Abstract
CrkL is a key signaling protein that mediates the leukemogenic activity of Bcr-Abl. CrkL is thought to adopt a structure that is similar to that of its CrkII homolog. The two proteins share high sequence identity and indistinguishable ligand binding preferences; yet they have distinct physiological roles. Here we show that the structures of CrkL and phosphorylated CrkL are drastically different than the corresponding structures of CrkII. As a result, the binding activities of the SH2 and SH3 domains in the two proteins are regulated in a distinct manner and to a different extent. The different structural architecture of CrkL and CrkII may account for their distinct functional roles. The data show that CrkL forms a constitutive complex with Abl thus explaining the strong preference of Bcr-Abl for CrkL. The results also highlight how the structural organization of the modular domains in adaptor proteins can control signaling outcome.
INTRODUCTION
The Crk family of adaptor proteins are important signaling molecules that function downstream of a wide number of receptors and regulate important cellular processes, including cell adhesion, motility, phagocytosis, differentiation, proliferation, transformation and apoptosis1–3. Crk proteins are implicated in many human cancers, including lung adenocarcinoma and glioblastoma4, prostate5, ovarian6, gastric7, and breast cancer8. The Crk family consists of two alternatively spliced protein forms, CrkI and CrkII, expressed by a single gene (crk)9 and the Crk-like (CrkL) protein expressed by a distinct gene (crkl)10. The CrkII (304 residues) and CrkL (303 residues) proteins each consists of a single SH2 domain, an N-terminal SH3 (SH3N) domain and a C-terminal SH3 domain (SH3C) (Supplementary Results, Supplementary Fig. 1). The SH3N and SH3C domains are tethered by a ~50 residue–long linker, which contains a tyrosine residue (Tyr221 in CrkII, Tyr207 in CrkL) that is phosphorylated by the Abl kinase11,12.
Crk proteins link activated receptors to specific downstream signaling cascades using their SH2 and SH3N domains, which selectively bind to pY-x-x-P and P-x-L-P-x-K motifs, respectively13,14. A large variety of proteins have been identified as binding partners of Crk proteins1,15. In contrast, the SH3C domain lacks the binding determinants of typical polyproline (PP)-II binding SH3 domains16 and was shown to function as an autoregulatory element in CrkII17–19 or even to promote certain signaling pathways20.
CrkL has received particular attention primarily because it is a preferred substrate of Bcr-Abl21,22, an oncogenic kinase that causes chronic myelogenous leukemia (CML)23. CrkL is indispensable for mediating the aberrant activity of Bcr-Abl24,25 and is constitutively phosphorylated in human CML cells26,27. In fact, the level of CrkL phosphorylation is being used as a predictor of clinical outcome in patients treated for CML28. In addition to the relevance of CrkL to cancer, deletion of the crkl gene causes cardiovascular and craniofacial defects resulting in embryonic lethality29. Interestingly, CrkL appears to be a rather unique adaptor protein since it has been reported to function in the cytoplasm, the nucleus and the extracellular milieu5.
Although CrkL and CrkII have been shown to compensate for each other in certain cases1, numerous studies have demonstrated that the two Crk proteins have distinct, non-overlapping functions1,29–31. Because of the high sequence identity between CrkL and CrkII (56%, up to 72% in the modular domains; Supplementary Fig. 1a) it is hypothesized that the two proteins adopt very similar structures. Taking also into account the identical binding preferences of their SH2 and SH3N domains, it has been difficult to account for the different functional roles and signaling output of the CrkL and CrkII proteins.
Here we report the structure of CrkL in its unliganded and unphosphorylated state as well as in the phosphorylated (pTyr207) form (pCrkL). The data show that the SH2 and SH3 modular domains in CrkL are organized in a drastically different architecture than in CrkII18,19: (i) the pTyr-binding site of SH2 is partially masked in CrkL whereas it is accessible in CrkII; (ii) the PPII-binding site of SH3N is accessible in CrkL but is completely occluded in CrkII; and, (iii) the SH3C domain is mobile and does not interact with any of the other domains in CrkL, whereas it interacts extensively with the SH2 and SH3N domains in CrkII thereby stabilizing its overall structure. We also show that upon Tyr207 phosphorylation, the linker region interacts in an intramolecular fashion with the CrkL SH2 domain, thereby inhibiting the binding of pTyr-ligands. Very interestingly, this intramolecular association has little effect on the overall structure of CrkL, in sharp contrast to CrkII wherein phosphorylation of Tyr211 results in SH3N autoinhibition. Notably, the present data demonstrate that CrkL forms a constitutive complex with Abl. Thus, despite the very high sequence identity of CrkL and CrkII, the structural organization of the two adaptors is drastically different and may account for their different functional roles, as well as the preference of Bcr-Abl to interact with CrkL rather than CrkII.
RESULTS
Structure determination of CrkL and pCrkL
Human CrkL (~34 kDa) consists of 303 residues. Gel filtration and multi-angle laser light scattering (MALLS) data show that the protein exists as a monomer in solution (Supplementary Fig. 2). Despite the relatively large size of CrkL, the NMR spectra are of outstanding sensitivity and resolution (Supplementary Fig. 3a). Assignment of CrkL was facilitated by preparing and assigning the isolated SH2 and SH3 modular domains (Supplementary Fig. 4a). Interestingly, overlay of the NMR spectra of the isolated domains with that of full-length CrkL revealed significant chemical shift differences only for a relatively small number of residues, which are located at the SH2 and SH3N domains, while no differences were observed for the SH3C domain (Supplementary Fig. 4b, c). The phosphorylated (pTyr207) CrkL (pCrkL) sample for NMR and structural characterization was prepared by adding catalytic amounts of the Abl kinase as detailed in Supplementary Methods (Supplementary Fig. 2d). Similarly to CrkL, the spectra of pCrkL are also of high quality allowing for complete backbone and side chain assignment (Supplementary Fig. 3b). The structures of both CrkL and pCrkL were determined by combining NOE, paramagnetic relaxation enhancement (PRE) and residual dipolar coupling (RDC) restraints (Supplementary Table 1).
