Abstract
Background. Epidemiological studies suggest that coffee consumption reduces the risk of cancer, but the molecular mechanisms of its chemopreventive effects remain unknown. Objective. To identify differentially expressed genes upon incubation of HT29 colon cancer cells with instant caffeinated coffee (ICC) or caffeic acid (CA) using whole-genome microarrays. Results. ICC incubation of HT29 cells caused the overexpression of 57 genes and the underexpression of 161, while CA incubation induced the overexpression of 12 genes and the underexpression of 32. Using Venn-Diagrams, we built a list of five overexpressed genes and twelve underexpressed genes in common between the two experimental conditions. This list was used to generate a biological association network in which STAT5B and ATF-2 appeared as highly interconnected nodes. STAT5B overexpression was confirmed at the mRNA and protein levels. For ATF-2, the changes in mRNA levels were confirmed for both ICC and CA, whereas the decrease in protein levels was only observed in CA-treated cells. The levels of cyclin D1, a target gene for both STAT5B and ATF-2, were downregulated by CA in colon cancer cells and by ICC and CA in breast cancer cells. Conclusions. Coffee polyphenols are able to affect cyclin D1 expression in cancer cells through the modulation of STAT5B and ATF-2.
1. Introduction
Polyphenols are the most abundant antioxidants in the diet. Their main dietary sources are fruits and plant-derived beverages such as fruit juices, tea, coffee, and red wine. Current evidence strongly supports a contribution of polyphenols to the prevention of cardiovascular diseases, cancers, and osteoporosis suggesting a role of these antioxidants in the prevention of neurodegenerative diseases and diabetes mellitus [1].
It is well established that polyphenol ingestion results in an increase of the plasma-antioxidant capacity. However, there is still some uncertainties about their efficiency to enhance the protection of cellular components, such as lipids or DNA, against oxidative stress in humans [2]. Polyphenols and other antioxidants were thought to protect cell constituents against oxidative damage by scavenging free radicals. However, this concept now appears to be an oversimplified view of their mode of action [3]. More likely, cells respond to polyphenols mainly through direct interactions with receptors or enzymes involved in signal transduction, which may result in modification of the redox status of the cell and may trigger a series of redox-dependent reactions [4]. This could also apply to the anticarcinogenic effects of polyphenols, which properties may be explained by many different mechanisms.
Hydroxycinnamic acids are a major class of polyphenols found in almost every plant [2]. The major representative of hydroxycinnamic acids is caffeic acid, which occurs in food mainly as an ester with quinic acid named chlorogenic acid (5-caffeoylquinic acid). Coffee is a major source of chlorogenic acid in the human diet; the daily intake in coffee drinkers is 0.5–1 g whereas coffee abstainers will usually ingest <100 mg/day. Studies have shown that approximately the 33% of ingested chlorogenic acid and the 95% of caffeic acid are absorbed intestinally [5]. Thus, about two-thirds of ingested chlorogenic acid reach the colon where it is probably metabolized to caffeic acid [6].
Bioavailability data suggest that the biological effects of chlorogenic acid would become apparent after its metabolism to caffeic acid, and hence the need of studying the effects of this acid. Chlorogenic acid and caffeic acid are antioxidants in vitro [7], and they might inhibit the formation of mutagenic and carcinogenic N-nitroso compounds since they are inhibitors of the N-nitrosation reaction in vivo [8]. Furthermore, chlorogenic acid can inhibit DNA damage in vitro [9] as it inhibits lipid peroxidation-induced DNA adduct formation [10] and suppresses reactive oxygen species-mediated nuclear factor (NF-κB), activator protein-1 (AP-1), and mitogen-activated protein kinase activation by upregulating antioxidant enzymes [11]. These studies suggested that coffee polyphenols are potent chemopreventive agents.
Recent meta-analyses demonstrate inverse associations between coffee intake and the risk of colon, liver, breast, and endometrial cancer [12–15]. Moreover, in prospective population-based cohort studies, the inverse association between coffee consumption and risk of cancer has been shown. The group of Naganuma [16] found that the consumption of at least one cup of coffee per day was associated with a 49% lower risk of upper gastrointestinal cancer in a Japanese population, while Wilson and collaborators [17] found that men who regularly drink coffee appeared to have a lower risk of developing a lethal form of prostate cancer. The lower risk was evident when consuming either regular or decaffeinated coffee. It has been proposed that the inverse association between coffee intake and colon cancer could be explained, at least in part, by the presence of chlorogenic acid in coffee [18]. Ganmaa et al. [19] observed a general protective effect of caffeine intake on breast cancer risk for both ER subtypes, but the effect was only found to be significant for ER-positive breast cancers. In this study, the association between caffeine and breast cancer was stronger among postmenopausal women with estrogen-receptor and progesterone-receptor-positive breast cancer than those with estrogen-receptor and progesterone-receptor negative breast cancer [19]. In another study, coffee drinking specifically reduced the risk of developing ER-negative breast cancer but not ER-positive breast cancer [20].
Although there is enough evidence from epidemiological data supporting that coffee seems to reduce the risk of certain cancers, the molecular mechanisms underlying the chemopreventive effects of coffee remain unknown. For this reason, the aim of our study was to determine the effect at the molecular level of coffee polyphenols at low concentrations equivalent to one cup of coffee, using as a model a human colon cancer cell line HT29 in a nutrigenomic approach. Furthermore, the effect of coffee polyphenols was also evaluated in breast cancer cells.
2. Materials and Methods
2.1. Materials and Chemicals
Cells were incubated with Instant Caffeinated Coffee (ICC) (regular lyophilized instant coffee) and Caffeic acid (CA, Sigma). Compounds were dissolved either in DMSO (CA), or sterile water (ICC), and stored at −20°C.
2.2. Cell Culture
Colon adenocarcinoma HT29 and breast cancer MCF-7 cell lines were routinely grown in Ham's F12 medium supplemented with 7% fetal bovine serum (FBS, both from Gibco) at 37°C in a 5% CO2 humidified atmosphere in 10 cm dish, or in 33 mm plate.
Cells were incubated with ICC or CA at concentrations equivalent to one cup of coffee. The concentrations used in cell incubations, 7 μg/mL in H2O mQ for ICC and 1.68 μg/mL in DMSO for CA, respectively, took into account the amount of these compounds in one cup of coffee and their distribution in a regular human body with 75% water content. These concentrations did not cause any cytotoxic effect in the cell incubations as determined by the MTT assay [21].
2.3. Microarrays
Gene expression was analyzed by hybridization to The GeneChip Human Genome U133A plus 2.0 microarrays from Affymetrix, containing 47,000 transcripts and variants. HT29 cells were incubated with ICC and CA for 24 h. Total RNA was prepared from triplicate samples using Speedtools Total RNA Extraction Kit (Biotools) following the recommendations of the manufacturer. RNA quality was tested by 2100 Bioanalyzer Eukaryote Total RNA Nano Series II (Agilent Technologies). Labeling, hybridization, and detection were carried out following the manufacturer's specifications at the IDIBAPS Genomic Service (Hospital Clínic, Barcelona).
2.4. Microarray Data Analyses
Quantification was carried out with GeneSpring GX v.11.5.1 software (Agilent Technologies), which allows multifilter comparisons using data from different experiments to perform the normalization, generation of lists, and the functional classification of the differentially expressed genes. The input data was subjected to preprocess baseline transformation using the Robust Multiarray Average summarization algorithm using the median of control samples. After grouping the triplicate of each experimental condition, list of differentially expressed genes could be generated by using volcano plot analysis. The expression of each gene is reported as the ratio of the value obtained after each condition relative to control condition after normalization and statistical analysis of the data. The corrected P value cutoff applied was of <0.05; then the output of this statistical analysis was filtered by fold expression, selecting specifically those genes that had a differential expression of at least 1.3-fold. Gene classification was established by the Gene Ontology database.
2.5. Common Genes between ICC and CA Treatments
Common genes were selected from the lists of differentially expressed genes for each treatment using Venn-Diagrams. The newly generated list contained both over and underexpressed genes.
2.6. Generation of Biological Association Networks
BANs were constructed with the aid of the Pathway Analysis within the GeneSpring v.11.5.1 (Agilent) as described in Selga et al. [22] with the list of common genes differentially expressed in both treatments. A filtered screening was processed by the program between our data and bibliographic interaction databases up to a total of 100 related genes. Network associations were confirmed in the literature.
