine rather than asparagine at positions 144 and 149 was responsible for the dependence of Ubp8 activity on the Sgf11-ZnF, we asked whether S144N or S149N substitutions in Ubp8 could overcome the deleterious effects of an Sgf11-ZnF deletion. We introduced the single mutations, as well as the double mutation, into Ubp8 and purified DUBm complexes containing the mutated Ubp8 proteins, the truncated Sgf111–72 fragment lacking the C-terminal zinc finger (Sgf11-ΔZnF) and wild-type Sgf11 and Sus1. In cleavage assays using ubiquitin-AMC (Ub-AMC) as the substrate, we found that the S144N or S149N mutation in Ubp8 increases the activity of the SAGA DUBm in the absence of Sgf11-ZnF by 5- or 7-fold, respectively (Figure 1F and Table 1). The double substitution Ubp8S144NS149N has an additive effect on DUBm activity in Ub-AMC cleavage assays, increasing kcat/KM 11-fold (Figure 1F and Table 1). Notably, the Sgf11-ZnF deletion and the activating mutations largely affect kcat, with KM values differing by less than two-fold among all complexes tested, indicating that the mutations and deletions primarily affect catalysis and not substrate binding (Table 1). The Ubp8 S144N and S149N substitutions do not increase activity in the context of DUBm containing intact Sgf11 (Table I), indicating that these substitutions do not simply increase the intrinsic activity of Ubp8. To verify that the observed effects of the stabilizing mutations were not substrate-specific, DUBm activity was assayed using K48-linked di-ubiquitin, which can also be cleaved by the DUBm. The overall effect of the mutations on cleavage of K48-linked diubiquitin (Figure 1G) is similar to that observed in Ub-AMC assays (Figure 2D). These data are consistent with the hypothesis that Sgf11 compensates for the absence of conserved residues that are important for maintaining an active conformation of Ubp8.
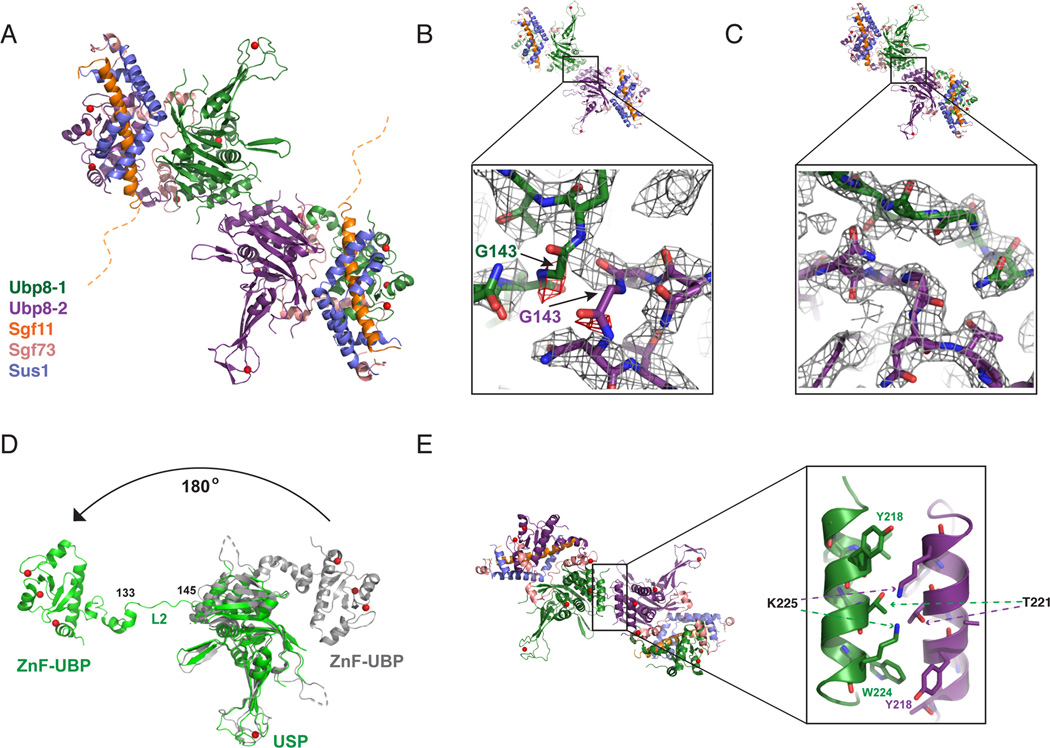
Figure 2. Structure of DUBm lacking the Sgf11-ZnF shows domain swapping.
(A) Structure of DUBm-ΔSgf11-ZnF/Ubp8-S144N shows domain swapping between two DUBm complexes. The two Ubp8 subunits in the complex are colored green and purple, respectively. Other subunits colored as indicated: Sus1 (blue), Sgf11 (orange) Sgf73 (salmon). The disordered Sgf11 linker is represented by a dashed orange line.
(B) Initial fit of two DUBm complexes to the electron density map before detection of domain swapping (2Fo-Fc contoured at 1σ, grey, Fo-Fc contoured at 3σ red).
(C) Fit of domain-swapped complexes to 2Fo-Fc electron density map.
(D) Conformation of Ubp8 in wild-type, intact DUBm (gray) and in domain-swapped complex (green). Residues 133–145 in loop L2 undergo a major conformational change that accompanies domain swapping.
(E) Helix in Ubp8 that is buried by the Sgf11-ZnF in the intact DUBm mediates dimerization contacts in the domain-swapped DUBm-ΔSgf11-ZnF.
Deletion of the Sgf11 zinc finger results in a domain-swapped DUB module dimer
To gain insight into the role of Sgf11 in maintaining the active conformation of Ubp8, we determined structures of the DUBm lacking the zinc finger of Sgf11, and containing either wild-type Ubp8 or the activating mutant Ubp8S144N. Both complexes crystallize in space group P212121 with 2 complexes in the asymmetric unit that are related by a non-crystallographic two-fold axis. The structures of DUBm-ΔSgf11-ZnF-Ubp8WT and DUBm-ΔSgf11-ZnF-Ubp8S144N were determined by molecular replacement using as a search model the coordinates of the DUBm (PDB ID: 3MHS, (Samara et al., 2010)), and refined to 2.8 Å and 2.7 Å resolution, respectively. Data and refinement statistics are given in Table 2.
Table 2.