Structural architecture of CrkL
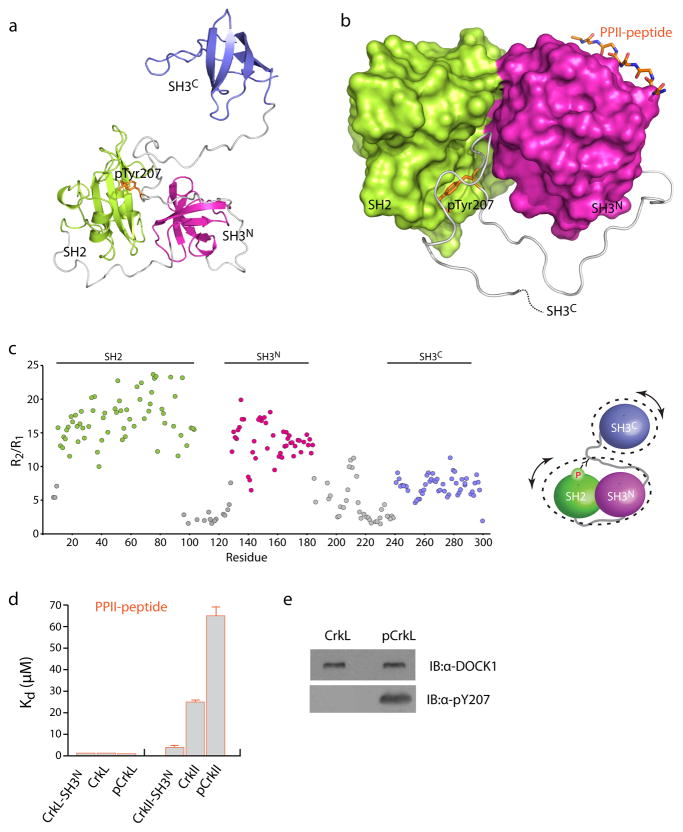
The lowest-energy structure of CrkL is shown in Figure 1a and the conformational ensemble is shown in Supplementary Figure 5a. The structures of the CrkL individual SH2 and SH3 domains are very similar to the corresponding domains of the CrkII protein (Supplementary Fig. 5). As expected on the basis of sequence conservation (Supplementary Fig. 1a), the binding pockets of SH2 and SH3N are almost identical in CrkL and CrkII (Supplementary Fig. 6), thus accounting for the same recognition preferences of CrkL and CrkII. The CrkII SH2 domain has a 17-residue-long insertion between β-strands D and E that forms a flexible loop (DE loop; Supplementary Fig. 6a). The DE loop is enriched in proline residues and has been shown to bind to the SH3 domain of the Abl kinase32. This loop is absent in CrkL SH2 (Supplementary Figs. 1a, 6a) and thus the binding mode of CrkL and CrkII to Abl is expected to be different. In agreement with previous studies33, the structural data show that the CrkL SH3C domain cannot bind polyproline II (PPII)–type sequences because of the lack of aromatic residues at its canonical binding site.
Figure 1. Structural and dynamic properties of CrkL.
(a) Structure of CrkL. The SH2, SH3N and SH3C domains are colored green, magenta, and blue, respectively. The linker regions are colored grey. The SH3C domain does not interact with the other domains. (b) Close-up view of the SH2–SH3N interface in CrkL. Only polar/charged residues mediate the interaction between the two domains. (c) Plot of the ratio of R2 over R1 15N relaxation rates of the CrkL backbone as a function of residue number. The R2/R1 ratio provides information about the tumbling of the molecule with higher values indicating slower tumbling. (d) Correlation times (τC) for the tumbling of CrkL. The SH2–SH3N module tumbles as a rigid unit whereas the SH3C domain tumbles much faster and independently of the other domains. (e) Residues undergoing substantial μs-ms time scale motions, as denoted by enhanced Rex values, are mapped on the structure of CrkL. Almost all residues located at the interface between the SH2 and SH3N domains exhibit relatively high Rex values indicating that the binding interface is dynamic.
The structural organization of CrkL is rather unique within the family of adaptor proteins: the SH2 and the first SH3 domain (SH3N) interact directly with each other, whereas the second SH3 domain (SH3C) does not interact with any of the modular domains (Fig. 1a). In agreement with the chemical shift analysis, the SH2–SH3N interaction is mediated by a small surface burying ~600 Å2. The binding surfaces on both SH2 and SH3N domains consist primarily of polar and charged residues and thus the interaction is mediated almost exclusively by polar contacts (Fig. 1b). Substitution of residues located at the SH2–SH3N interface disrupts the interaction between the two domains in CrkL (Supplementary Fig. 4c).
Dynamic properties of CrkL
In order to determine the motional properties of CrkL we used NMR relaxation methodologies34. We measured the 1H-15N nuclear Overhauser effect (NOE), the longitudinal relaxation rate R1 and the transverse relaxation rate R2 (Fig. 1c and Supplementary Fig. 7a). The R1 and R2 relaxation rates are sensitive to and thus report on changes of the diffusion properties of the protein34. If only residues located at rigid parts of the molecule are considered, then the ratio R2/R1 provides a good estimate of the correlation time35. The relaxation analysis demonstrates that the SH2 and SH3N domains tumble as a unit, with a correlation time τc of ~11.0 ns, whereas the SH3C domain tumbles much faster and in an independent fashion with a τc of ~7.8 ns (Fig. 1d). Therefore, the relaxation data are in agreement with the structural data (Fig. 1a) showing that the SH2 and SH3N domains interact with each other while the SH3C domain does not interact with any of the other domains. The present results indicate that SH3C play no role in regulating the activity of the SH2 and SH3N domains in CrkL, in sharp contrast to CrkII proteins wherein SH3C was shown to act as a regulatory structural element by stabilizing the autoinhibitory conformation18,19.
Further analysis of the relaxation data demonstrates that most of the residues located at the SH2–SH3N interface exhibit enhanced motions on the micro-to-millisecond (μs-ms) timescale (Fig. 1e). This observation suggests that the two domains move relatively to each other giving rise to a dynamic binding interface.