2.7. RT Real-Time PCR
Total RNA was extracted from HT29 cells using Ultraspec (Biotex) in accordance with the manufacturer's instructions.
Complementary DNA was synthesized as described in Selga et al. [23] and the cDNA product was used for amplification by real time PCR. STAT5B and ATF-2 mRNA levels were determined in an ABI Prism 7000 Sequence Detection System (Applied Biosystems) using 3 μL of the cDNA reaction and the assays-on-demand Hs00560035_m1 for STAT5B, Hs00153179_ml for ATF-2, and Hs00356991_m1 for APRT (all from Applied Biosystems). APRT mRNA was used as an endogenous control. The reaction was performed following the manufacturers recommendations. Fold changes in gene expression were calculated using the standard ΔΔCt method.
2.8. Western Blot
Whole extracts were obtained from 2.5 × 106 control or treated cells according to Selga et al. [23]. Five μL of the extract was used to determine protein concentration by the Bradford assay (Bio-Rad). The extracts were frozen in liquid N2 and stored at −80°C. Total extracts (50 μg) were resolved on SDS-polyacrylamide gels and transferred to PVDF membranes (Immobilon P, Millipore) using a semidry electroblotter.
The SNAP i.d. protein detection system technology (Millipore) was used to probe the membranes. This system applies vacuum through the membrane to actively drive reagents to protein locations, unlike the traditional technique of diffusion over the membrane as a reagent transport. Table 1 compiles the antibodies used in the different determinations.
Table 1.
| Antibody | Molecular weight (KDa) | Dilution used | Supplier |
|---|---|---|---|
| STAT5B | 95 | 1: 200 | sc-835, Santa Cruz Biotechnology Inc. |
| ATF-2 | 72 | 1: 200 | sc-6233, Santa Cruz Biotechnology Inc. |
| Cyclin D1 | 38 | 1: 200 | sc-8396, Santa Cruz Biotechnology Inc. |
| β-actin | 42 | 1: 200 | A2066, Sigma |
| Tubulin | 60 | 1: 100 | CP06, Calbiochem |
Signals were detected by secondary horseradish peroxidase-conjugated antibody, either anti-rabbit (1 : 5000 or 1 : 10000 dilution; Dako) or anti-mouse (1 : 2500 dilution, Amersham NIF 824) and enhanced chemiluminescence using the ECL method, as recommended by the manufacturer (Amersham). Chemiluminescence was detected with ImageQuant LAS 4000 Mini technology (GE Healthcare).
2.9. Statistical Methods
For the RT-PCR and Western blot analyses, values are expressed as the mean ± SE of three different experiments. Data were evaluated by unpaired Student's t test, and analyses were performed using the PASW Statistics v. 18.0.0. software.
3. Results
3.1. Effect of ICC and CA Incubations in HT29 Gene Expression
The expression profile of over 47,000 transcripts and variants included in the microarray HG U133 plus 2.0 from Affymetrix was compared between HT29 control cells and cells incubated with either CA or ICC, at nontoxic concentrations for 24 h. GeneSpring GX software v.11.5.1 was used to analyze the results. A list of differentially expressed genes by 1.3-fold with a P value cutoff of <0.05 was generated as described in Methods. When HT29 cells were incubated with ICC, 57 genes were overexpressed whereas 161 genes were underexpressed. Among the overexpressed genes, 24% belonged to the Transcription factors category and 19% to Cell cycle or to Biosynthetic processes. Within the underexpressed genes, the category corresponding to cell cycle was the most affected (53% of the genes) followed by Transcription factors (19%) and Biosynthetic processes (12%). Upon incubation with CA, 12 genes were overexpressed whereas 32 genes were underexpressed. Among the overexpressed genes, 33% belonged to the Transcription factors category, 25% to Cell cycle, and 16,7% to Biosynthetic processes or immune response. Within the underexpressed genes, again the category corresponding to Cell cycle was the most affected (30% of the genes) followed by Biosynthetic processes (15%) and Transcription factors (12%). The lists of differentially expressed genes are presented as Tables 2, 3, 4, and 5. The data presented in this work have been deposited in the Gene Expression Omnibus (GEO, http://www.ncbi.nlm.nih.gov/geo/) and are accessible through GEO series accession number [GSM867162].
Table 2.
List of overexpressed genes in HT29 cells upon incubation with instant caffeinated coffee.
| Gene symbol | Gene title | P value | FC absolute | Regulation |
|---|---|---|---|---|
| CALM3 | Calmodulin 3 (phosphorylase kinase, delta) | 0.016 | 1.3 | Up |
| CDC42EP1 | CDC42 effector protein (Rho GTPase binding) 1 | 0.027 | 1.3 | Up |
| FOXN3 | Forkhead box N3 | 0.022 | 1.3 | Up |
| KIR2DL1 | Killer cell immunoglobulin-like receptor, two domains, long cytoplasmic tail, 1 | 0.023 | 1.3 | Up |
| ORAI2 | ORAI calcium release-activated calcium modulator 2 | 0.011 | 1.3 | Up |
| RAPGEF1 | Rap guanine nucleotide exchange factor (GEF) 1 | 0.022 | 1.3 | Up |
| STH | Saitohin | 0.031 | 1.3 | Up |
| SLC39A3 | Solute carrier family 39 (zinc transporter), member 3 | 0.028 | 1.3 | Up |
| ZNF397OS | Zinc finger protein 397 opposite strand | 0.024 | 1.3 | Up |
| ZP4 | Zona pellucida glycoprotein 4 | 0.046 | 1.3 | Up |
| FGFRL1 | Fibroblast growth factor receptor-like 1 | 0.035 | 1.31 | Up |
| ITGA9 | Integrin, alpha 9 | 0.002 | 1.31 | Up |
| IRAK1 | Interleukin-1 receptor-associated kinase 1 | 0.038 | 1.31 | Up |
| OBSL1 | Obscurin-like 1 | 0.008 | 1.31 | Up |
| RPS17L4 | Ribosomal protein S17-like 4 | 0.026 | 1.31 | Up |
| STAT5B | Signal transducer and activator of transcription 5B | 0.007 | 1.31 | Up |
| TRABD | TraB domain containing | 0.043 | 1.31 | Up |
| MYO9B | Myosin IXB | 0.041 | 1.32 | Up |
| NME7 | Nonmetastatic cells 7, protein expressed in (nucleoside-diphosphate kinase) | 0.037 | 1.32 | Up |
| RPS6KA4 | Ribosomal protein S6 kinase, 90 kDa, polypeptide 4 | 0.014 | 1.32 | Up |
| SIRPA | Signal-regulatory protein alpha | 0.019 | 1.32 | Up |
| TBX20 | T-box 20 | 0.035 | 1.32 | Up |
| TCF20 | Transcription factor 20 (AR1) | 0.022 | 1.32 | Up |
| ALDH3B1 | Aldehyde dehydrogenase 3 family, member B1 | 0.005 | 1.33 | Up |
| BGN | Biglycan | 0.029 | 1.33 | Up |
| GNB4 | Guanine nucleotide binding-protein (G protein), b-polypeptide 4 | 0.044 | 1.33 | Up |
| IFNA17 | Interferon, alpha 17 | 0.026 | 1.33 | Up |
| KY | Kyphoscoliosis peptidase | 0.013 | 1.33 | Up |
| SCARF1 | Scavenger receptor class F, member 1 | 0.025 | 1.33 | Up |
| SERPINB8 | Serpin peptidase inhibitor, clade B (ovalbumin), member 8 | 0.01 | 1.33 | Up |
| FST | Follistatin | 0.025 | 1.34 | Up |
| MOGAT1 | Monoacylglycerol O-acyltransferase 1 | 0.009 | 1.34 | Up |
| PPARGC1A | Peroxisome proliferator-activated receptor gamma, coactivator 1 alpha | 0.015 | 1.34 | Up |
| SUCLG2 | Succinate-CoA ligase, GDP-forming, beta subunit | 0.011 | 1.34 | Up |
| SULT1B1 | Sulfotransferase family, cytosolic, 1B, member 1 | 0.018 | 1.34 | Up |
| TBX10 | T-box 10 | 0.011 | 1.34 | Up |
| ZNF503 | Zinc finger protein 503 | 0.022 | 1.34 | Up |
| HBA1 | Hemoglobin, alpha 1 | 0.04 | 1.35 | Up |
| MEPE | Matrix, extracellular phosphoglycoprotein with ASARM motif | 0.001 | 1.35 | Up |
| PPP1CB | Protein phosphatase 1, catalytic subunit, beta isoform | 0.03 | 1.35 | Up |
| ARV1 | ARV1 homolog (S. cerevisiae) | 0.011 | 1.36 | Up |
| BCL3 | B-cell CLL/lymphoma 3 | 0.034 | 1.36 | Up |
| CTRC | Chymotrypsin C (caldecrin) | 0.045 | 1.36 | Up |
| EPOR | Erythropoietin receptor | 0.008 | 1.37 | Up |
| HMGA1 | High-mobility group AT-hook 1 | 0.039 | 1.37 | Up |
| IL19 | Interleukin 19 | 0.018 | 1.38 | Up |
| ABCC12 | ATP-binding cassette, subfamily C (CFTR/MRP), member 12 | 6.00E-04 | 1.39 | Up |
| RAI1 | Retinoic acid induced 1 | 0.017 | 1.39 | Up |
| KLF5 | Kruppel-like factor 5 (intestinal) | 0.028 | 1.4 | Up |
| CBWD1 | COBW domain containing 1 | 0.044 | 1.41 | Up |
| ASAH3 | N-acylsphingosine amidohydrolase (alkaline ceramidase) 3 | 0.039 | 1.43 | Up |
| ABHD14B | Abhydrolase domain containing 14B | 0.03 | 1.45 | Up |
| TLN1 | Talin 1 | 0.049 | 1.45 | Up |
| ARHGAP23 | Rho GTPase-activating protein 23 | 0.024 | 1.65 | Up |
| HINT3 | Histidine triad nucleotide binding protein 3 | 0.002 | 1.77 | Up |
| ARHGDIA | Rho GDP dissociation inhibitor (GDI) alpha | 0.034 | 1.83 | Up |
| CALR | Calreticulin | 0.007 | 1.93 | Up |
The table shows the list of overexpressed genes by 1.3-fold with a P value <0.05 obtained in cells treated with instant caffeinated coffee and includes the gene symbol for all genes, and their associated description. The ratio columns correspond to the absolute fold change in expression relative to the control group and the type of regulation (up: upregulation).