X-ray crystallographic data and refinement statistics
| Data Collection Statistics | DUBm-ΔSgf11-ZnF/ Ubp8WT |
DUBm-ΔSgf11-ZnF/ Ubp8S144N |
DUBm-Ubp8S144N |
|---|---|---|---|
| Energy (eV) | 12000 | 12000 | 12658 |
| Resolution limit (A) | 2.8 | 2.7 | 2.03 |
| Unique reflections | 36287 | 39990 | 61,049 |
| Redundancy | 10.9 (6.0) | 7.8 (6.2) | 14.8 (14.7) |
| Completeness (%) | 96.7 (94.7) | 91.9 (84.9) | 100 (100) |
| Average I/s (I) | 10.8 (1.2) | 12.2 (2.3) | 28.8 (4.73) |
| Rsym (%) | 21.7 (93.0) | 15.1 (63.7) | 7.0 (88) |
| Refinement Statistics | |||
| Space group and unit cell (Å) | P212121 a=76.5 b=79.4 c=267 |
P212121 a=76.0 b=80.0 c=274 |
P212121 a=83.6 b=105 c=107 |
| Molecules per asymmetric unit | 2 | 2 | 1 |
| Rwork (%) | 20.8 | 20.0 | 18.8 |
| Rfree (%) | 28.8 | 27.6 | 23 |
| Rmsd bonds (Å) | 0.013 | 0.015 | 0.024 |
| Rmsd angles (o) | 1.386 | 1.205 | 1.89 |
| Protein atoms | 10558 | 10668 | 5,709 |
| Water molecules | 49 | 48 | 437 |
| Zinc (II) ions | 14 | 14 | 7 |
| Average B (Å2) | 18.8 | 22.7 | 16.0 |
While the overall architecture of each complex appears similar to that of the WT DUBm, we unexpectedly found that the two complexes participate in domain swapping interactions, with the N-terminal ZnF-UBP domain incorporated into one DUBm while the USP domain is incorporated into the second complex (Figure 2A). The domain swapping was initially noted in electron density maps by the absence of density for loop L2 residue G143 and the presence of unaccounted-for density bridging the two complexes (Figures 2B and 2C). The domain swapping is the result of a dramatic conformational change in loop L2 residues 133–145, which peel away from the USP domain and bridge the junction between opposing complexes (Figure 2D). The repositioned Ubp8 ZnF-UBP domain is related to its position in the wild-type structure by a 180° rotation (Figure 2D). Only one of the cross-over loops is ordered in the DUBm-ΔSgf11-ZnF complex containing wild-type Ubp8, while both loops are ordered in the DUBm-ΔSgf11-ZnF-Ubp8S144N complex (Figure 3A and 3B), With the exception of the marked changes in the residues connecting the two domains of Ubp8, the wild-type DUBm superposes with each domain-swapped complex with an RMSD of about 1.3 Å. In addition to the tethering of the two complexes via the loop L2 residues, the two Ubp8 USP domains contact one another with a helix (residues 214–226) that is partially buried by the Sgf11-ZnF in the intact DUBm (Figure 2E).
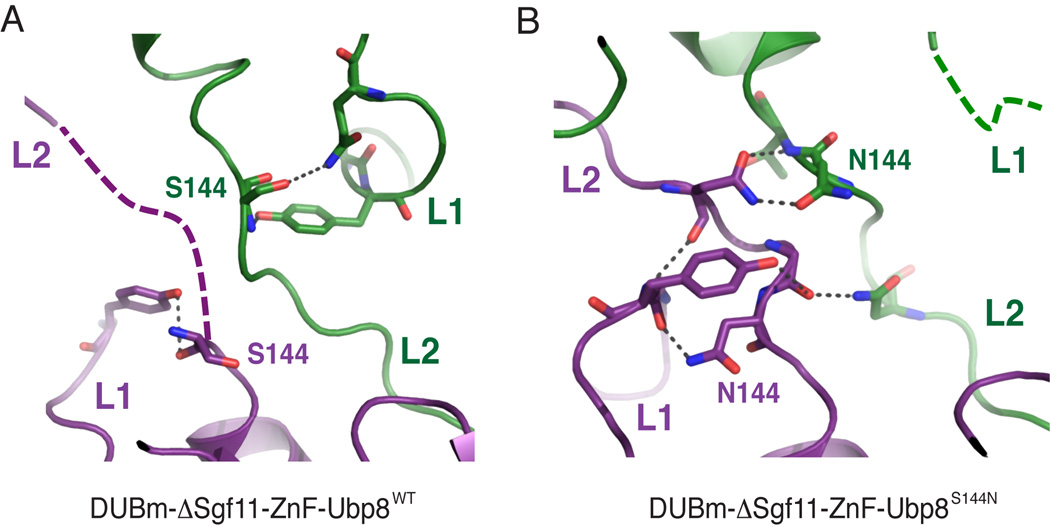
Figure 3. Ubp8 loops L1 and L2 in the domain-swapped structures.
(A) Detailed view of Ubp8 crossover loops (L2) and loop L1 in the domain-swapped DUBmΔSgf11-ZnF-Ubp8WT structure. Respective Ubp8 monomers are colored purple and green.
(B) View as in (A) of the DUBm-ΔSgf11-ZnF-Ubp8S144N complex. See also Tables S1 and S2.
The extended linker (residues 46–72) that connects the N-terminal helix of Sgf11 to its zinc finger in the wild-type complex (Figure 1A) is disordered in the ΔZnF structures, indicating that the zinc finger is required to anchor the flexible linker to the surface of the USP domain of Ubp8 (Figure 2A). Loop L1 of Ubp8, which is disordered in the intact apo DUBm complex (Samara et al., 2010; Kohler et al., 2010), and becomes ordered upon contacting the C-terminal tail of ubiquitin (Samara et al., 2010), is unexpectedly well-ordered in both molecules in the DUBm-ΔSgf11-ZnF complex. The ordering of loop L1 appears to be due to stabilizing contacts with S144 in loop L2, which are made possible by the conformational change in loop L2 (Figure 3A). Loop L1 is ordered in just one molecule of the DUBm-ΔSgf11-ZnF-Ubp8S144N complex as a result of hydrogen bonding interactions with N144 in the opposing loop L2 (Figure 3B). The N144 residue in the second Ubp8 protein adopts a different conformation and forms hydrogen bonds that bridge the L2 loops of the two opposing Ubp8 proteins (Figure 3B). In the absence of contacts with N144, loop L1 of the second Ubp8 molecule becomes disordered (Figure 3B).