The binding site of the SH2 domain in CrkL is occluded
Structural analysis of the SH2–SH3N interface reveals that although the pTyr-binding site of the SH2 domain is largely accessible, the SH3N domain masks the binding sites for the residues immediately downstream of the pTyr residue (Fig. 2). Thus, the structural data raise the intriguing possibility that the SH2–SH3N intramolecular arrangement in CrkL may inhibit the binding activity of SH2 for phosphotyrosine ligands. To test this hypothesis, we used isothermal titration calorimetry (ITC) to directly measure the binding energetics of a phosphorylated peptide containing a consensus sequence for CrkL SH2 (CrkL-pTyr207-peptide) to isolated SH2 and full-length CrkL (Supplementary Fig. 8a). The CrkL-pTyr207-peptide binds to isolated SH2 with a moderate affinity (Kd ~7 μM), whereas the affinity for SH2 in the context of full-length CrkL is more than 3-fold weaker (Kd ~23 μM) (Fig. 2c). Thus, the intramolecular arrangement in CrkL gives rise to an autoinhibitory mechanism that partially occludes the SH2 domain thereby modulating its activity for phosphotyrosine ligands.
Figure 2. Binding of pTyr- and PPII-peptide ligands to CrkL and CrkII.
(a, b) Structure of the SH2–SH3N module in (a) CrkL and (b) CrkII. The pTyr-peptide and PPII-peptide are shown as they have been previously determined to bind to the isolated SH2 (PDB 1JU5) and SH3N domains (PDB 1CKA), respectively. The pTyr-peptide binding site in CrkL is partially masked but is completely accessible in CrkII. Conversely, the PPII-peptide binding site in CrkL is completely accessible but is entirely masked in CrkII. (c) Dissociation constants (Kd) of pTyr-peptide and PPII-peptide complexes with CrkL (Supplementary Fig. 8a) and CrkII. The Kd values of PPII-peptide binding to CrkII were obtained from ref. 19.
In contrast to CrkL, the SH2 domain in CrkII appears not to be inhibited (Fig. 2b)19. Indeed, our ITC experiments show that a phosphorylated peptide containing a consensus sequence for CrkII SH2 (CrkII-pTyr221-peptide) binds with the same affinity to both the isolated SH2 and full-length CrkII (Fig. 2c). It was previously proposed that the SH2 domain of CrkII and CrkL have different sequence specificity, with CrkL reported to bind a phosphorylated peptide derived from the FGF receptor (FGFR-pTyr-peptide) with a ~30-fold higher affinity than CrkII36. However, ITC data show that the intrinsic affinity of the isolated SH2 domains of CrkII and CrkL for phosphotyrosine ligands (Fig. 2c), even for the FGFR-pTyr-peptide (Supplementary Fig. 9a), is very similar and the two domains have identical specificity properties, as expected on the basis of the structures of the SH2-pTyr complexes (Supplementary Fig. 9b). Taken together, the present data show that any phosphotyrosine ligand will bind preferably to full-length CrkII over full-length CrkL by a factor of ~3–4 (Fig. 2c).
The binding site of the SH3N domain in CrkL is accessible
Structural analysis of CrkII proteins have shown that the canonical PPII-binding site of the SH3N domain is almost completely occluded (Fig. 2b)18,19. As a result, PPII ligands, such as the Abl kinase, bind to full-length CrkII with a ~10-fold lower affinity than in the isolated SH3N (Fig. 2c)17,19,37. In sharp contrast to CrkII, the present structural data demonstrate that the PPII-binding site of SH3N in CrkL is completely accessible (Fig. 2a). Indeed, ITC data (Supplementary Fig. 8b) show that a PPII-peptide containing the consensus sequence for SH3N binds to isolated SH3N and full-length CrkL with the same affinity (Fig. 2c). NMR analysis of the titration of the PPII-peptide to CrkL demonstrates that no steric clashes occur between the SH3N-bound peptide and the rest of the protein indicating that PPII-peptide binding to CrkL is unrestricted. In contrast, PPII-peptide titration to CrkII results in drastic conformational rearrangement and relieve of the autoinhibitory conformation17. Taken together, our results show that a PPII ligand will bind preferably to CrkL over CrkII by a factor of ~10 (Fig. 2c).
CrkL Tyr207 phosphorylation results in SH2 inhibition
Tyr207 in CrkL is phosphorylated by the Abl kinase12,38. The pTyr207-x-x-Pro210 region in the CrkL linker is a target sequence for CrkL SH213. NMR analysis of pCrkL (Supplementary Fig. 3b) shows that the CrkL SH2 domain interacts with pTyr207. MALLS and gel filtration data demonstrate that pCrkL remains a monomer in solution (Supplementary Fig. 2a, b). The data collectively show that upon phosphorylation of CrkL Tyr207, the SH2 domain interacts in an intramolecular fashion with pTyr207, similarly to CrkII19,39.
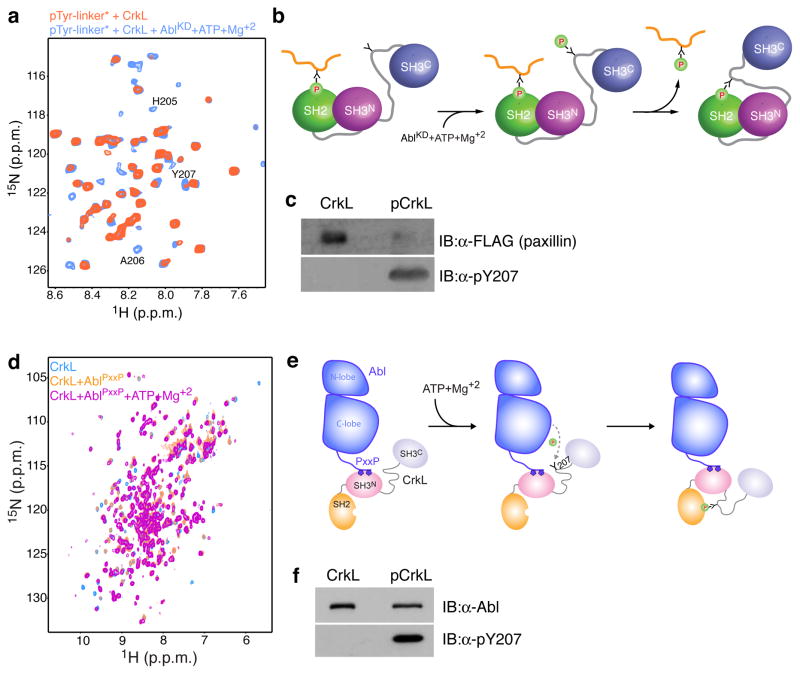
We used NMR to determine the solution structure of pCrkL. In agreement with the NMR chemical shift analysis, the structural data show that the phosphorylated linker is bound to the canonical pTyr-binding cleft of the SH2 domain (Fig. 3). The interactions between the linker and the SH2 domain are virtually identical to those reported previously for the structure of CrkII SH2 and a phosphopeptide (Supplementary Fig. 9b)32. Interestingly, the structural rearrangement elicited to CrkL by the phosphorylation of Tyr207 and the ensuing intramolecular binding to SH2 is minimal. The SH2–SH3N interface adjusts slightly so as to accommodate the binding of the pTyr-linker region to SH2 (Fig. 3b and Supplementary Fig. 5c), whereas the SH3C domain tumbles independently (Fig. 3c) as in the unphosphorylated form.