Table 3.
List of underexpressed genes in HT29 cells upon incubation with instant coffee.
| Gene symbol | Gene title | P value | FC absolute | Regulation |
|---|---|---|---|---|
| ACBD5 | Acyl-coenzyme A binding domain containing 5 | 0.017 | 1.3 | Down |
| CXADR | Coxsackie virus and adenovirus receptor | 0.015 | 1.3 | Down |
| FANCD2 | Fanconi anemia, complementation group D2 | 0.047 | 1.3 | Down |
| FRYL | FRY-like | 0.039 | 1.3 | Down |
| NUB1 | Negative regulator of ubiquitin-like proteins 1 | 0.029 | 1.3 | Down |
| PBRM1 | Polybromo 1 | 0.004 | 1.3 | Down |
| PRKACB | Protein kinase, cAMP-dependent, catalytic, beta | 0.033 | 1.3 | Down |
| RIF1 | RAP1 interacting factor homolog (yeast) | 0.012 | 1.3 | Down |
| SLC39A6 | Solute carrier family 39 (zinc transporter), member 6 | 0.022 | 1.3 | Down |
| TMEM170 | Transmembrane protein 170 | 0.032 | 1.3 | Down |
| WDR26 | WD repeat domain 26 | 0.028 | 1.3 | Down |
| RNGTT | RNA guanylyltransferase and 5′-phosphatase | 0.04 | 1.3 | Down |
| CTDSPL2 | CTD small phosphatase like 2 | 0.03 | 1.3 | Down |
| ZC3H11A | Zinc finger CCCH-type containing 11A | 0.014 | 1.3 | Down |
| TMOD3 | Tropomodulin 3 (ubiquitous) | 0.0171 | 1.3 | Down |
| CPD | Carboxypeptidase D | 0.002 | 1.31 | Down |
| CBL | Cas-Br-M ecotropic retroviral transforming sequence | 0.008 | 1.31 | Down |
| CDC42SE2 | CDC42 small effector 2 | 0.022 | 1.31 | Down |
| CLN5 | Ceroid-lipofuscinosis, neuronal 5 | 0.001 | 1.31 | Down |
| DDX3X | DEAD (Asp-Glu-Ala-Asp) box polypeptide 3, X-linked | 0.027 | 1.31 | Down |
| FGFR1OP2 | FGFR1 oncogene partner 2 | 0.049 | 1.31 | Down |
| LRRFIP1 | Leucine-rich repeat (in FLII) interacting protein 1 | 0.026 | 1.31 | Down |
| PDCD4 | Programmed cell death 4 | 0.005 | 1.31 | Down |
| REPS2 | RALBP1-associated Eps domain containing 2 | 0.046 | 1.31 | Down |
| SLC7A6 | Solute carrier family 7, member 6 | 0.002 | 1.31 | Down |
| TFRC | Transferrin receptor (p90, CD71) | 0.038 | 1.31 | Down |
| TMEM19 | Transmembrane protein 19 | 0.024 | 1.31 | Down |
| AGPS | Alkylglycerone phosphate synthase | 0.001 | 1.31 | Down |
| SLC4A7 | Solute carrier family 4, member 7 | 0.028 | 1.31 | Down |
| SPTAN1 | Spectrin, alpha, nonerythrocytic 1 (alpha-fodrin) | 0.02 | 1.31 | Down |
| GPD2 | Glycerol-3-phosphate dehydrogenase 2 (mitochondrial) | 0.033 | 1.31 | Down |
| BICD1 | Bicaudal D homolog 1 (Drosophila) | 0.008 | 1.31 | Down |
| FBXW11 | F-box and WD repeat domain containing 11 | 0.025 | 1.31 | Down |
| BCLAF1 | BCL2-associated transcription factor 1 | 0.025 | 1.32 | Down |
| CDH1 | Cadherin 1, type 1, E-cadherin (epithelial) | 0.011 | 1.32 | Down |
| CLK4 | CDC-like kinase 4 | 0.049 | 1.32 | Down |
| PTAR1 | Protein prenyltransferase alpha subunit repeat containing 1 | 0.027 | 1.32 | Down |
| SMEK2 | SMEK homolog 2, suppressor of mek1 (Dictyostelium) | 0.012 | 1.32 | Down |
| CEPT1 | Choline/ethanolamine phosphotransferase 1 | 0.038 | 1.32 | Down |
| SAR1A | SAR1 gene homolog A (S. cerevisiae) | 0.033 | 1.32 | Down |
| PDGFC | Platelet-derived growth factor C | 0.02 | 1.32 | Down |
| NFAT5 | Nuclear factor of activated T-cells 5, tonicity responsive | 0.045 | 1.32 | Down |
| FRS2 | Fibroblast growth factor receptor substrate 2 | 0.03 | 1.32 | Down |
| BMS1P5 | BMS1 pseudogene 5 | 0.036 | 1.33 | Down |
| GLS | Glutaminase | 5.00E-04 | 1.33 | Down |
| LMAN1 | Lectin, mannose binding, 1 | 7.00E-04 | 1.33 | Down |
| ARHGAP18 | Rho GTPase-activating protein 18 | 8.00E-04 | 1.33 | Down |
| ARHGAP5 | Rho GTPase-activating protein 5 | 0.006 | 1.33 | Down |
| CCNE2 | Cyclin E2 | 0.036 | 1.33 | Down |
| SPCS3 | Signal peptidase complex subunit 3 homolog (S. cerevisiae) | 0.008 | 1.33 | Down |
| NCOA2 | Nuclear receptor coactivator 2 | 0.005 | 1.33 | Down |
| SRPRB | Signal recognition particle receptor, B subunit | 0.018 | 1.33 | Down |
| TLK1 | Tousled-like kinase 1 | 0.04 | 1.33 | Down |
| NCOA3 | Nuclear receptor coactivator 3 | 0.048 | 1.33 | Down |
| STRN3 | Striatin, calmodulin-binding protein 3 | 2.00E-04 | 1.33 | Down |
| AP1G1 | Adaptor-related protein complex 1, gamma 1 subunit | 0.004 | 1.34 | Down |
| B3GALNT2 | Beta-1,3-N-acetylgalactosaminyltransferase 2 | 0.034 | 1.34 | Down |
| PPHLN1 | Periphilin 1 | 2.00E-04 | 1.34 | Down |
| SNX13 | Sorting nexin 13 | 0.001 | 1.34 | Down |
| TMED2 | Transmembrane emp24 domain-trafficking protein 2 | 0.041 | 1.34 | Down |
| BRWD1 | Bromodomain and WD repeat domain containing 1 | 0.011 | 1.34 | Down |
| HLA-B | Major histocompatibility complex, class I, B | 0.028 | 1.34 | Down |
| CHP | Calcium-binding protein P22 | 0.002 | 1.34 | Down |
| MTMR9 | Myotubularin-related protein 9 | 0.026 | 1.34 | Down |
| DCUN1D4 | DCN1, defective in cullin neddylation 1, domain containing 4 | 0.031 | 1.34 | Down |
| ARL6IP2 | ADP-ribosylation factor-like 6 interacting protein 2 | 0.02 | 1.35 | Down |
| GLIS3 | GLIS family zinc finger 3 | 0.01 | 1.35 | Down |
| LARP4 | La ribonucleoprotein domain family, member 4 | 0.019 | 1.35 | Down |
| PTPLB | Protein tyrosine phosphatase-like member b | 0.036 | 1.35 | Down |
| TRAM1 | Translocation-associated membrane protein 1 | 0.002 | 1.35 | Down |
| TMEM64 | Transmembrane protein 64 | 0.001 | 1.35 | Down |
| CBFB | Core-binding factor, beta subunit | 0.005 | 1.35 | Down |
| SELT | Selenoprotein T | 0.002 | 1.35 | Down |
| PEX13 | Peroxisome biogenesis factor 13 | 0.011 | 1.35 | Down |