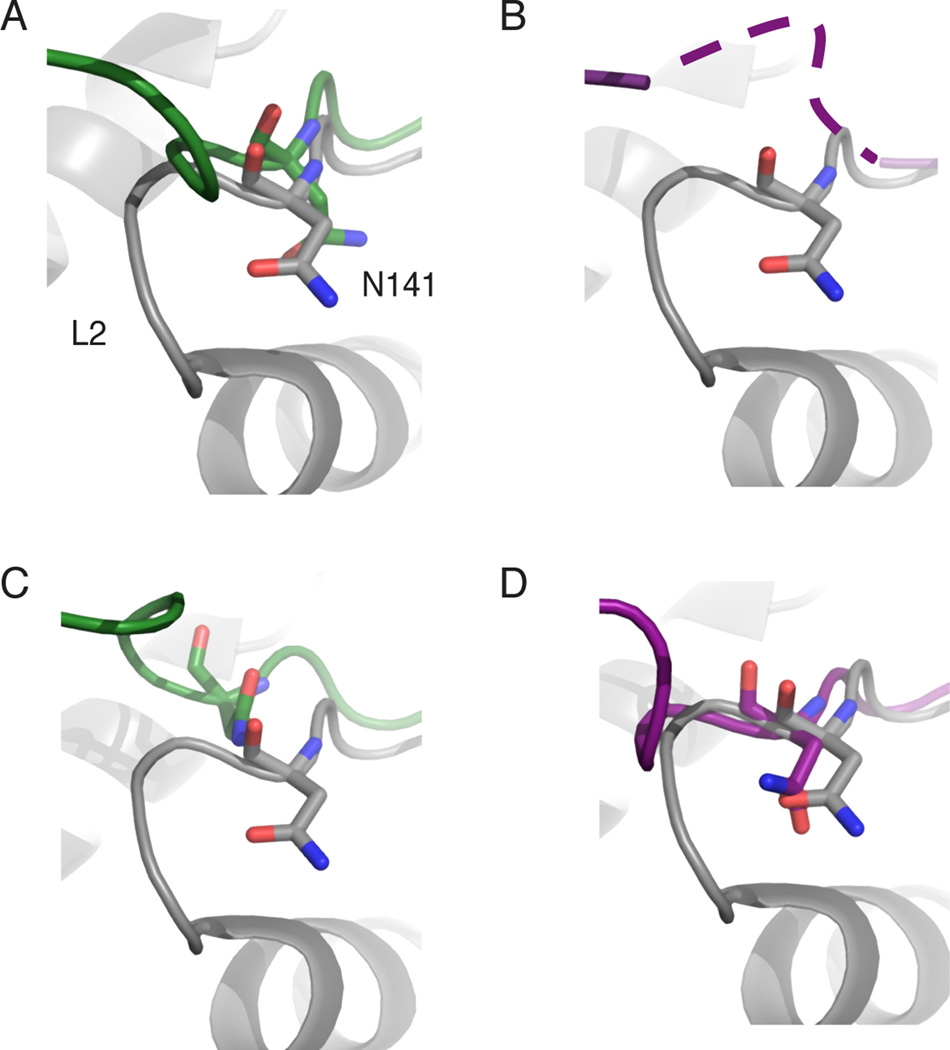
The structures show that the dramatic alteration of the loop L2 conformation that accompanies domain swapping changes the alignment of active site residues, potentially accounting for the loss of activity in these versions of the DUBm. The predicted oxyanion hole residue, N141, is located in loop L2 and is thus repositioned as a result of domain swapping (Figure 4A–D). As a consequence, N141 from one Ubp8 molecule is either incorrectly positioned in the active site of the second Ubp8 molecule relative to its position in the wild-type DUBm or disordered (Figure 4A–D). In either case, the residue is not in an orientation to interact with the oxyanion intermediate that forms during catalysis.
Figure 4. Effect of domain swapping on the oxyanion hole residue, N141.
(A) Superposition of one Ubp8 monomer in the domain-swapped DUBm complex lacking the Sgf11-ZnF (green) with Ubp8 in intact DUBm complex (gray) showing altered position of N141.
(B) Superposition as in (A) showing the second Ubp8 monomer (purple) in the domain-swapped complex in which N141 disordered.
(C, D) Superpositions as in (A) showing Ubp8 in the domain-swapped DUBm-ΔSgf11-ZnF complex containing Ubp8S144N. Coloring as in (A)
To confirm that the repositioning of loop L2 and misalignment of N141 occurs because of the absence of the Sgf11-ZnF and not because of the S144N mutation, we determined the structure of DUBm-apo with full-length Sgf11 and Ubp8S144N to 2.0 Å resolution with an R/Rfree of 18.9%/22.7% (Table 2). The structure of the DUBm containing Ubp8S144N is virtually identical to that of the wild-type apo DUBm structure, including the active site conformation and the disorder of loop L1 (Supplementary Figure S2).
The DUBm-Sgf11(ΔZnF)/Ubp8WT and DUBm-Sgf11(ΔZnF)/Ubp8S144N complexes are dimers in solution
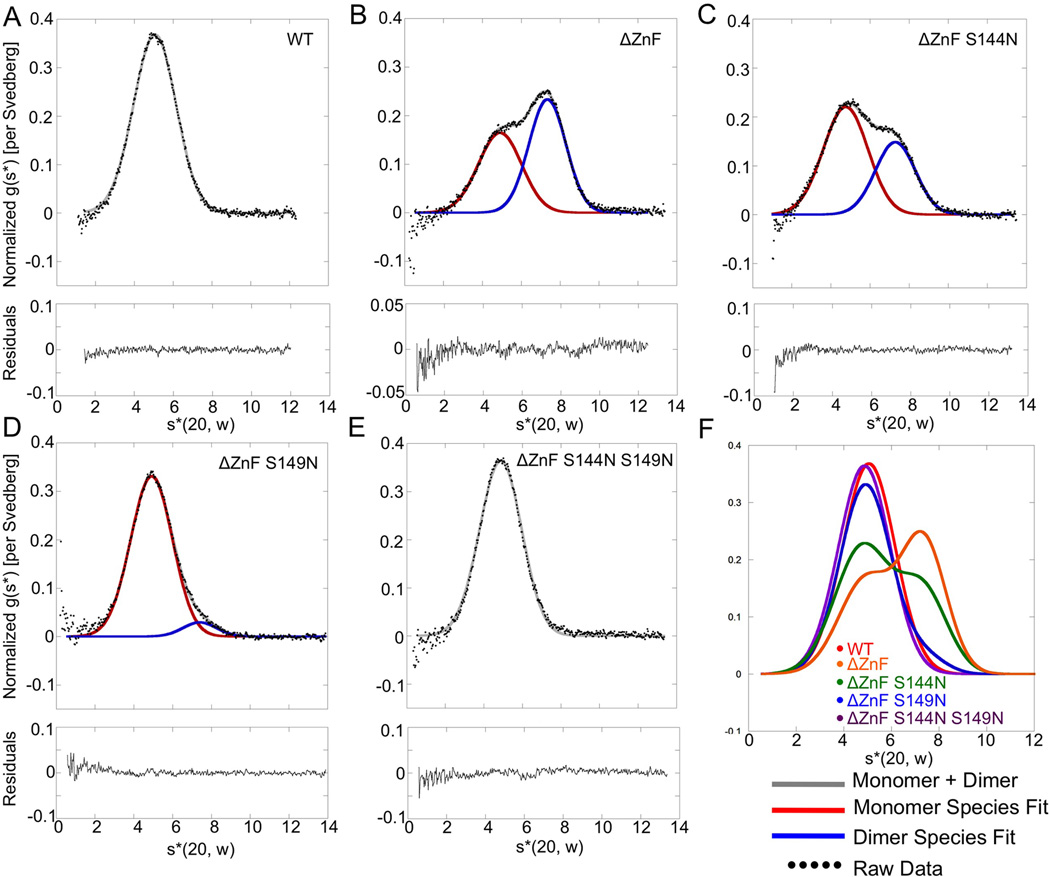
To test whether the domain-swapped dimerization of the DUBm occurs in solution, we used analytical ultracentrifugation to assess the dimerization state of the Sgf11-ZnF deletion mutants. Sedimentation velocity (Figure 5) and equilibrium sedimentation (Supplementary Figure S3) experiments show that the intact DUBm behaves as a monomer (Figure 5A), while DUBm lacking the Sgf11-ZnF (DUBm-ΔSgf11-ZnF-Ubp8WT) exists in a monomer-dimer equilibrium (Figure 5B) with a Kd around 17 µM (Supplementary Figure S3, Table S1 and S2). The proportion of monomer is somewhat higher when Ubp8 contains the S144N substitution (Fig. 5C), with a Kd for the monomer-dimer equilibrium of 28 µM (Supplementary Figure S3, Table S1 and S2). By contrast, the S149N substitution alone and the double S144N/ S149N substitution in Ubp8 restore the monomeric behavior of DUBm complexes lacking the Sgf11 zinc finger: DUBm-ΔSgf11-ZnF-Ubp8S149N and DUBm-ΔSgf11-ZnF-Ubp8S144N/S149N (Figures 5D–F, Supplementary Figure S3). The monomeric states adopted by the DUBm lacking the Sgf11-ZnF have a molecular weight and sedimentation coefficient similar to the monomeric wild-type DUBm, suggesting that their shape is similar to that of the intact wild-type DUBm in solution. Overall, the analytical ultracentrifugation studies show that the domain-swapped dimer that is observed in the crystal forms in solution and is in equilibrium with monomeric DUBm. Taken together, our results indicate that deletion of the Sgf11-ZnF destabilizes the association of the Ubp8 catalytic domain with the remainder of the DUBm, thus disrupting enzymatic activity, and that activating mutations in Ubp8 partially restore activity by stabilizing the properly folded, monomeric DUBm.