Figure 3. Structural and dynamic properties of pCrkL.
(a) Structure of pCrkL. Phosphorylated Tyr207 (pTyr207) is shown as orange sticks. (b) Close-up view of the pTyr207-binding site. The SH2–SH3N interface adjusts slightly to accommodate the binding of the linker to SH2. (c) Plot of the ratio of R2 over R1 relaxation rates of pCrkL as a function of residue number. As in CrkL, the SH2–SH3N module in pCrkL tumbles as a unit whereas the SH3C domain tumbles much faster and independently of the other domains. (d) Kd values of PPII-peptide complexes with CrkL (Supplementary Fig. 8a) and CrkII19 variants. (e) Pull-down of CrkL and pCrkL with DOCK1, an SH3N binding physiological partner of CrkL (Supplementary Fig. 12).
The intramolecular binding of the phosphorylated linker (pTyr207) to the SH2 domain is expected to prevent SH2 from interacting with other phosphorylated ligands. To test this hypothesis we monitored by NMR the effect of phosphorylating CrkL that is already bound to a phosphorylated peptide encompassing the linker sequence (pTyr-linker; Fig. 4a). Indeed, the NMR data show that following the addition of catalytic amounts of Abl kinase domain (AblKD) the phosphorylated linker binds intramolecularly to SH2 and as a result the pTyr-linker is displaced (Fig. 4b). Pull-down of CrkL and pCrkL with Tyr-phosphorylated paxillin, a focal adhesion protein that interacts specifically with CrkL SH23, show that paxillin forms a complex only with the unphosphorylated CrkL (Fig. 4c). Collectively, the data show that Tyr207 phosphorylation of CrkL by Abl results in intramolecular binding of the linker with the SH2 domain thereby giving rise to CrkL SH2 inhibition.
Figure 4. Effect of Tyr207 phosphorylation on CrkL folding and its association with Abl kinase.
(a) 1H-15N HSQC NMR spectra of the linker region of CrkL containing the phosphorylated Tyr207 (pTyr-linker) in the presence of CrkL (orange) and after the addition of catalytic amounts of AblKD and ATP-Mg+2 (blue). The pTyr-linker is 15N-labeled whereas CrkL and AblKD are unlabeled. (b) Analysis of the NMR experiments in (a) shows that the pTyr-linker binds to the SH2 domain of CrkL. Phosphorylation of Tyr207 in CrkL induces the intramolecular association of pTyr207 and SH2. As a result, the pTyr-linker is displaced. (c) Pull-down of CrkL and pCrkL with paxillin, an SH2 binding physiological partner of CrkL (Supplementary Fig. 12). (d) 1H-15N HSQC NMR spectra of free CrkL (blue), in complex with AblPxxP (orange) and after adding ATP-Mg+2 (magenta). AblPxxP is a construct that encompasses the kinase domain and the first PxxP motif that binds to CrkL. (e) Analysis of the NMR experiments in (d) shows that CrkL forms a 1:1 complex with AblPxxP using its SH3N domain. Phosphorylation of Tyr207 elicits the intramolecular association of pTyr207 and SH2 but the intramolecular folding in CrkL has no effect on the CrkL–AblPxxP complex, which remains tightly associated. (f) Pull-down of CrkL and pCrkL with full-length Abl (form 1b) (Supplementary Fig. 12).
pCrkL interacts with signaling partners via SH3N
Upon Tyr phosphorylation, the SH2 domain in both CrkL (Fig. 4a–c) and CrkII11,39 are inhibited for pTyr-ligand binding. CrkII has been shown to undergo a major conformational change upon phosphorylation that results in PPII-ligand binding inhibition to SH3N19. However, the structural rearrangement elicited by phosphorylation and the ensuing intramolecular binding in CrkL is minimal (Fig. 3a, b and Supplementary Fig. 5c). As a result, the PPII-binding site of the SH3N domain in phosphorylated CrkL is completely accessible (Fig. 3b). In agreement with the structural data, ITC experiments (Supplementary Fig. 8b) show that a PPII-peptide binds to the unphosphorylated CrkL (or the isolated SH3N) and pCrkL with very similar affinities (Fig. 3d). Pull-down of CrkL and pCrkL with DOCK1, a guanine exchange factor (GEF) that activates Rac1 and binds specifically to the SH3N domain, shows that DOCK1 associates strongly with both CrkL and pCrkL (Fig. 3e).
Abl binds to the SH3N domain of CrkL and CrkII using a consensus PxxP motif located C-terminally to its kinase domain40. An important implication of SH3N inhibition in CrkII is that Abl binding to and phosphorylation of CrkII will result in pCrkII–Abl complex dissociation41. To test the emerging hypothesis that pCrkL will remain tightly bound to Abl, we titrated AblPxxP, an Abl construct encompassing the kinase domain and the first PxxP Crk-binding motif, to labeled CrkL (Fig. 4d). NMR analysis indicates that AblPxxP binds to the SH3N domain of CrkL (Fig. 4e). Addition of ATP-Mg+2 results in Abl-mediated phosphorylation of Tyr207. NMR analysis (Fig. 4d) shows that pCrkL adopts the intramolecularly folded conformation but forms a tight complex with AblPxxP (Fig. 4e). In agreement, pull-down of CrkL and pCrkL with full-length Abl kinase shows that Abl forms complexes with CrkL that are not dependent on the CrkL phosphorylation state (Fig. 4f).