| TNKS2 | TRF1-interacting ankyrin-related ADP-ribose polymerase 2 | 0.034 | 1.35 | Down |
| TMPO | Thymopoietin | 0.001 | 1.35 | Down |
| LIN7C | Lin-7 homolog C (C. elegans) | 0.007 | 1.35 | Down |
| MTA2 | Metastasis-associated 1 family, member 2 | 0.013 | 1.36 | Down |
| TMEM168 | Transmembrane protein 168 | 0.035 | 1.36 | Down |
| CREBZF | CREB/ATF bZIP transcription factor | 0.016 | 1.36 | Down |
| OSTF1 | Osteoclast-stimulating factor 1 | 0.002 | 1.36 | Down |
| WDR57 | WD repeat domain 57 (U5 snRNP specific) | 0.001 | 1.36 | Down |
| GLT25D1 | Glycosyltransferase 25 domain containing 1 | 0.008 | 1.36 | Down |
| NAPG | N-ethylmaleimide-sensitive factor attachment protein, gamma | 0.015 | 1.36 | Down |
| CCDC126 | Coiled-coil domain containing 126 | 0.039 | 1.37 | Down |
| LASS6 | LAG1 homolog, ceramide synthase 6 | 0.005 | 1.37 | Down |
| MYSM1 | Myb-like, SWIRM and MPN domains 1 | 0.021 | 1.37 | Down |
| CYP51A1 | Cytochrome P450, family 51, subfamily A, polypeptide 1 | 0.007 | 1.37 | Down |
| PDE4DIP | Phosphodiesterase 4D interacting protein (myomegalin) | 0.024 | 1.37 | Down |
| SAP30L | SAP30-like | 0.012 | 1.37 | Down |
| PTPRJ | Protein tyrosine phosphatase, receptor type, J | 0.011 | 1.37 | Down |
| PGGT1B | Protein geranylgeranyltransferase type I, beta subunit | 9.00E-04 | 1.37 | Down |
| ASPH | Aspartate beta-hydroxylase | 0.011 | 1.37 | Down |
| SEMA3C | Sema domain, (semaphorin) 3C | 0.036 | 1.38 | Down |
| WDR76 | WD repeat domain 76 | 0.016 | 1.38 | Down |
| ATP13A3 | ATPase-type 13A3 | 0.002 | 1.38 | Down |
| LMBR1 | Limb region 1 homolog (mouse) | 0.014 | 1.38 | Down |
| GLUD1 | Glutamate dehydrogenase 1 | 0.001 | 1.39 | Down |
| GSTCD | Glutathione S-transferase, C-terminal domain containing | 0.029 | 1.39 | Down |
| SPTLC1 | Serine palmitoyltransferase, subunit 1 | 0.02 | 1.39 | Down |
| U2AF1 | U2 small nuclear RNA auxiliary factor 1 | 9.00E-04 | 1.39 | Down |
| UHMK1 | U2AF homology motif (UHM) kinase 1 | 0.007 | 1.39 | Down |
| ARGLU1 | Arginine and glutamate-rich 1 | 6.00E-04 | 1.39 | Down |
| ANKRD12 | Ankyrin repeat domain 12 | 0.03 | 1.39 | Down |
| PPP3R1 | Protein phosphatase 3, regulatory subunit B, alpha isoform | 0.023 | 1.39 | Down |
| XRN1 | 5′-3′ exoribonuclease 1 | 0.019 | 1.4 | Down |
| CLSPN | Claspin homolog (Xenopus laevis) | 0.013 | 1.4 | Down |
| CXADRP1 | Coxsackie virus and adenovirus receptor pseudogene 1 | 0.034 | 1.4 | Down |
| G3BP1 | GTPase-activating protein- (SH3 domain) binding protein 1 | 0.002 | 1.4 | Down |
| TMEM30A | Transmembrane protein 30A | 0.01 | 1.4 | Down |
| CLCN3 | Chloride channel 3 | 0.035 | 1.41 | Down |
| STK4 | Serine/threonine kinase 4 | 0.039 | 1.41 | Down |
| ZNF644 | Zinc finger protein 644 | 0.02 | 1.41 | Down |
| TCP11L1 | T-complex 11 (mouse)-like 1 | 0.014 | 1.41 | Down |
| SFRS6 | Splicing factor, arginine/serine-rich 6 | 0.031 | 1.41 | Down |
| NPL | N-acetylneuraminate pyruvate lyase | 0.006 | 1.41 | Down |
| G3BP2 | GTPase-activating protein- (SH3 domain) binding protein 2 | 0.001 | 1.42 | Down |
| HNRNPU | Heterogeneous nuclear ribonucleoprotein U | 0.01 | 1.42 | Down |
| TBL1XR1 | Transducin (beta)-like 1 X-linked receptor 1 | 0.001 | 1.42 | Down |
| PHTF2 | Putative homeodomain transcription factor 2 | 0.002 | 1.42 | Down |
| ADAM10 | ADAM metallopeptidase domain 10 | 0.011 | 1.43 | Down |
| ADAM9 | ADAM metallopeptidase domain 9 (meltrin gamma) | 0.01 | 1.43 | Down |
| MALAT1 | Metastasis-associated lung adenocarcinoma transcript 1 | 0.04 | 1.43 | Down |
| SCARB2 | Scavenger receptor class B, member 2 | 0.001 | 1.43 | Down |
| CANX | Calnexin | 0.043 | 1.43 | Down |
| CASP2 | Caspase 2, apoptosis-related cysteine peptidase | 0.033 | 1.43 | Down |
| TRPS1 | Trichorhinophalangeal syndrome I | 0.005 | 1.44 | Down |
| ZFX | Zinc finger protein, X-linked | 0.033 | 1.44 | Down |
| SGPL1 | Sphingosine-1-phosphate lyase 1 | 0.04 | 1.44 | Down |
| PTPN11 | Protein tyrosine phosphatase, nonreceptor type 11 | 0.045 | 1.44 | Down |
| SFRS11 | Splicing factor, arginine/serine-rich 11 | 0.045 | 1.45 | Down |
| B3GNT5 | Beta-1,3-N-acetylglucosaminyltransferase 5 | 0.021 | 1.45 | Down |
| MAP3K1 | Mitogen-activated protein kinase kinase kinase 1 | 0.019 | 1.45 | Down |
| SNHG4 | Small nucleolar RNA host gene (nonprotein coding) 4 | 0.004 | 1.46 | Down |
| PARD6B | Par-6 partitioning defective 6 homolog beta (C. elegans) | 0.04 | 1.46 | Down |
| ROD1 | ROD1 regulator of differentiation 1 (S. pombe) | 0.001 | 1.46 | Down |
| SPTBN1 | Spectrin, beta, nonerythrocytic 1 | 0.02 | 1.48 | Down |
| TXNDC1 | Thioredoxin domain containing 1 | 0.013 | 1.48 | Down |
| ATF2 | Activating transcription factor 2 | 0.005 | 1.48 | Down |
| RDX | Radixin | 0.043 | 1.48 | Down |
| SCAMP1 | Secretory carrier membrane protein 1 | 0.009 | 1.48 | Down |
| PTAR1 | Protein prenyltransferase alpha subunit repeat containing 1 | 0.018 | 1.49 | Down |
| RC3H2 | Ring finger and CCCH-type zinc finger domains 2 | 0.0037 | 1.49 | Down |
| ADAM17 | ADAM metallopeptidase domain 17 | 0.007 | 1.49 | Down |
| FAM76B | Family with sequence similarity 76, member B | 0.014 | 1.5 | Down |
| ITGB8 | Integrin, beta 8 | 1.00E-04 | 1.5 | Down |
| TRIM23 | Tripartite motif-containing 23 | 0.005 | 1.5 | Down |
| CASC5 | Cancer susceptibility candidate 5 | 0.019 | 1.52 | Down |
| SLC16A1 | Solute carrier family 16, member 1 | 0.002 | 1.52 | Down |