Figure 5. Assays of wild-type and mutant DUBm oligomerization by velocity sedimentation.
(A) Normalized g(s*) analysis for sedimentation of intact wild-type DUBm. Data are fit to a single Gaussian. Residuals shown below.
(B) Normalized g(s*) analysis for DUBm-ΔSgf11-ZnF-Ubp8WT. Black dots indicate raw data, which were fit to a sum of two Gaussians (gray) corresponding to sedimentation of monomeric (red) and dimeric (blue) DUBm.
(C) Analysis as in (B) of DUBm-ΔSgf11-ZnF-Ubp8S144N
(D) Analysis as in (B) of DUBm-ΔSgf11-ZnF-Ubp8S149N
(E) Analysis as in (B) of DUBm-ΔSgf11-ZnF-Ubp8S144N/S149N
(F) Superposition of fitted curved from panels A–F; color key in panel inset. See also Figure S3.
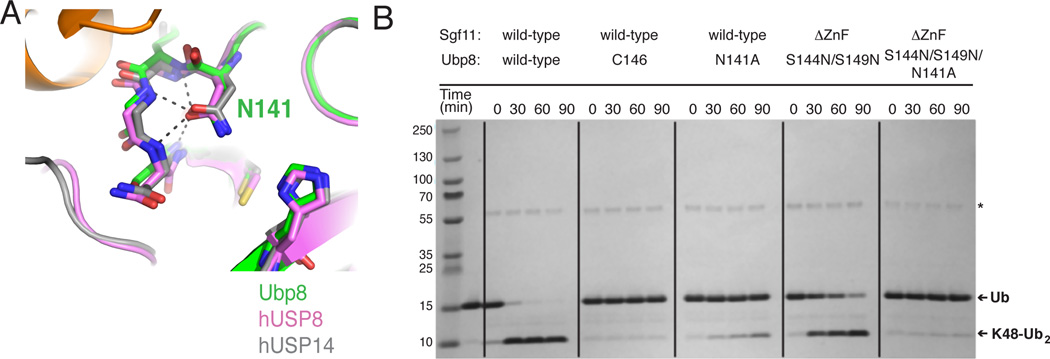
Loop 2 residue N141 is important for catalysis
The activating S144N and S149N substitutions in Ubp8 both lie near the asparagine residue, N141 that has been proposed to stabilize the oxyanion intermediate in USP isopeptidases (Hu et al., 2002; W. Zhang et al., 2011). This residue adopts virtually the identical conformation in the structure of Ubp8 in the intact DUB module, as well as in structures of the human USP enzymes, USP8 (Avvakumov et al., 2006) and USP14 (Hu et al., 2005), (Figure 6A). In the structure of Ubp8 in the intact DUBm, the backbone amide groups in loop L2 position N141 by donating four hydrogen bonds to its amide carbonyl (Figure 6A) thus orienting the amino group of N141 to stabilize the oxyanion intermediate. To test whether the activating mutations correctly reconstitute the Ubp8 active site, we compared the effects of an N141A substitution in both wild-type, intact DUBm and in DUBm-ΔSgf11-ZnF-Ubp8S144N/S149N. Assays of K48 di-ubiquitin cleavage indicate that the N141A substitution has a comparable effect in both wild-type and mutant complexes (Figure 6B). Activity is significantly decreased in both, although not to the extent of mutation of the active site cysteine, DUBm-Ubp8C146A, which abrogates activity (Figure 6B). The similar effects of the Ubp8-N141 substitution on both the wild-type DUBm and activated deletion mutant, DUBm-ΔSgf11-ZnF-Ubp8S144N/S149N is consistent with the idea that the N141 side chain is positioned similarly in both complexes. It is thus likely that N141 forms wild-type backbone interactions with loop L2 residues 142–145 as seen in the monomeric DUBm (Figure 6A), suggesting that loop L2 has been stabilized by both asparagine mutations.
Figure 6. Ubp8-N141 is important for DUBm catalysis.
(A) Superpositions showing hydrogen bonding of conserved oxyanion hole residue (N141 in Ubp8) to backbone amides. Apo-Ubp8 (2MHH, gray), hUSP8 (2GFO, pink), and hUSP14 (2AYN, gray).
(B) Effect of Ubp8 active site mutations on DUBm activity Gel shows time course for cleavage of K48-linked diubiquitin for the intact DUBm-WT, DUBm-Ubp8C146A, DUBm-Ubp8N141A, DUBm-ΔSgf11-ZnF-Ubp8S144N/S149N, and DUBm-ΔSgf11-ZnF-Ubp8S144N/S149N/N141A.
Discussion
There are now a number of examples of deubiquitinating enzymes that depend upon interactions with partner proteins for full activity (Cohn et al., 2009, 2007; Kee et al., 2010; Scheuermann et al., 2010; Ventii & Wilkinson, 2008), but the mechanisms by which DUB activity is potentiated has been unclear. In this study, we sought a structural explanation for the observation that deletion of the Sgf11-ZnF domain, which binds immediately adjacent to the Ubp8 active site, significantly disrupts the activity of the SAGA DUB module. The effect of the Sgf11-ZnF deletion in our assays is on kcat (Table 1), indicating that Sgf11 plays a role in organizing the active site rather than in promoting binding of the substrate ubiquitin. We find that the Sgf11-ZnF domain plays an unexpected role in maintaining the catalytic activity of Ubp8 by ensuring the proper organization and folding of the four-protein complex. Our structural and biophysical studies show that the absence of the Sgf11-ZnF destabilizes the incorporation of Ubp8 into the DUBm, causing the ZnF-UBP domain of Ubp8 to peel away from the DUBm and adopt an extended conformation that promotes domain swapping and dimerization. The domain swapping arises from a dramatic change in the conformation of residues 133–145, which are located in the loop L2 that is contacted by the Sgf11-ZnF in the intact DUBm. The result of this conformational change is a displacement of N141, a residue that is important for stabilizing the oxyanion reaction intermediate. Gain-of-function mutations S144N, S149N and S144N/S149N in the active site region of Ubp8 increased DUBm activity in the absence of the Sgf11-ZnF. The S149N mutation in particular favors the compact, monomeric form of the DUBm, while the S144N partially shifts the equilibrium towards the monomer, thus accounting at least in part for the activating effect of these mutations. The role of the Sgf11-ZnF in stabilizing the conformation of loop L2, and thus the active site, provides a potential model for the role that other proteins, such as UAF1, may play in increasing the catalytic rate of other USP class deubiquitinating enzymes (Faesen et al., 2011; Villamil et al., 2012)
While the observed displacement of the oxyanion residue N141 is most dramatic in the domain-swapped dimer, destabilization of loop L2 alone would be sufficient to alter the position of N141 and thus impact catalysis. The concentration of Ubp8 in yeast, and presumably that of the SAGA DUBm, is roughly 0.1 µM (Ghaemmaghami et al., 2003), although the effective concentration of DUBm in the nucleus, and in light of molecular crowding, is likely significantly higher. While the proportion of domain-swapped dimer versus destabilized monomer is thus difficult to predict, either form of the mutant DUBm would have decreases catalytic efficiency.