DISCUSSION
CrkL is an adaptor protein that regulates important cellular processes ranging from cell adhesion and motility to phagocytosis and apoptosis. CrkL has been recently identified as an essential gene in cancer cell proliferation25 and has been shown to be indispensable for mediating the leukemogenic activity of Bcr-Abl24,25. CrkL is constitutively phosphorylated in human CML cells26,27 and the level of CrkL phosphorylation is being used as a predictor of clinical outcome in patients treated for CML28. Despite the important role of CrkL, the lack of its structure has impeded a proper understanding of its function.
Because CrkL shares high sequence identity with CrkII, it has been thought that the structures of the two proteins are very similar. The structural data reported here demonstrate that the structural organization of the two proteins is drastically different. The CrkII structure19 is stabilized by a hydrophobic segment in the linker region, part of which is quite different in CrkL (Supplementary Fig. 10). Moreover, several contacts exist between the SH2 DE-loop and SH3C in CrkII. In contrast, the DE-loop is not present in CrkL SH2. The autoinhibited structure of pCrkII is stabilized by a linker region that is totally different in CrkL (Supplementary Fig. 10b). Thus, despite the high sequence identity (up to 72% in the structured regions), few key sequence differences between CrkL and CrkII appear to modulate the overall structural organization of the two proteins (Supplementary Fig. 10). The distinct structural architectures of CrkL and CrkII determine the signaling input and output giving rise to distinct physiological functions for the two proteins. These results further highlight the notion that adaptors regulate signaling in a dynamic way and do not simply serve to wire signaling components in a passive manner42.
In the resting state, the pTyr-binding site of the SH2 in CrkL is inhibited, whereas the one in CrkII is not and as a result the binding of phosphorylated ligands to CrkII will be favored over binding to CrkL (Fig. 2). The differential SH2 binding activity modulation in CrkL vs CrkII can have important implications, such as in the binding of the p130 Crk-associated substrate (p130CAS), a scaffold protein mediating integrin signaling (Fig. 5)3. Although p130CAS has multiple phosphorylation sites, it is conceivable that the number of such sites may be limited either because of the action of phosphatases or cell conditions43. In this case, competition will favor CrkII over CrkL binding to p130CAS (Fig. 5i).
Figure 5. CrkL versus CrkII in integrin signaling.
(i) Integrin activation elicits p130CAS phosphorylation by tyrosine kinases (TK) and as a result CrkL and CrkII are recruited. (ii) GEFs (for example, DOCK1 and C3G) associate with CrkL and CrkII via their SH3N domain giving rise to efficient localized activation (iii) of small GTPases (for example, Rac and Rap) at the membrane. (iv) Abl-induced phosphorylation of CrkL and CrkII forces their dissociation from p130CAS and thus results in signaling suppression. The distinct structural organization of CrkL and CrkII modulates the interactions with their physiological partners to a different extent. The blue and orange shaded regions in SH2 and SH3N denote the pTyr- and PPII-binding sites, respectively. See text for details.
Eventual association of CrkL/CrkII with p130CAS will have minimal effect on the overall structure of either CrkL (Figs. 1a and 3a) or CrkII19. Therefore, the PPII-binding site of SH3N in CrkII will be inhibited in the p130CAS-bound CrkII41, whereas the corresponding site in CrkL will be accessible. As a result, guanine nucleotide exchange factors (GEFs), such as DOCK1 and C3G, will bind to CrkL with a much higher affinity than to CrkII (Figs. 2c and 5ii). In this case the CrkL-mediated complex is expected to activate more efficiently downstream GTPases giving rise to stronger signaling outcome than the CrkII-mediated complex (Fig. 5iii). Enhanced association between CrkL and DOCK1 will increase cell migration and invasion3 and may explain previous observations that CrkL has a much higher oncogenic potential than CrkII in fibroblasts21.
In both CrkL and CrkII complexes, phosphorylation of Tyr207 (Fig. 3) and Tyr22111,19,39, respectively, by Abl will cause intramolecular folding and displacement of the pCrk proteins from p130CAS (Fig. 4a–c), resulting in negative regulation of cell migration (Fig. 5iv)19. Interestingly, pCrkL may form a constitutive complex with DOCK1, in contrast to pCrkII, since in pCrkL the SH3N domain is not inhibited (Fig. 3e). Thus, when phosphatases act to dephosphorylate CrkL and CrkII, the p130CAS CrkL DOCK1 complex will assemble more readily than the corresponding CrkII complex.
A particularly striking result in the present work is that the phosphorylated form of CrkL remains active and may interact via its uninhibited SH3N domain with various ligands, including the Abl kinase (Fig. 4d–f). The interaction of CrkL with Bcr-Abl stimulates substantially the leukemogenic activity of the oncoprotein21,44. The present data demonstrate that CrkL binds and remains tightly bound to Abl even after the kinase has phosphorylated CrkL (Fig. 4e, f), explaining why CrkL is a preferred substrate for Bcr-Abl. This is in sharp contrast to pCrkII, which has been shown to be an entirely inactive protein11. Therefore, although the intrinsic affinity of the SH3N domains of CrkL and CrkII for the PxxP motif of Abl is almost identical (Fig. 2c), the overall structural organization of the proteins confers an advantage to CrkL since the binding of CrkII to Abl is suppressed.
Although the SH3C domain has been shown to function as an autoregulatory element in CrkII17–19,37, in CrkL SH3C does not interact with any of the other domains and thus its role remains elusive. Functional data however indicate that CrkL SH3C is indispensable for fibroblast transformation and hematopoietic cell adhesion38. It is likely that SH3C mediates its function by interacting with an as yet identified partner or by being tyrosine phosphorylated, as shown recently for CrkII45.