| FNBP1 | Formin-binding protein 1 | 0.037 | 1.53 | Down |
| PRKAR1A | Protein kinase, cAMP-dependent, regulatory, type I, alpha | 9.00E-04 | 1.53 | Down |
| B4GALT1 | Beta 1,4-galactosyltransferase, polypeptide 1 | 0.035 | 1.55 | Down |
| MDM4 | Mdm4 p53-binding protein homolog (mouse) | 0.011 | 1.58 | Down |
| FGD4 | FYVE, RhoGEF, and PH domain containing 4 | 0.001 | 1.59 | Down |
| UBA6 | Ubiquitin-like modifier activating enzyme 6 | 8.00E-04 | 1.62 | Down |
| ZDHHC21 | Zinc finger, DHHC-type containing 21 | 0.036 | 1.64 | Down |
| REEP3 | Receptor accessory protein 3 | 7.00E-04 | 1.65 | Down |
| SSR3 | Signal sequence receptor, gamma | 0.014 | 1.65 | Down |
| ZDHHC20 | Zinc finger, DHHC-type containing 20 | 0.003 | 1.66 | Down |
| EIF2S3 | Eukaryotic translation initiation factor 2, subunit 3 gamma | 0.001 | 1.7 | Down |
| HNRNPH1 | Heterogeneous nuclear ribonucleoprotein H1 | 0.011 | 1.79 | Down |
| ATL3 | Atlastin 3 | 0.001 | 2.02 | Down |
The table shows the list of underexpressed genes by 1.3-fold with a P value <0.05 obtained in cells treated with instant caffeinated coffee and includes the gene symbol for all genes, and their associated description. The ratio columns correspond to the absolute fold change in expression relative to the control group and the type of regulation (down: downregulation).
Table 4.
List of overexpressed genes in HT29 cells upon incubation with caffeic acid.
| Gene symbol | Gene title | P value | FC absolute | Regulation |
|---|---|---|---|---|
| SULT1B1 | Sulfotransferase family, cytosolic, 1B, member 1 | 0.02 | 1.3 | Up |
| BCL6B | B-cell CLL/lymphoma 6, member B (zinc finger protein) | 3.00E-04 | 1.3 | Up |
| KCNJ5 | Potassium inwardly-rectifying channel, subfamily J, member 5 | 0.01 | 1.31 | Up |
| EPOR | Erythropoietin receptor | 0.02 | 1.32 | Up |
| DNAJC21 | DnaJ (Hsp40) homolog, subfamily C, member 21 | 0.049 | 1.33 | Up |
| STAT5B | Signal transducer and activator of transcription 5B | 0.012 | 1.33 | Up |
| FST | Follistatin | 0.021 | 1.37 | Up |
| CD84 | CD84 molecule | 0.033 | 1.37 | Up |
| THRA | Thyroid hormone receptor, alpha | 0.017 | 1.37 | Up |
| MAPK8IP3 | Mitogen-activated protein kinase 8 interacting protein 3 | 0.028 | 1.4 | Up |
| SIAE | Sialic acid acetylesterase | 0.01 | 2.42 | Up |
| HINT3 | Histidine triad nucleotide-binding protein 3 | 0.033 | 2.6 | Up |
The table shows the list of overexpressed genes by 1.3-fold with a P value <0.05 obtained in cells treated with caffeic acid and includes the gene symbol for all genes, their associated description. The ratio columns correspond to the absolute fold change in expression relative to the control group and the type of regulation (up: upregulation).
Table 5.
List of underexpressed genes in HT29 cells upon incubation with caffeic acid.
| Gene symbol | Gene title | P value | FC absolute | Regulation |
|---|---|---|---|---|
| MFSD7 | Major facilitator superfamily domain containing 7 | 1.00E-04 | 1.3 | Down |
| MSI2 | Musashi homolog 2 (Drosophila) | 0.027 | 1.3 | Down |
| CDA | Cytidine deaminase | 2.00E-04 | 1.31 | Down |
| DEFB1 | Defensin, beta 1 | 0.026 | 1.31 | Down |
| PIP5K1A | Phosphatidylinositol-4-phosphate 5-kinase, type I, alpha | 0.027 | 1.31 | Down |
| ZDHHC20 | Zinc finger, DHHC-type containing 20 | 0.005 | 1.31 | Down |
| ZDHHC21 | Zinc finger, DHHC-type containing 21 | 0.016 | 1.31 | Down |
| SLC4A7 | Solute carrier family 4, member 7 | 0.0249 | 1.32 | Down |
| CEACAM1 | Carcinoembryonic antigen-related cell adhesion molecule 1 | 0.0459 | 1.32 | Down |
| PDZRN3 | PDZ domain containing RING finger 3 | 0.002 | 1.32 | Down |
| WDR62 | WD repeat domain 62 | 0.005 | 1.32 | Down |
| FAM76B | Family with sequence similarity 76, member B | 0.036 | 1.32 | Down |
| TCF21 | Transcription factor 21 | 0.029 | 1.33 | Down |
| TBL1XR1 | Transducin (beta)-like 1 X-linked receptor 1 | 6.00E-04 | 1.33 | Down |
| CLK4 | CDC-like kinase 4 | 0.021 | 1.33 | Down |
| CYP2A13 | Cytochrome P450, family 2, subfamily A, polypeptide 13 | 0.009 | 1.34 | Down |
| CXCR4 | Chemokine (C-X-C motif) receptor 4 | 0.0488 | 1.34 | Down |
| ATF2 | Activating transcription factor 2 | 0.0158 | 1.35 | Down |
| PDE10A | Phosphodiesterase 10A | 0.03 | 1.35 | Down |
| METT10D | Methyltransferase 10 domain containing | 0.003 | 1.35 | Down |
| PRMT2 | Protein arginine methyltransferase 2 | 7.00E-04 | 1.36 | Down |
| GLS | Glutaminase | 5.70E-04 | 1.37 | Down |
| SLC38A5 | Solute carrier family 38, member 5 | 0.043 | 1.37 | Down |
| TINAG | Tubulointerstitial nephritis antigen | 0.043 | 1.38 | Down |
| AQP1 | Aquaporin 1 (Colton blood group) | 0.0221 | 1.4 | Down |
| JMJD6 | Jumonji domain containing 6 | 0.004 | 1.4 | Down |
| SAP30L | SAP30-like | 0.021 | 1.4 | Down |
| FGD4 | FYVE, RhoGEF, and PH domain containing 4 | 0.026 | 1.52 | Down |
| S100A2 | S100 calcium-binding protein A2 | 0.005 | 1.53 | Down |
| CTSZ | Cathepsin Z | 0.045 | 1.53 | Down |
| SLC4A4 | Solute carrier family 4, member 4 | 9.00E-04 | 1.54 | Down |
| AGR3 | Anterior gradient homolog 3 (Xenopus laevis) | 0.011 | 1.69 | Down |
The table shows the list of underexpressed genes by 1.3-fold with a P value <0.05 obtained in cells treated with caffeic acid and includes the gene symbol for all genes, their associated description. The ratio columns correspond to the absolute fold change in expression relative to the control group and the type of regulation (down: downregulation).