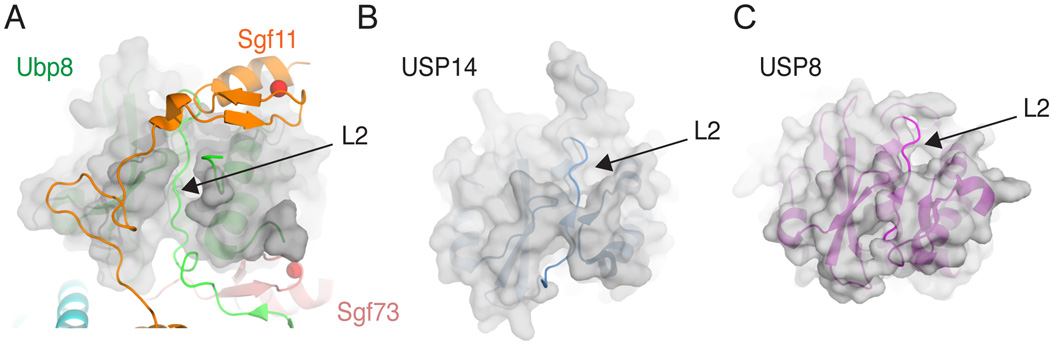
A comparison of Ubp8 to other USP family enzymes shows that Ubp8 lacks structural features that stabilize the active conformation of loop L2 in other DUBs. Ubp8 therefore depends upon interactions with the Sgf11-ZnF to stabilize loop L2 and thereby maintain the enzyme in an active conformation. Monomeric USP family DUBs such as USP8 (Avvakumov et al., 2006) and USP14 (Hu et al., 2005), whose active site residues are in a catalytically competent configuration in the absence of substrate, have additional structural features that partly bury residues in loop L2, further stabilizing its association with the USP domain. In Ubp8, residues 136–139 are partly solvent-exposed on the Ubp8 surface, while the corresponding residues are buried in USP8 and USP14 (Figure 7). Burial of these residues thus stabilizes the association of this strand with the USP domain, which in turn keeps the oxyanion hole asparagine residue (N141 in Ubp8) in position for catalysis. In these enzymes, a protein such as Sgf11 is not required to maintain the conformation of the L2 loop. In addition, the asparagine corresponding to Ubp8 residue S149 forms hydrogen bonds and van der Waals interactions with loop L2 that favor the position of the loop seen in the intact, monomeric DUBm (Figure 1E). It is not clear what features of the USP domain may stabilize this loop in USP22, the human homologue of yeast Ubp8. While USP22 contains an asparagine in place of Ubp8 residue S149, which is expected to help stabilize loop L2, it may similarly lack features that bury loop L2 in other USP domains. While the lack of sequence conservation in this region of USP domains makes it difficult to make structural predictions, we speculate that loop L2 of USP22 similarly depends upon interactions with the Sgf11 homologue, ATXN7L3, to adopt a catalytically competent conformation.
Figure 7. Ubp8 lacks conserved structural elements that could stabilize loop L2.
(A) View of intact DUBm with transparent surface depicted for all Ubp8 (green) USP domain residues except loop L2 in Ubp8 (green). Interactions with the Sgf11 (orange) and Sgf73 (pink) zinc fingers help to maintain the observed conformation of loop L2.
(B) View of USP14 (2AYN, blue) with transparent surface shown for all residues C-terminal to loop L2. Additional residues in the USP domain bury part of loop L2 and form stabilizing beta sheet interactions.
(C) View of hUSP8 (2GFO, magenta) with transparent surface shown for all residues excluding loop L2.
The S144N substitution was predicted to hydrogen bond with loop L1 of Ubp8, which is disordered in the apo DUBm but contacts both the C-terminal tail of ubiquitin and the Sgf11-ZnF in the complex with ubiquitin aldehyde (Samara et al., 2010). This loop is analogous to the “switching loop” of USP7/HAUSP, which undergoes a substrate-induced conformational change (Hu et al., 2002) and mediates allosteric activation of HAUSP (Faesen et al., 2011). Loop L1 was not, however, ordered in the structure of the intact DUBm containing the Ubp8-S144N mutant (Figure S1) or in the domain-swapped dimer (Figure 3B). It therefore seems unlikely that this loop plays a role in Ubp8 activation analogous to that in USP7 (Faesen et al., 2011). It remains possible that this residue could form stabilizing interactions within the monomeric form of the DUBm lacking the Sgf11-ZnF. The modest effect of this mutation on shifting the equilibrium towards monomeric DUBm lacking the Sgf11-ZnF as compared to S149N suggests that the S144N substitution makes a further contribution to activity that remains to be determined.
While our studies revealed an unexpected role for Sgf11 in maintaining the overall folding of the DUBm into a compact complex, it is possible that the Sgf11-ZnF also contributes to Ubp8 activity by stabilizing the catalytically competent configuration of the active site through interactions with loops L2. We were unable to obtain crystals of the monomeric form of the DUBm lacking the Sgf11-ZnF and containing the Ubp8 S149N mutation or the S149N/S144N mutations, so this question could not be addressed directly in our study. The activated double mutant is still 10-fold less active than the wild-type intact DUBm, indicating that Sgf11 makes an additional contribution to full enzymatic activity besides its role in maintaining the properly folded monomeric DUBm.
The DUBm subunit, Sgf73, may similarly play a dual role in maintaining the correctly folded conformation of the complex, as well as contributing in other ways to activation. A portion of Sgf73, including a zinc finger, lies between the two lobes of the DUBm, while the remainder forms part of the ZnF-UBP (assembly) lobe. Small deletions of Sgf73 that remove the Sgf73 zinc finger disrupt the incorporation of Sgf73 into the DUBm and completely abrogate enzymatic activity (Kohler et al., 2008). Without Sgf73 to hold the two lobes of the DUBm together, it is possible that the two Ubp8 domains separate via conformational changes in the linker including loop L2 that joins the two Ubp8 domains, as has been suggested (Kohler et al., 2010). This could both transmit conformational changes to the active site and potentially disrupt the association of the Sgf11-ZnF as well. Indeed, the Sgf73 zinc finger contacts loop L2 directly (Figure 7A), and could thereby play a role in stabilizing the compact form of the DUBm. As with Sgf11, the binding of Sgf73 to the USP domain likely makes further contributions to enzymatic activity, possibly by affecting the folding of the ubiquitin binding pocket or contributing in other ways to the integrity of the catalytic domain.
