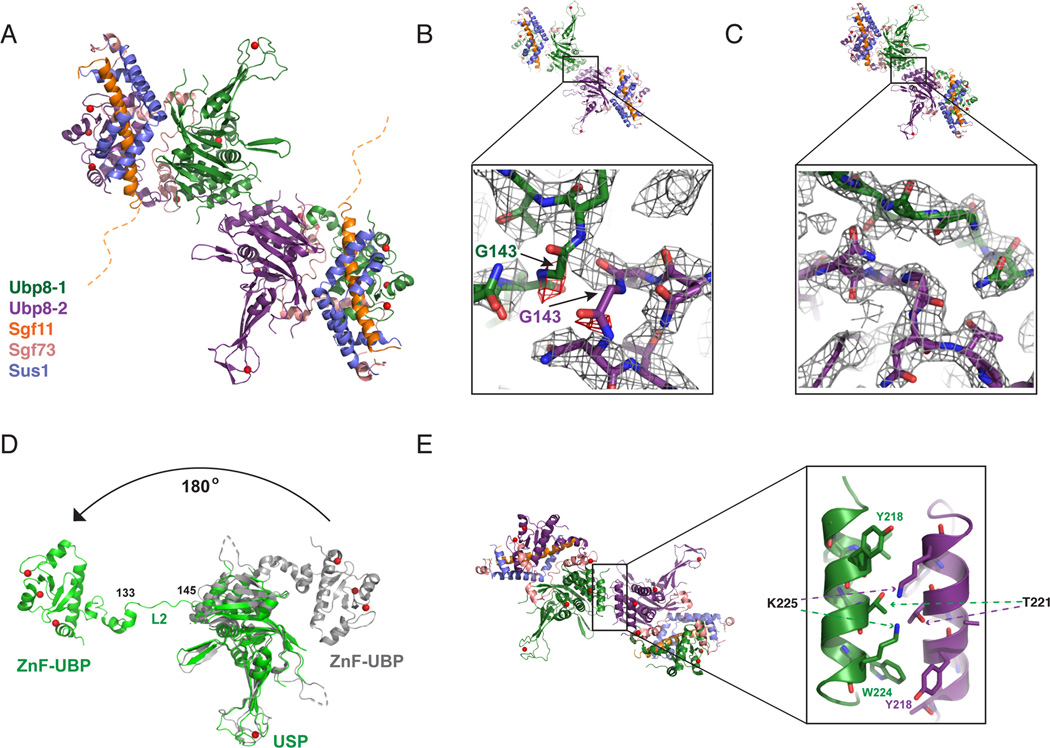
Figure 2. Structure of DUBm lacking the Sgf11-ZnF shows domain swapping.
(A) Structure of DUBm-ΔSgf11-ZnF/Ubp8-S144N shows domain swapping between two DUBm complexes. The two Ubp8 subunits in the complex are colored green and purple, respectively. Other subunits colored as indicated: Sus1 (blue), Sgf11 (orange) Sgf73 (salmon). The disordered Sgf11 linker is represented by a dashed orange line.
(B) Initial fit of two DUBm complexes to the electron density map before detection of domain swapping (2Fo-Fc contoured at 1σ, grey, Fo-Fc contoured at 3σ red).
(C) Fit of domain-swapped complexes to 2Fo-Fc electron density map.
(D) Conformation of Ubp8 in wild-type, intact DUBm (gray) and in domain-swapped complex (green). Residues 133–145 in loop L2 undergo a major conformational change that accompanies domain swapping.
(E) Helix in Ubp8 that is buried by the Sgf11-ZnF in the intact DUBm mediates dimerization contacts in the domain-swapped DUBm-ΔSgf11-ZnF.