SH2 and SH3 domains have long been thought to mediate sequence-specific interactions involving PxxP and pY motifs, respectively. However, there is now a growing list of examples indicating that these signaling domains can interact with sequences that do not conform to these general rules46. Interestingly, our data demonstrate that the two domains can engage each other in a completely unconventional manner. SH2–SH3 interactions have been previously observed in Itk kinase47 and the SAP–Fyn complex48. Comparison of the binding mode in these systems and in CrkL demonstrates the great versatility of the SH2 and SH3 domains in mediating interactions in cell signaling (Supplementary Fig. 11).
METHODS
Protein preparation
The following human CrkL constructs were prepared: SH2 (residues 1–105), SH3N (residues 120–188) linker-SH3C (residues 184–303), SH2-SH3N (residues 1–188), pTyr-linker (188–230), and full-length CrkL (residues 1–303). The constructs were cloned into the pet42a vector using the NcoI and Xho1 restriction sites. A Tev protease cleavage site was introduced between the histidine tag and the protein. For NMR studies, the samples were dialyzed in NMR buffer (50 mM KPi (pH 6.8), 140 mM NaCl and 1 mM β-mercaptoethanol) and concentrated using Amicon cell units (Millipore). All fragments are monomeric in solution at concentrations used for the NMR studies (typically 0.6–1.0 mM), as indicated by gel filtration and light scattering. Phosphorylated CrkL and pTyr-linker were prepared by the addition of catalytic amounts of AblKD. Phosphorylation efficiency is over 99% as judged by mass spectrometry and NMR. The sequence of the CrkL-pTyr207-, CrkII-pTyr221-, PPII- and FGFR-pTyr-peptides used are EPAHApYAQPTT, PEPGPpYAQPSV, YLQAPELPTKTRTS, and AGVSEpYELPEDPR, respectively. The MS-determined molecular mass of the peptides were 1393.1, 1221.2, 1605.3, and 1541.4, respectively. All peptides were >99% pure. More details about the preparation of AblKD, AblKD-PxxP and NMR samples are provided in Supplementary Methods.
NMR spectroscopy
NMR experiments were performed on Varian 800- and 600-MHz and Bruker 700- and 600-MHz spectrometers. Complete backbone and side chain assignment for the CrkL proteins studied was achieved using standard triple-resonance pulse sequences. NOEs were measured using two- and three-dimensional 13C and 15N edited NOESY experiments using mixing times of 80 and 100 ms, respectively. All experiments were performed at 32 °C.
Paramagnetic Relaxation Enhancement Measurements
Nitroxide spin labels (MTSL; Toronto Research Chemicals Inc.) were introduced via cysteine-specific modification of engineered CrkL derivatives containing single-solvent-accessible cysteine residues. Mutants and MTSL derivatives that did not perturb the CrkL structure, as assessed by 1H-15N HSQC spectra, were used for measuring PRE rates. PRE-derived distances were determined from 1H-15N-HSQC spectra of CrkL by measuring peak intensities before (paramagnetic) and after (diamagnetic) reduction of the nitroxide spin label with ascorbic acid. More details are provided in Supplementary Information.
Residual Dipolar Coupling (RDC) Measurements
Alignment of the proteins for RDC measurements was achieved using poly(ethylene glycol)/alcohol mixtures49. More experimental details are provided in Supplementary Information.
Relaxation measurement and analysis
Three relaxation parameters were measured for all backbone amides of CrkL: the {1H}-15N NOE, the longitudinal relaxation rate R1 and the transverse relaxation rate R2. Experimental details about data collection and analysis are provided in Supplementary Information.
Calorimetry
All calorimetric titrations were performed on a iTC200 microcalorimeter (GE), as described in Supplementary Methods.
Structure calculation and refinement
Structure calculations were performed with Xplor-NIH. The 13Cα, 13Cβ, 13C′, Hα, 15N and NH chemical shifts served as input for the TALOS+ program50 to extract dihedral (φ and ψ) angles. The initial structure was calculated using NOEs, PREs, dihedral angles and hydrogen bonds. RDC restraints were included in the final stages of the calculation. Ramachandran statistics are as follows: most favored regions: 90%; allowed regions: 8%; disallowed regions: 2%. The summary of NMR restraints and structure refinement statistics is presented in Supplementary Table 1.
Pull-down assays
Phosphorylated GST or GST-CrkL was obtained by incubating purified GST or GST-CrkL (~1μM) with purified AblKD (~0.1 μM) overnight at room temperature in a buffer containing 50 mM Tris (pH 7.5), 150 mM NaCl, 10 mM MgCl2, 1 mM DTT and 5 mM ATP. Purified GST-CrkL was incubated in the same buffer without AblKD and ATP overnight for the unphosphorylated sample. Lactose-free glutathione agarose beads (Sigma) were then washed once with PBS containing 0.1 % Triton-X-100 and then incubated with the aforementioned samples for 60 minutes at 4 °C. Beads were then spun down and washed twice with PBS containing 0.1 % Triton-X-100. Beads with bound phosphorylated GST or phosphorylated/unphosphorylated GST-CrkL were each incubated with lysates of 293T cells untransfected or transfected with Flag-Paxillin or human Abl1b for 2 hours at 4 °C. Beads were then spun down and washed twice with PBS containing 0.1% Triton-X-100. Samples were then boiled in SDS sample buffer and analyzed by western blotting with anti-Flag, anti-DOCK180, anti-Abl, anti-CrkL and anti-pCrkL(Y207) antibodies.
Supplementary Material
Acknowledgments
We thank S. Karamanou for performing the MALLS experiments and D. Duffield for collecting the mass spectrometry data. This work was supported by the Department of Defense (R.B.B.) and the U.S. National Institute of Health (GM80308 to C.G.K.).
Footnotes
Author contributions
W.J., T.S., S.G., R.A.B., and C.G.K. designed research; W.J., T.S., M.-T.P., and S.G. performed research; W.J., T.S., M.-T.P., S.G., R.A.B., and C.G.K. analyzed data; W.J., T.S., and C.G.K. wrote the manuscript.
Competing financial interests
The authors declare no competing financial interests.
Accession codes
Protein Data Bank: Coordinates for the CrkL and pCrkL have been deposited with accession codes 2LQN and 2LQW, respectively.