What is the biological advantage of the dependence of Ubp8 activity on its incorporation into the SAGA DUBm, a property that is shared by the human homologue, USP22 (Kohler et al., 2010; Lang et al., 2011)? The deubiquitination of H2B-Ub is tightly regulated during the transcription cycle by the SAGA complex, which is recruited to the actively transcribed gene and thus targets Ubp8 to the appropriate location. The dependence of Ubp8 on the other SAGA subunits mitigates against non-specific deubiquitination of other substrates, including poly-ubiquitin chains. In addition to its role in DUBm assembly that we have demonstrated in this study, Sgf11 is thought to facilitate interactions with the nucleosome via a cluster of conserved arginine residues in the Sgf11-ZnF, which form a basic patch that can interact with the negatively charged DNA or the conserved acidic patch on H2A-H2B dimers (Bonnet et al., 2010; Makde, England, Yennawar, & Tan, 2010; Samara et al., 2010). Similarly, Sgf73 (Kohler et al., 2008; Lee et al., 2009) and its human homologue ATXN7 (Bonnet et al., 2010) have also been implicated in binding to nucleosomes, an interaction that is required for in vivo DUBm activity (Bonnet et al., 2010). Thus, both Sgf11 and Sgf73 have dual roles in coupling activity to substrate recognition and specificity.
The importance of the positioning of loop L2 in Ubp8 is an “Achilles heel” of the USP domain that could be exploited in drug design. Small molecules that either bind within the loop L2 binding groove or stabilize the active conformation of loop L2 could function as inhibitors or activators, respectively. Since the residues joining helices 214 and 226 of the USP domain that abut the loop L2 binding groove are not well-conserved among USP enzymes, these molecules could in principle be specific for particular deubiquitinating enzymes, thus serving as useful tools in cell-based assays and potentially as therapeutic agents.
Experimental Procedures
Protein expression and purification
Rosetta 2(DE3)pLysS (EMD Millipore, Merck KGaA, Darmstadt, Germany) cells were transformed with three plasmids encoding 1) Ubp8WT, Ubp8S144N, Ubp8N141A Ubp8S149, Ubp8S144NS149N, or Ubp8S144NS149NN141A (pET-32a, EMD Millipore), 2) Sus1 (pRSF-1, EMD Millipore), and 3) Sgf11WT or Sgf11ΔZnF (1–72) (pCDFDuet-1-MCSI, EMD Millipore), which was cloned into the same vector as Sgf731–96 (pCDFDuet-1-MCSII, EMD Millipore). All versions of the DUBm complex were co-expressed and purified using the previously published protocol for the expression and purification of wild-type DUBm (Samara et al., 2010).
Protein crystallization
Crystals were grown by the hanging drop method in VDX48 plates with sealant (Hampton Research, Aliso Viejo, CA). DUBm-ΔSgf11-ZnF-Ubp8S144N and DUBm-ΔSgf11-ZnF-Ubp8WT crystals grew in 14% PEG3350, 0.18–0.22 M tri-ammonium citrate, and 8–14% glycerol using a 1:1 ratio of protein to well solution. Crystals formed within 3 days at 18°C and were flash-frozen in liquid nitrogen in the presence of mother liquor supplemented with 20% glycerol as a cryo-protectant. The crystals grew to a size of about 50 µm × 500 µm.
Data collection, structure determination and refinement
Diffraction data from DUBm-ΔSgf11-ZnF-Ubp8WT and DUBm-ΔSgf11-ZnF-Ubp8S144N crystals were collected at the GM/CA CAT beamline (23ID-B) at the Advanced Photon Source. Crystals were frozen in cryo-protectants and stored in pucks that were loaded into a sample exchange robot (ALS/Berkeley style), and data were collected on a MARMosiac 300 CCD detector. The structure of each complex was determined by molecular replacement with MolRep (Vagin & Teplyakov, 1997) using the coordinates of the wild-type DUBm as the search model (PDB ID: 3MHS). The DUBm-ΔSgf11-ZnF-Ubp8WT and DUBm-ΔSgf11-ZnF-Ubp8S144N structures were refined with PHENIX (Adams et al., 2002) and Refmac (Murshudov, Vagin, & Dodson, 1997). Coot (Emsley, Lohkamp, Scott, & Cowtan, 2010) was used for manual model building. The DUBm-ΔSgf11-ZnF-Ubp8WT complex contains 1307 ordered residues, and the DUBm-ΔSgf11-ZnF-Ubp8S144N complex contains 1289 ordered residues out of a total of 1478 in the crystal and 14 zinc atoms. Pymol (DeLano, 2002) was used to generate all structure figures.
K48-di-ubiquitin assays
K48-diubiquitin stock solution was calculated to be 1.45 mM by absorbance at λ= 280 nm using an εdi-Ub = 2980 M−1cm−1. Di-ubiquitin was diluted with DUBm assay buffer to yield a final reaction concentration of either 40 µM or 100 µM in a reaction volume of 50 µL. A 2.5 µL volume of enzyme at 5.8 µM (0.5 mg/mL) or 0.58 µM (0.05 mg/mL) was added at 25 °C, and 47.5 µL of di-ubiquitin was added and mixed by pipetting 10 times. A 10 µL sample was removed for the 0 min time-point that was quenched with 5 µL LDS buffer. Similar samples were obtained for 30 min, 60 min and 120 min. 7.5 µL of each sample was loaded into a 4–12% gradient Bis-Tris NU-PAGE gel (Invitrogen, Grand Island, NY) for SDS-PAGE, and the gels were stained with Coomassie Brilliant Blue.
Enzyme kinetics
Steady-state kinetic measurements were performed using ubiquitin-AMC dissolved in 100% DMSO (Boston Biochem, Cambridge, MA) as the substrate. Deubiquitinating (DUB) assays were performed in 384-well black polystyrene microplates at 30°C in a POLARstar Omega plate reader (BMG Labtech, Cary, NC) using an excitation wavelength of 385 nm and emission wavelength of 460 nm. DUB reactions were performed in assay buffer containing 150 mM NaCl, 5 mM DTT, 50 mM HEPES pH 7.6, 25 µM ZnCl2 and 7.5% DMSO. First, DUBm was titrated into a fixed concentration of Ub-AMC (2 µM) in order to find a concentration range where initial rate was independent of enzyme concentration (vi/ [Enzyme] = constant; data not shown). DUBm concentrations were determined spectrophotometrically using ε280 = 77,240 M−1 cm−1. For steady-state kinetic measurements, wild-type DUBm was held at a concentration of 25 nM, S144N at 25 nM, S149N at 20nM, ΔZnF at 400 nM, ΔZnFS144N at 400 nM, ΔZnFS149N at 300 nM, and ΔZnFS144N/ S149N at 150 nM. Ubiquitin-AMC concentration was varied between 0.23 and 17 µM, which kept the total amount of DMSO below 7.5%. Enzyme and assay buffer were mixed together in a total reaction volume of 30 µL, allowed to equilibrate at 30°C for ten minutes inside the plate reader, and then Ub-AMC added to initiate the reaction. The release of AMC was followed at 460 nm and the first 45–300 seconds of data were used to fit initial rates. In order to convert intensity units to concentration of Ub-AMC hydrolyzed, the reactions were allowed to incubate for 60 minutes and the total fluorescence measured at 30 second intervals for 5 minutes. When the total fluorescence was invariant with time, fluorescence versus [Ub-AMC] was plotted to generate a calibration curve.