3.2. Generation of Biological Association Networks
A Biological Association Network (BAN) was constructed using the Pathway Analysis within GeneSpring v.11.5.1 as described in Methods using as the starting list the common genes differentially expressed upon incubation with CA and ICC. This list included five overexpressed genes and twelve underexpressed genes (Table 6). In the generated network, signal transducer and activator of transcription 5B (STAT5B) and activating transcription factor 2 (ATF-2) appeared as highly interconnected nodes (Figure 1). These two main nodes were selected for further validations. STAT5B was overexpressed with respect to the control by 23.8% in cells treated with ICC and by 33.4% in cells treated with CA, whereas ATF-2 was found underexpressed in HT29 incubated with ICC (32.5% decrease compared to the control) and with CA (26% decrease).
Table 6.
Common differentially expressed genes in HT29 treated-cells.
| Gene symbol | FC absolute ICC | P value | Regulation | FC absolute CA | P value | Regulation |
|---|---|---|---|---|---|---|
| FST | 1.343 | 0.025 | Up | 1.375 | 0.022 | Up |
| SULT1B1 | 1.349 | 0.018 | Up | 1.304 | 0.020 | Up |
| EPOR | 1.372 | 0.008 | Up | 1.321 | 0.021 | Up |
| HINT3 | 2.410 | 0.040 | Up | 2.607 | 0.033 | Up |
| STAT5B | 1.312 | 0.007 | Up | 1.334 | 0.012 | Up |
| GLS | 1.335 | 0.001 | Down | 1.370 | 0.001 | Down |
| PPP3R1 | 1.397 | 0.023 | Down | 1.423 | 0.026 | Down |
| ATF2 | 1.481 | 0.005 | Down | 1.354 | 0.016 | Down |
| SLC4A7 | 1.314 | 0.029 | Down | 1.322 | 0.025 | Down |
| MARCH3 | 1.330 | 0.016 | Down | 1.319 | 0.005 | Down |
| TBL1XR1 | 1.426 | 0.001 | Down | 1.332 | 0.001 | Down |
| SAP30L | 1.375 | 0.013 | Down | 1.405 | 0.021 | Down |
| FGD4 | 1.593 | 0.001 | Down | 1.523 | 0.027 | Down |
| ZDHHC20 | 1.665 | 0.004 | Down | 1.314 | 0.005 | Down |
| ZDHHC21 | 1.642 | 0.037 | Down | 1.318 | 0.016 | Down |
| FAM76B | 1.506 | 0.014 | Down | 1.325 | 0.037 | Down |
| CLK4 | 1.326 | 0.049 | Down | 1.339 | 0.021 | Down |
Common differentially expressed genes in HT29 treated-cells with a P value <0.05 and a minimum fold of 1.3. Column ICC correspond to cells treated with instant caffeinated coffee and column CA corresponds to cells treated with caffeic acid. Overexpressed genes are indicated on the upper part of the table, whereas underexpressed genes are depicted in the lower part. The genes in bold, STAT5B and ATF-2, were chosen for further analysis.
Figure 1.
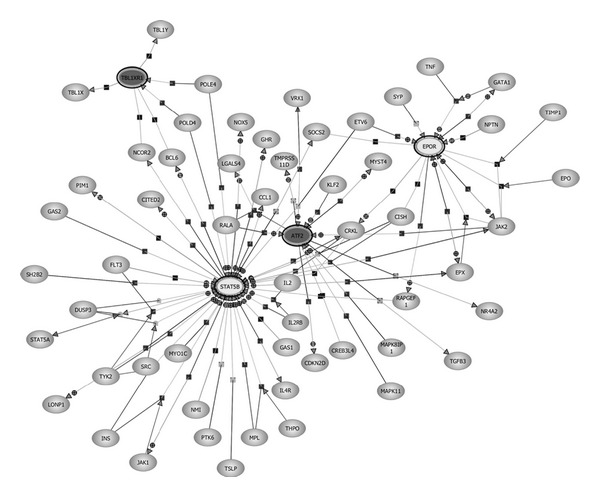
Biological association network (BAN) of differentially expressed genes in common between CA and ICC. The list of common genes between both treatments was used to construct a BAN with the Pathway Analysis software within GeneSpring v.11.5.1. An expanded network was constructed by setting an advanced filter that included the categories of binding, expression, metabolism, promoter binding, protein modification, and regulation. Only proteins are represented. The BAN shows the node genes STAT5B and ATF-2 that were further studied.
3.3. Validation of STAT5B and ATF-2 Changes at the mRNA and Protein Levels
STAT5B overexpression in HT29 cells upon incubation with CA and ICC was confirmed at the mRNA (1.16- and 1.3-fold compared to the control, respectively) and protein levels (1.5- and 1.2-fold compared to the control, respectively) (Figures 2(a) and 2(c)). In the case of ATF-2, the changes in mRNA levels were confirmed for both CA and ICC (0.88- and 0.86-fold compared to the control, respectively), whereas the decrease in protein levels was only observed in CA-treated cells (0.62-fold compared to the control) (Figures 2(b) and 2(d)).
Figure 2.
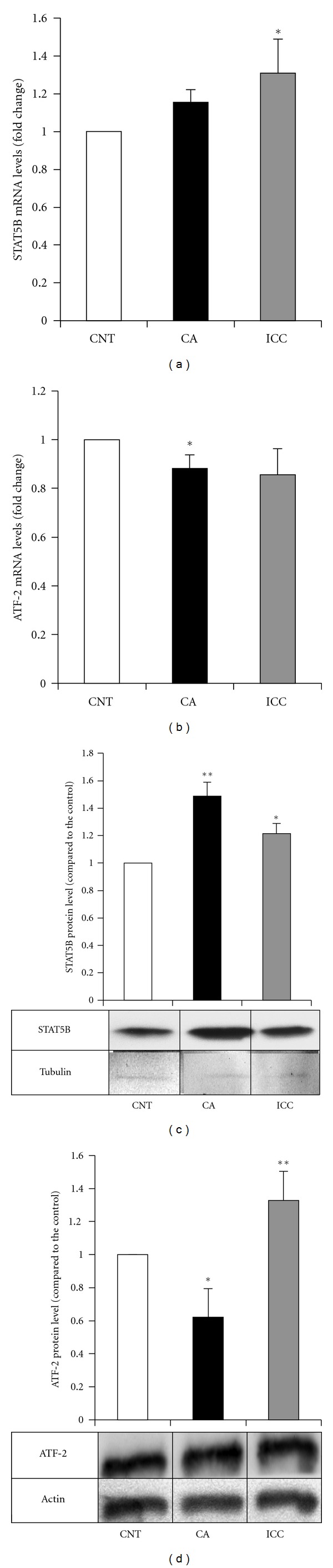
Quantitation of mRNA and protein levels for STAT5B and ATF-2 in HT29 cells. The mRNA levels of STAT5B (a) and ATF-2 (b) were determined in control HT29 cells (empty bars) and cells treated with caffeic acid (CA, filled bars) and instant caffeinated coffee (ICC, grey bars) by RT real-time PCR as described in Methods. Results are expressed in fold changes compared to the control and are the mean ± SE of 3 different experiments. *P < 0.05 compared with the corresponding control. The protein levels of STAT5B (c) and ATF-2 (d) were determined in control HT29 cells (empty bars) and cells treated with caffeic acid (CA, filled bars) and instant caffeinated coffee (ICC, grey bars) by Western blot. Blots were reprobed with an antibody against β-actin or tubulin to normalize the results. Results represent the mean ± SE of 3 different experiments. *P < 0.05 and **P < 0.01 compared with the corresponding control.
3.4. Expression of Cyclin D1 upon Incubation with ICC and CA
Cyclin D1 is overexpressed at the mRNA and protein level in over 50% of the breast cancers either in the presence or absence of gene amplification, and it is one of the most commonly overexpressed proteins in breast cancer [24, 25]. Cyclin D1 transcription is regulated by STAT5 [26–29] and ATF-2 [30–32].
We analyzed the levels of cyclin D1 by western blot in MCF-7 and HT29 cells upon incubation with ICC and CA. As shown in Figure 3(a), incubation of MCF-7 cells with either CA and ICC led to a drastic decrease in the levels of cyclin D1 protein, together with an increase in the levels of STAT5B, but not to a decrease in the levels of ATF-2. In HT29 cells, incubation with CA did not affect cyclin D1 levels, whereas the presence of ICC led to an increase in cyclin D1 levels 3 (b).