References
- 1.Birge RB, Kalodimos C, Inagaki F, Tanaka S. Crk and CrkL adaptor proteins: networks for physiological and pathological signaling. Cell Commun Signal. 2009;7:13. doi: 10.1186/1478-811X-7-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Isakov N. A new twist to adaptor proteins contributes to regulation of lymphocyte cell signaling. Trends Immunol. 2008;29:388–396. doi: 10.1016/j.it.2008.04.006. [DOI] [PubMed] [Google Scholar]
- 3.Cabodi S, del Pilar Camacho-Leal M, Di Stefano P, Defilippi P. Integrin signalling adaptors: not only figurants in the cancer story. Nat Rev Cancer. 2010;10:858–870. doi: 10.1038/nrc2967. [DOI] [PubMed] [Google Scholar]
- 4.Kim YH, et al. Genomic and functional analysis identifies CRKL as an oncogene amplified in lung cancer. Oncogene. 2010;29:1421–1430. doi: 10.1038/onc.2009.437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mintz PJ, et al. An unrecognized extracellular function for an intracellular adapter protein released from the cytoplasm into the tumor microenvironment. Proc Natl Acad Sci USA. 2009;106:2182–2187. doi: 10.1073/pnas.0807543105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wang H, et al. The role of Crk/Dock180/Rac1 pathway in the malignant behavior of human ovarian cancer cell SKOV3. Tumour Biol. 2010;31:59–67. doi: 10.1007/s13277-009-0009-9. [DOI] [PubMed] [Google Scholar]
- 7.Feng R, et al. miR-126 functions as a tumour suppressor in human gastric cancer. Cancer Lett. 2010;298:50–63. doi: 10.1016/j.canlet.2010.06.004. [DOI] [PubMed] [Google Scholar]
- 8.Noren NK, Foos G, Hauser CA, Pasquale EB. The EphB4 receptor suppresses breast cancer cell tumorigenicity through an Abl-Crk pathway. Nat Cell Biol. 2006;8:815–825. doi: 10.1038/ncb1438. [DOI] [PubMed] [Google Scholar]
- 9.Matsuda M, et al. Two species of human CRK cDNA encode proteins with distinct biological activities. Mol Cell Biol. 1992;12:3482–3489. doi: 10.1128/mcb.12.8.3482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hoeve ten J, Morris C, Heisterkamp N, Groffen J. Isolation and chromosomal localization of CRKL, a human crk-like gene. Oncogene. 1993;8:2469–2474. [PubMed] [Google Scholar]
- 11.Feller SM, Knudsen B, Hanafusa H. c-Abl kinase regulates the protein binding activity of c-Crk. EMBO J. 1994;13:2341–2351. doi: 10.1002/j.1460-2075.1994.tb06518.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.de Jong R, Hoeve ten J, Heisterkamp N, Groffen J. Tyrosine 207 in CRKL is the BCR/ABL phosphorylation site. Oncogene. 1997;14:507–513. doi: 10.1038/sj.onc.1200885. [DOI] [PubMed] [Google Scholar]
- 13.Songyang Z, et al. SH2 domains recognize specific phosphopeptide sequences. Cell. 1993;72:767–778. doi: 10.1016/0092-8674(93)90404-e. [DOI] [PubMed] [Google Scholar]
- 14.Wu X, et al. Structural basis for the specific interaction of lysine-containing proline-rich peptides with the N-terminal SH3 domain of c-Crk. Structure. 1995;3:215–226. doi: 10.1016/s0969-2126(01)00151-4. [DOI] [PubMed] [Google Scholar]
- 15.Cheerathodi M, Ballif BA. Identification of CrkL-SH3 Binding Proteins from Embryonic Murine Brain: Implications for Reelin Signaling during Brain Development. J Proteome Res. 2011;10:4453–4462. doi: 10.1021/pr200229a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Muralidharan V, et al. Solution Structure and Folding Characteristics of the C-Terminal SH3 Domain of c-Crk-II. Biochemistry. 2006;45:8874–8884. doi: 10.1021/bi060590z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sarkar P, Reichman C, Saleh T, Birge RB, Kalodimos CG. Proline cis-trans isomerization controls autoinhibition of a signaling protein. Mol Cell. 2007;25:413–426. doi: 10.1016/j.molcel.2007.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sarkar P, Saleh T, Tzeng SR, Birge RB, Kalodimos CG. Structural basis for regulation of the Crk signaling protein by a proline switch. Nat Chem Biol. 2011;7:51–57. doi: 10.1038/nchembio.494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kobashigawa Y, et al. Structural basis for the transforming activity of human cancer- related signaling adaptor protein CRK. Nat Struct Mol Biol. 2007;14:503–510. doi: 10.1038/nsmb1241. [DOI] [PubMed] [Google Scholar]
- 20.Antoku S, Mayer BJ. Distinct roles for Crk adaptor isoforms in actin reorganization induced by extracellular signals. J Cell Sci. 2009;122:4228–4238. doi: 10.1242/jcs.054627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Senechal K, Halpern J, Sawyers C. The CRKL adaptor protein transforms fibroblasts and functions in transformation by the BCR-ABL oncogene. J Biol Chem. 1996;271:23255–23261. doi: 10.1074/jbc.271.38.23255. [DOI] [PubMed] [Google Scholar]
- 22.Sattler M, Salgia R. Role of the adapter protein CRKL in signal transduction of normal hematopoietic and BCR/ABL-transformed cells. Leukemia. 1998;12:637–644. doi: 10.1038/sj.leu.2401010. [DOI] [PubMed] [Google Scholar]
- 23.Colicelli J. ABL tyrosine kinases: evolution of function, regulation, and specificity. Sci Signal. 2010;3:re6. doi: 10.1126/scisignal.3139re6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Seo JH, et al. A specific need for CRKL in p210BCR-ABL-induced transformation of mouse hematopoietic progenitors. Cancer Res. 2010;70:7325–7335. doi: 10.1158/0008-5472.CAN-10-0607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Luo B, et al. Highly parallel identification of essential genes in cancer cells. Proc Natl Acad Sci USA. 2008;105:20380–20385. doi: 10.1073/pnas.0810485105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nichols GL, et al. Identification of CRKL as the constitutively phosphorylated 39-kD tyrosine phosphoprotein in chronic myelogenous leukemia cells. Blood. 1994;84:2912–2918. [PubMed] [Google Scholar]