Sedimentation velocity analytical ultracentrifugation
Sedimentation velocity experiments were performed in an An60Ti rotor at 40,000 rpm at 15°C in a Beckman-Coulter XL-I analytical ultracentrifuge. DUB module samples were dialyzed overnight at 4°C into buffer containing 150 mM NaCl, 20 mM HEPES pH 8.0, 5 mM DTT, and 20 µM ZnCl2 and then loaded into two-sector cells with 12 mm path length and sapphire windows using the dialysis buffer as the reference. All DUB module constructs were run at a concentration of 12.9 µM based on an extinction coefficient at 280 nm of 77,240 M−1 cm−1. Absorbance scans were collected at 280 nm on two-minute intervals with a 0.003 cm radial step, and a single sample was run per rotor. Normalized g(s*) data were analyzed using the time-derivative method implemented in the program DCDT+ version 2.3.2 (Philo, 2006) and fit either to a single Gaussian or two Gaussians, representing a single or two sedimenting species, respectively (Stafford, 1997). The same set of scans was used to analyze each data set. Molecular mass, species concentration, and sedimentation coefficient were fit for each sedimenting species. The data were converted to s*(20,w) using protein partial specific volumes and solvent viscosities calculated from the program SEDNTERP (Laue et al., 1992). These values, corrected for temperature, were also used for analysis of sedimentation equilibrium data.
Supplementary Material
Acknowledgements
We thank Mario Bianchet and Chris Berndsen for helpful comments and suggestions, and Anthony DiBello for providing di-ubiquitin chains and wild-type DUB module. Supported by NIH grant R01GM095822. Coordinates have been deposited in the Protein Data Bank with deposition numbers 4FK5, 4FJC and 4FIP.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Adams PD, Grosse-Kunstleve RW, Hung LW, Ioerger TR, McCoy AJ, Moriarty NW, Read RJ, Sacchettini JC, Sauter NK, Terwilliger TC. PHENIX: building new software for automated crystallographic structure determination. Acta Crystallogr. Sect. D: Biol. Crystallogr. 2002;58:1948–1954. doi: 10.1107/s0907444902016657. [DOI] [PubMed] [Google Scholar]
- Avvakumov GV, Walker JR, Xue S, Finerty PJ, Mackenzie F, Newman EM, Dhe-Paganon S. Amino-terminal dimerization, NRDP1-rhodanese interaction, and inhibited catalytic domain conformation of the ubiquitin-specific protease 8 (USP8) J. Biol. Chem. 2006;281:38061–38070. doi: 10.1074/jbc.M606704200. [DOI] [PubMed] [Google Scholar]
- Baker SP, Grant PA. The SAGA continues: expanding the cellular role of a transcriptional co-activator complex. Oncogene. 2007;26:5329–5340. doi: 10.1038/sj.onc.1210603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonnet J, Wang YH, Spedale G, Atkinson RA, Romier C, Hamiche A, Pijnappel WW, Timmers HT, Tora L, Devys D, et al. The structural plasticity of SCA7 domains defines their differential nucleosome-binding properties. EMBO Rep. 2010;11:612–618. doi: 10.1038/embor.2010.98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohn MA, Kee Y, Haas W, Gygi SP, D’Andrea AD. UAF1 is a subunit of multiple deubiquitinating enzyme complexes. J. Biol. Chem. 2009;284:5343–5351. doi: 10.1074/jbc.M808430200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohn MA, Kowal P, Yang K, Haas W, Huang TT, Gygi SP, D’Andrea AD. A UAF1-containing multisubunit protein complex regulates the Fanconi anemia pathway. Mol. Cell. 2007;28:786–797. doi: 10.1016/j.molcel.2007.09.031. [DOI] [PubMed] [Google Scholar]
- Daniel JA, Torok MS, Sun ZW, Schieltz D, Allis DC, Yates JR, Grant PA. Deubiquitination of histone H2B by a yeast acetyltransferase complex regulates transcription. J. Biol. Chem. 2004;279:1867–1871. doi: 10.1074/jbc.C300494200. [DOI] [PubMed] [Google Scholar]
- DeLano WL. The PyMOL Molecular Graphics System. 2002 [Google Scholar]
- Edelmann MJ, Iphöfer A, Akutsu M, Altun M, di Gleria K, Kramer HB, Fiebiger E, Dhe-Paganon S, Kessler BM. Structural basis and specificity of human otubain 1-mediated deubiquitination. Biochem. J. 2009;418:379–390. doi: 10.1042/BJ20081318. [DOI] [PubMed] [Google Scholar]
- Emsley P, Lohkamp B, Scott WG, Cowtan K. Features and development of Coot. Acta Crystallogr., Sect. D: Biol. Crystallogr. 2010;66:486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faesen AC, Dirac AMG, Shanmugham A, Ovaa H, Perrakis A, Sixma TK. Mechanism of USP7/HAUSP Activation by Its C-Terminal Ubiquitin-like Domain and Allosteric Regulation by GMP-Synthetase. Mol. Cell. 2011;44:147–159. doi: 10.1016/j.molcel.2011.06.034. [DOI] [PubMed] [Google Scholar]
- Ghaemmaghami S, Huh W, Bower K, Howson R, Belle A, Dephoure N, O’Shea E, Weissman J. Global analysis of protein expression in yeast. Nature. 2003;425:737–741. doi: 10.1038/nature02046. [DOI] [PubMed] [Google Scholar]
- Henry KW, Wyce A, Lo WS, Duggan LJ, Emre NCT, Kao CF, Pillus L, Shilatifard A, Osley MA, Berger SL. Transcriptional activation via sequential histone H2B ubiquitylation and deubiquitylation, mediated by SAGA-associated Ubp8. Genes Dev. 2003;17:2648–2663. doi: 10.1101/gad.1144003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu M, Li P, Li M, Li W, Yao T, Wu JW, Gu W, Cohen RE, Shi Y. Crystal structure of a UBP-family deubiquitinating enzyme in isolation and in complex with ubiquitin aldehyde. Cell. 2002;111:1041–1054. doi: 10.1016/s0092-8674(02)01199-6. [DOI] [PubMed] [Google Scholar]
- Hu M, Li P, Song L, Jeffrey PD, Chenova TA, Wilkinson KD, Cohen RE, Shi Y. Structure and mechanisms of the proteasome-associated deubiquitinating enzyme USP14. EMBO J. 2005;24:3747–3756. doi: 10.1038/sj.emboj.7600832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hussain S, Zhang Y, Galardy P. DUBs and cancer: The role of deubiquitinating enzymes as oncogenes, non-oncogenes and tumor suppressors. Cell Cycle. 2009;8:1688–1697. doi: 10.4161/cc.8.11.8739. [DOI] [PubMed] [Google Scholar]
- Hwang W, Venkatasubrahmanyam S, Ianculescu A, Tong A, Boone C, Madhani H. A conserved RING finger protein required for histone H2B monoubiquitination and cell size control. Mol. Cell. 2003;11:261–266. doi: 10.1016/s1097-2765(02)00826-2. [DOI] [PubMed] [Google Scholar]
- Ingvarsdottir K, Krogan NJ, Emre N, Wyce A, Thompson NJ, Emili A, Hughes TR, Greenblatt JF, Berger SL. H2B ubiquitin protease Ubp8 and Sgf11 constitute a discrete functional module within the Saccharomyces cerevisiae SAGA complex. Mol. Cell. Biol. 2005;25:1162–1172. doi: 10.1128/MCB.25.3.1162-1172.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kee Y, Yang K, Cohn MA, Haas W, Gygi SP, D’Andrea AD. WDR20 regulates activity of the USP12 x UAF1 deubiquitinating enzyme complex. J. Biol. Chem. 2010;285:11252–11257. doi: 10.1074/jbc.M109.095141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohler A, Schneider M, Cabal GG, Nehrbass U, Hurt E. Yeast Ataxin-7 links histone deubiquitination with gene gating and mRNA export. Nat. Cell Biol. 2008;10:707–715. doi: 10.1038/ncb1733. [DOI] [PubMed] [Google Scholar]