Figure 3.
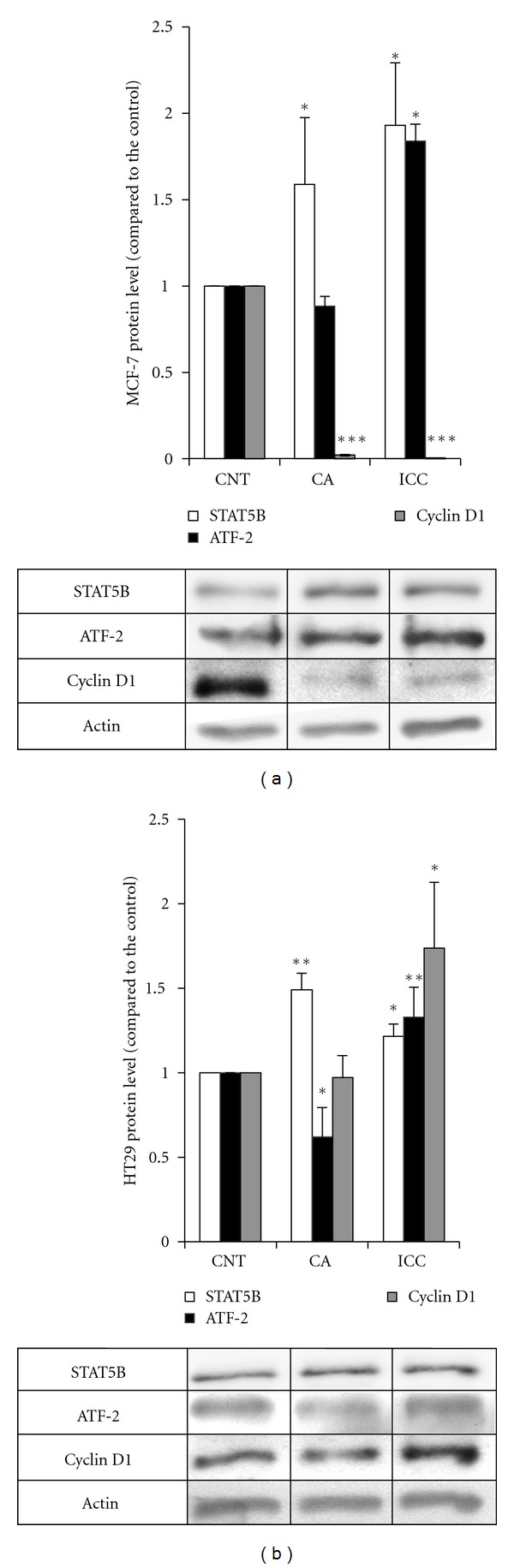
Expression of cyclin D1 upon incubation with ICC and CA in HT29 and MCF-7 cells. (a) Quantitation of STAT5b (empty bars), ATF-2 (filled bars), and cyclin D1 (grey bars) protein levels in MCF-7 cells. The protein levels were determined in control MCF-7 cells (CNT) and cells treated with caffeic acid (CA) and instant coffee (ICC) by Western blot. Blots were reprobed with an antibody against β-actin to normalize the results. Results represent the mean ± SE of 3 different experiments. *P < 0.05 and ***P < 0.001 compared with the corresponding control. (b) Quantitation of STAT5b (empty bars), ATF-2 (filled bars), and cyclin D1 (grey bars) protein levels in HT29 cells. The protein levels were determined in control HT29 cells (CNT) and cells treated with caffeic acid (CA) and instant coffee (ICC) by Western blot. Blots were reprobed with an antibody against β-actin to normalize the results. Results represent the mean ± SE of 3 different experiments.*P < 0.05 and **P < 0.01 compared with the corresponding control.
4. Discussion
In this work we analyzed the gene expression profile of human cancer cells treated with either ICC or CA. Caffeic acid was chosen since it is the main representative of hydroxycinnamic acids. Using microarrays we identified the differential expression of specific genes involved in several biological pathways. The changes in mRNA expression of two outlier genes, STAT5B and ATF-2, observed in the microarrays were confirmed by RT real-time PCR, and the changes in protein levels were also analyzed by Western blot. The selection of STAT5B and ATF-2 was made according to the results obtained in the construction of a biological association network. Finally, the modulation of cyclin D1, a target of STAT5B and ATF-2 transcription factors, upon incubation with coffee polyphenols was also established.
We show that ICC and the amount of CA of one cup of coffee are able to induce STAT5B mRNA and protein levels in HT29 cells. STAT5 was originally described as a prolactin-induced mammary gland factor [33]. The cloning of two closely related STAT5 cDNAs, from both mouse and human cDNA libraries, showed two distinct genes, STAT5A and STAT5B that encoded two STAT5 proteins [34–37].
In addition to prolactin, STAT5 proteins are activated by a wide variety of cytokines and growth factors, including IL-2, IL-3, IL-5, IL-7, IL-9, IL-15, granulocyte-macrophage colony-stimulating factor, erythropoietin, growth hormone, thrombopoietin, epidermal growth factor, and platelet-derived growth factor. The key function of STAT5B is to mediate the effects of growth hormone [38, 39]. Modulation of STAT5 levels or transcriptional activity has already been described in cells treated with natural compounds such as nobiletin, a citrus flavonoid [40], thea flavins [41], and silibinin, a natural polyphenolic flavonoid which is a major bioactive component of silymarin isolated from Silybum marianum [42]. Furthermore, it has been reported that butein, the major biologically active polyphenolic component of the stems of Rhus verniciflua, downregulated the expression of STAT3-regulated gene products such as Bcl-xL, Bcl-2, cyclin D1, and Mcl-1 [43].
STAT5B participates in diverse biological processes, such as growth development, immunoregulation, apoptosis, reproduction, prolactin pathway, and lipid metabolism. STAT5B deficiency is a recently identified disease entity that involves both severe growth hormone-resistant growth failure and severe immunodeficiency [44–46]. The induction of STAT5B expression upon incubation with CA and ICC could represent a nutritional tool to upregulate this transcription factor and suggests novel research strategies for natural therapies in Crohn's disease and inflammatory bowel disease in which STAT5B appears to maintain the mucosal barrier integrity and tolerance [47, 48]. In colorectal cancer both STAT5a and STAT5b play important roles in progression and downregulation of both STAT5A and STAT5B results in a gradual decrease in cell viability, predominantly attributed to G1 cell cycle arrest, and apoptotic cell death [49]. In this context the increase in STAT5B caused by ICC and CA would have a negative effect on colorectal cancer patients, as it would trigger cell proliferation and survival.
In human breast cancer, STAT5A/B has been shown a dual role in the mammary gland as an initiator of tumor formation as well as a promoter of differentiation of established tumors. STAT3, STAT5A, and STAT5B are overexpressed or constitutively activated in breast cancer [50–52] and active STAT5A/B in human breast cancer predicted favorable clinical outcome [53]. Prolactin receptor signal transduction through the Jak2-STAT5 pathway has been considered to be essential for proliferation and differentiation of normal mammary epithelial cells [54–56]. It has been shown that the levels of NUC-pYSTAT5 decreased as breast cancer progressed from normal to in situ, to invasive, and then to nodal metastases [57]. Additionally Peck et al. [57] found that the absence of detectable NUC-pYStat5 in tumors of patients how where under antiestrogen therapy was associated with poor breast cancer-specific survival. We analyzed STAT5B modulation through the PRL pathway in response to coffee polyphenols in a breast cancer cell line. The MCF-7 cell line was chosen because expression of the prolactin receptor is more often found in estrogen receptor-positive breast tumors [58]. In our conditions, incubation with CA and ICC led to an increase in STAT5B protein levels in MCF-7 cells, and this result could be the basis for a possible inclusion of coffee polyphenols in the diet of breast cancer patients.
ATF-2 is a member of the ATF-cAMP response element-binding protein (CREB) family of transcription factors that can bind to the cAMP response element (CRE) found in many mammalian gene promoters [59, 60]. ATF-2 exhibits both oncogenic and tumor suppressor functions [61]. CREs are found in several genes involved in the control of the cell cycle, for example, the cyclin D1 gene, and ATF-2 binding to this sequence stimulates the transcription of cyclin D1 [30, 31]. ATF-2 mediated cyclin D1 promoter induction can be stimulated by a number of growth-promoting agents, such as estrogen [31], hepatocyte growth factor [62], and regenerating gene product [63]. ATF-2 has been correlated with proliferation, invasion, migration, and resistance to DNA-damaging agents in breast cancer cell lines.