- 27.Hoeve ten J, Arlinghaus RB, Guo JQ, Heisterkamp N, Groffen J. Tyrosine phosphorylation of CRKL in Philadelphia+ leukemia. Blood. 1994;84:1731–1736. [PubMed] [Google Scholar]
- 28.Lucas CM, et al. BCR-ABL1 tyrosine kinase activity at diagnosis, as determined via the pCrkL/CrkL ratio, is predictive of clinical outcome in chronic myeloid leukaemia. Br J Haematol. 2010;149:458–460. doi: 10.1111/j.1365-2141.2009.08066.x. [DOI] [PubMed] [Google Scholar]
- 29.Guris DL, Fantes J, Tara D, Druker BJ, Imamoto A. Mice lacking the homologue of the human 22q11.2 gene CRKL phenocopy neurocristopathies of DiGeorge syndrome. Nat Genet. 2001;27:293–298. doi: 10.1038/85855. [DOI] [PubMed] [Google Scholar]
- 30.Wang J, et al. Crk and CrkL present with different expression and significance in epithelial ovarian carcinoma. Mol Carcinog. 2011;50:506–515. doi: 10.1002/mc.20745. [DOI] [PubMed] [Google Scholar]
- 31.Mihrshahi R, Brown MH. Downstream of tyrosine kinase 1 and 2 play opposing roles in CD200 receptor signaling. J Immunol. 2010;185:7216–7222. doi: 10.4049/jimmunol.1002858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Donaldson LW, Gish G, Pawson T, Kay LE, Forman-Kay JD. Structure of a regulatory complex involving the Abl SH3 domain, the Crk SH2 domain, and a Crk-derived phosphopeptide. Proc Natl Acad Sci USA. 2002;99:14053–14058. doi: 10.1073/pnas.212518799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Harkiolaki M, Gilbert RJC, Jones EY, Feller SM. The C-terminal SH3 domain of CRKL as a dynamic dimerization module transiently exposing a nuclear export signal. Structure. 2006;14:1741–1753. doi: 10.1016/j.str.2006.09.013. [DOI] [PubMed] [Google Scholar]
- 34.Mittermaier AK, Kay LE. Observing biological dynamics at atomic resolution using NMR. Trends Biochem Sci. 2009;34:601–611. doi: 10.1016/j.tibs.2009.07.004. [DOI] [PubMed] [Google Scholar]
- 35.Tjandra N, Feller S, Pastor R, Bax A. Rotational diffusion anisotropy of human ubiquitin from N-15 NMR relaxation. J Am Chem Soc. 1995;117:12562–12566. [Google Scholar]
- 36.Seo JH, Suenaga A, Hatakeyama M, Taiji M, Imamoto A. Structural and Functional Basis of a Role for CRKL in a Fibroblast Growth Factor 8-Induced Feed-Forward Loop. Mol Cell Biol. 2009;29:3076–3087. doi: 10.1128/MCB.01686-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Cho JH, et al. Tuning protein autoinhibition by domain destabilization. Nat Struct Mol Biol. 2011;18:550–555. doi: 10.1038/nsmb.2039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Senechal K, Heaney C, Druker B, Sawyers C. Structural requirements for function of the Crkl adapter protein in fibroblasts and hematopoietic cells. Mol Cell Biol. 1998;18:5082–5090. doi: 10.1128/mcb.18.9.5082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rosen MK, et al. Direct demonstration of an intramolecular SH2-phosphotyrosine interaction in the Crk protein. Nature. 1995;374:477–479. doi: 10.1038/374477a0. [DOI] [PubMed] [Google Scholar]
- 40.Ren R, Ye ZS, Baltimore D. Abl protein-tyrosine kinase selects the Crk adapter as a substrate using SH3-binding sites. Genes Dev. 1994;8:783–795. doi: 10.1101/gad.8.7.783. [DOI] [PubMed] [Google Scholar]
- 41.Huang X, Wu D, Jin H, Stupack D, Wang JYJ. Induction of cell retraction by the combined actions of Abl-CrkII and Rho-ROCK1 signaling. J Cell Biol. 2008;183:711–723. doi: 10.1083/jcb.200801192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Pawson T. Dynamic control of signaling by modular adaptor proteins. Curr Opin Cell Biol. 2007;19:112–116. doi: 10.1016/j.ceb.2007.02.013. [DOI] [PubMed] [Google Scholar]
- 43.Sawada Y, et al. Force sensing by mechanical extension of the Src family kinase substrate p130Cas. Cell. 2006;127:1015–1026. doi: 10.1016/j.cell.2006.09.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kardinal C, et al. Cell-penetrating SH3 domain blocker peptides inhibit proliferation of primary blast cells from CML patients. Faseb J. 2000;14:1529–1538. doi: 10.1096/fj.14.11.1529. [DOI] [PubMed] [Google Scholar]
- 45.Sriram G, et al. Phosphorylation of Crk on tyrosine 251 in the RT loop of the SH3C domain promotes Abl kinase transactivation. Oncogene. 2011;30:4645–4655. doi: 10.1038/onc.2011.170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Li S. Specificity and versatility of SH3 and other proline-recognition domains: structural basis and implications for cellular signal transduction. Biochem J. 2005;390:641–653. doi: 10.1042/BJ20050411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Severin A, Joseph RE, Boyken S, Fulton DB, Andreotti AH. Proline isomerization preorganizes the Itk SH2 domain for binding to the Itk SH3 domain. J Mol Biol. 2009;387:726–743. doi: 10.1016/j.jmb.2009.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Chan B, et al. SAP couples Fyn to SLAM immune receptors. Nat Cell Biol. 2003;5:155–160. doi: 10.1038/ncb920. [DOI] [PubMed] [Google Scholar]
- 49.Ruckert M, Otting G. Alignment of biological macromolecules in novel nonionic liquid crystalline media for NMR experiments. J Am Chem Soc. 2000;122:7793–7797. [Google Scholar]
- 50.Shen Y, Delaglio F, Cornilescu G, Bax A. TALOS+: a hybrid method for predicting protein backbone torsion angles from NMR chemical shifts. J Biomol NMR. 2009;44:213–223. doi: 10.1007/s10858-009-9333-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.