- Kohler A, Zimmerman E, Schneider M, Hurt E, Zheng N. Structural Basis for Assembly and Activation of the Heterotetrameric SAGA Histone H2B Deubiquitinase Module. Cell. 2010;8:1–12. doi: 10.1016/j.cell.2010.04.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komander D, Clague MJ, Urbé S. Breaking the chains: structure and function of the deubiquitinases. Nat. Rev. Mol. Cell Biol. 2009;10:550–563. doi: 10.1038/nrm2731. [DOI] [PubMed] [Google Scholar]
- Koutelou E, Hirsch CL, Dent SYR. Multiple faces of the SAGA complex. Curr. Opin. Cell Biol. 2010;22:374–382. doi: 10.1016/j.ceb.2010.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lang G, Bonnet J, Umlauf D, Karmodiya K, Koffler J, Stierle M, Devys D, Tora L. The tightly controlled deubiquitination activity of the human SAGA complex differentially modifies distinct gene regulatory elements. Mol. Cell. Biol. 2011;31:3734–3744. doi: 10.1128/MCB.05231-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laue TM, Shah BD, Ridgeway TM, Pelletier SL, Harding SE, Horton JC, Rowe AJ. In: Computer-aided interpretation of analytical sedimentation data for proteins. Harding S, Rowe A, Horton J, editors. Cambridge: Royal Society of Chemistry; 1992. [Google Scholar]
- Lee KK, Swanson SK, Florens L, Washburn MP, Workman JL. Yeast Sgf73/Ataxin-7 serves to anchor the deubiquitination module into both SAGA and Slik (SALSA) HAT complexes. Epigenetics Chromatin. 2009;2:1–12. doi: 10.1186/1756-8935-2-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makde RD, England JR, Yennawar HP, Tan S. Structure of RCC1 chromatin factor bound to the nucleosome core particle. Nature. 2010;467:562–566. doi: 10.1038/nature09321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murshudov G, Vagin A, Dodson E. Refinement of Macromolecular Structures by the Maximum-Likelihood Method. Acta Crystallogr., Sect. D: Biol. Crystallogr. 1997;53:240–255. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
- Philo JS. Improved methods for fitting sedimentation coefficient distributions derived by time-derivative techniques. Anal. Biochem. 2006;354:238–246. doi: 10.1016/j.ab.2006.04.053. [DOI] [PubMed] [Google Scholar]
- Renatus M, Parrado SG, D’Arcy A, Eidhoff U, Gerhartz B, Hassiepen U, Pierrat B, Riedl R, Vinzenz D, Worpenberg S, et al. Structural basis of ubiquitin recognition by the deubiquitinating protease USP2. Structure. 2006;14:1293–1302. doi: 10.1016/j.str.2006.06.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reyes-Turcu FE, Horton JR, Mullally JE, Heroux A, Cheng X, Wilkinson KD. The ubiquitin binding domain ZnF UBP recognizes the C-terminal diglycine motif of unanchored ubiquitin. Cell. 2006;124:1197–1208. doi: 10.1016/j.cell.2006.02.038. [DOI] [PubMed] [Google Scholar]
- Reyes-Turcu FE, Wilkinson KD. Polyubiquitin binding and dissassembly by deubiquitinating enzymes. Chem Rev. 2010;109:1495–1508. doi: 10.1021/cr800470j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodríguez-Navarro S. Insights into SAGA function during gene expression. EMBO Rep. 2009;10:843–850. doi: 10.1038/embor.2009.168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samara NL, Datta AB, Berndsen CE, Zhang X, Yao T, Cohen RE, Wolberger C. Structural insights into the assembly and function of the SAGA deubiquitinating module. Science. 2010;328:1025–1029. doi: 10.1126/science.1190049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheuermann JC, de Ayala Alonso AG, Oktaba K, Ly-Hartig N, McGinty RK, Fraterman S, Wilm M, Muir TW, Müller J. Histone H2A deubiquitinase activity of the Polycomb repressive complex PR-DUB. Nature. 2010;465:243–247. doi: 10.1038/nature08966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sowa ME, Bennett EJ, Gygi SP. Defining the human deubiquitinating enzyme interaction landscape. Cell. 2009;138:389–403. doi: 10.1016/j.cell.2009.04.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stafford WF. Sedimentation velocity spins a new weave for an old fabric. Curr. Pharm. Biotechnol. 1997;8:14–24. doi: 10.1016/s0958-1669(97)80152-8. [DOI] [PubMed] [Google Scholar]
- Vagin A, Teplyakov A. MOLREP: an automated program for molecular replacement. J. Appl. Cryst. 1997;30:1022–1025. [Google Scholar]
- Ventii KH, Wilkinson KD. Protein partners of deubiquitinating enzymes. Biochem. J. 2008;414:161–175. doi: 10.1042/BJ20080798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villamil M, Chen J, Liang Q, Zhuang Z. A noncanonical cysteine protease USP1 is activated through active site modulation by USP1-associated factor 1. Biochemistry. 2012;51:2829–2839. doi: 10.1021/bi3000512. [DOI] [PubMed] [Google Scholar]
- Weake VM, Workman JL. Inducible gene expression: diverse regulatory mechanisms. Nat. Rev. Genet. 2010;11:426–437. doi: 10.1038/nrg2781. [DOI] [PubMed] [Google Scholar]
- Wiener R, Zhang X, Wang T, Wolberger C. The mechanism of OTUB1-mediated inhibition of ubiquitination. Nature. 2012;483:618–622. doi: 10.1038/nature10911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wyce A, Xiao T, Whelan KA, Kosman C, Walter W, Eick D, Hughes TR, Krogan NJ, Strahl BD, Berger SL. H2B ubiquitylation acts as a barrier to Ctk1 nucleosomal recruitment prior to removal by Ubp8 within a SAGA-related complex. Mol. Cell. 2007;27:275–288. doi: 10.1016/j.molcel.2007.01.035. [DOI] [PubMed] [Google Scholar]
- Ye Y, Akutsu M, Reyes-Turcu F, Enchev RI, Wilkinson KD, Komander D. Polyubiquitin binding and cross-reactivity in the USP domain deubiquitinase USP21. EMBO Rep. 2011;12:0–7. doi: 10.1038/embor.2011.17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang W, Sulea T, Tao L, Cui Q, Purisima EO, Vongsamphanh R, Lachance P, Lytvyn V, Qi H, Li Y, et al. Insights into catalysis by ubiquitin specific proteases. Biochemistry. 2011;50:4775–4785. doi: 10.1021/bi101958h. [DOI] [PubMed] [Google Scholar]
- Zhao Y, Lang G, Ito S, Bonnet J, Metzger E, Sawatsubashi S, Suzuki E, Le Guezennec X, Stunnenberg HG, Krasnov A, et al. A TFTC/STAGA module mediates histone H2A and H2B deubiquitination, coactivates nuclear receptors, and counteracts heterochromatin silencing. Mol. Cell. 2008;29:92–101. doi: 10.1016/j.molcel.2007.12.011. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.