The downregulation of ATF-2 expression after CA and ICC incubation in HT29 cells reported here is in accordance with the observed decrease in activity of ATF-2 in gastric cells when incubating with chlorogenic acid, the precursor of caffeic acid [64]. Surprisingly, the validation of the protein levels showed the upregulation of ATF-2 protein with ICC, but not with CA, both in HT29 and MCF-7 cells. This differential behavior could be due to other ICC components besides CA. In this direction Rubach et al. [64] reported a different response in ATF-2 activity after incubation of a gastric cell line with different coffee compounds. The presence of pyrogallol, catechol, βN-alkanoylhydroxytryptamides, and N-methylpyridinium increased ATF-2 activity, whereas chlorogenic acid and caffeine decrease it [64]. In our conditions incubation of HT29 cells with ICC caused a modest decrease in ATF-2 mRNA levels. However this effect was not translated at the protein level. We hypothesize that ICC contains other polyphenols in addition to caffeic acid that are able to increase ATF-2 protein levels through an increase of the translation of its mRNA, the increase of stability of the protein or an inhibition of its degradation. In this direction several plant polyphenols such as (-)-epigallocatechins-3-gallate (EGCG), genistein, luteolin, apigenin, chrysin, quercetin, curcumin, and tannic acid have been described to possess proteasome-inhibitory activity [65, 66].
The regulation of ATF-2 transcriptional activity, mostly at the level of its phosphorylation status, has been described upon treatment of cancer cells with several natural compounds. In MCF-7 cells, the anticancer agent 3,30-Diindolylmethane, derived from Brassica vegetables, activates both JNK and p38 pathways, resulting in c-Jun and ATF-2 phosphorylation, and the increase of binding of the c-Jun–ATF-2 homodimers and heterodimers to the proximal regulatory element of IFN-γ promoter [67]. Biochanin-A, an isoflavone, existing in red clover, cabbage and alfalfa, has an inhibitory and apoptogenic effect on certain cancer cells by blocking the phosphorylation of p38 MAPK and ATF-2 in a dose-dependent fashion [68]. The JNK stress-activated pathway is one of the major intracellular signal transduction cascades involved in intestinal inflammation [69, 70], and upregulation of ATF-2 has been shown in Crohn's disease [71, 72]. Thus CA could represent potential therapeutical properties in different states of intestinal inflammation due to its combined effects on STAT5B and ATF-2 in HT29 cells.
Finally, the modulation of cyclin D1, a target of STAT5B and ATF-2 transcription factors, upon incubation with coffee polyphenols was established in colon and breast cancer cells. Cyclin D1 overexpression is common in colorectal cancer, but the findings regarding its prognostic value are conflicting. In a recent study, positive expression of cyclin D1 protein was detected in 95 of 169 colonic adenocarcinoma specimens, and increased cyclin D1 levels were associated with poorer prognosis [73]. Furthermore, there was a significant correlation between the positive expression of p-Stat5 and cyclin D1 in patients with colonic adenocarcinoma. However, in a second study, cyclin D1 overexpression was associated with improved outcome in a total of 386 patients who underwent surgical resection for colon cancer, classified as TNM stage II or III. Belt et al. [74] showed that low p21, high p53, low cyclin D1, and high AURKA were associated with disease recurrence in stage II and III colon cancer patients. In this context the effect of ICC on cyclin D1 levels could represent either a positive or a negative effect in colon cancer cells, depending on tumor progression. The increase in cyclin D1 levels could represent a marker of better outcome since it has been recently established that cyclin D1 expression is strongly associated with prolonged survival in male colorectal cancer and that lack of cyclin D1 is associated with a more aggressive phenotype in male patients [75]. However, several natural compounds such as anthocyanins, anthocyanidins, apigenin, luteolin, and fisetin have all been described to induce experimentally cell-cycle arrest and apoptosis through the decrease of cyclin D1 levels in HT29 cells [76–80]. In accordance to these data, the increase observed in cyclin D1 levels in HT29 cells upon incubation with ICC could probably be the consequence of the presence of different compounds other than polyphenols in ICC.
In MCF-7 breast cancer cells, cyclin D1 was downregulated upon incubation with coffee polyphenols. The rationale for the choice of MCF-7 cell line was based on the observation that although cyclin D1 overexpression is present across multiple histologic subtypes of breast cancer, it has been shown that the large majority of cyclin D1–overexpressing breast cancers are ER positive [24, 25, 81]. Cyclin D1 overexpression has been reported between 40 and 90% of cases of invasive breast cancer, while gene amplification is seen in about 5–20% of tumors [24, 81–83]. In cyclin D1-driven cancers, blocking cyclin D1 expression by targeting the cyclin D1 gene, RNA, or protein should increase the chances for therapeutic success. Cell culture studies have raised the possibility that certain compounds might act in this way [84, 85] and approaches to blocking cyclin D1 expression using antisense, siRNA, or related molecules specifically target the driving molecular lesion itself [86–88]. It is believed that compounds that modulate cyclin D1 expression could have a role in the prevention and treatment of human neoplasias. For instance, flavopiridol, a synthetic flavonoid based on an extract from an Indian plant for the potential treatment of cancer, induces a rapid decline in cyclin D1 steady-state protein levels [89]. Taking all these results together, inhibition of cyclin D1 expression appears to be a good approach for cancer treatment. In this direction our observation that coffee and caffeic acid are able to drastically reduce the expression of cyclin D1 in breast cancer cells could suggest that some coffee components could be used as a coadjuvant therapeutic tool in the treatment of breast cancer.
Acknowledgments
This work was supported by Grants CDTI 050618, SAF2008-0043, SAF2011-23582 (Ministerio de Educación y Ciencia de España), and ISCIII-RTICc RD06/0020 (Redes Temáticas de Investigación Cooperativa en Salud) RD06/0020/0046. Our research group holds the ‘‘quality distinction” from the ‘‘Generalitat de Catalunya” SGR2009-00118. C. Oleaga was a recipient of a fellowship from the FEC (Federación Española del Café). All the authors have no conflict of interests.
Abbreviations
- APRT:
Adenine phosphoribosyltransferase
- ATF-2:
Activating transcription factor
- BAN:
Biological association network
- CA:
Caffeic acid
- DMSO:
Dimethyl sulfoxide
- DEPC:
Diethyl pyrocarbonate
- ICC:
Instant caffeinated coffee
- RT-PCR:
Reverse transcription-polymerase chain reaction
- STAT5B:
Signal transducer and activator of transcription 5B.
References
- 1.Scalbert A, Manach C, Morand C, Rémésy C, Jiménez L. Dietary polyphenols and the prevention of diseases. Critical Reviews in Food Science and Nutrition. 2005;45(4):287–306. doi: 10.1080/1040869059096. [DOI] [PubMed] [Google Scholar]
- 2.Scalbert A, Johnson IT, Saltmarsh M. Polyphenols: antioxidants and beyond. The American Journal of Clinical Nutrition. 2005;81(1):215S–217S. doi: 10.1093/ajcn/81.1.215S. [DOI] [PubMed] [Google Scholar]
- 3.Azzi A, Davies KJ, Kelly F. Free radical biology—terminology and critical thinking. FEBS Letters. 2004;558(1–3):3–6. doi: 10.1016/s0014-5793(03)01526-6. [DOI] [PubMed] [Google Scholar]
- 4.Halliwell B, Rafter J, Jenner A. Health promotion by flavonoids, tocopherols, tocotrienols, and other phenols: direct or indirect effects? Antioxidant or not? The American Journal of Clinical Nutrition. 2005;81(1):268S–276S. doi: 10.1093/ajcn/81.1.268S. [DOI] [PubMed] [Google Scholar]
- 5.Olthof MR, Hollman PC, Katan MB. Chlorogenic acid and caffeic acid are absorbed in humans. Journal of Nutrition. 2001;131(1):66–71. doi: 10.1093/jn/131.1.66. [DOI] [PubMed] [Google Scholar]
- 6.Olthof MR, Hollman PC, Buijsman MN, van Amelsvoort JM, Katan MB. Chlorogenic acid, quercetin-3-rutinoside and black tea phenols are extensively metabolized in humans. Journal of Nutrition. 2003;133(6):1806–1814. doi: 10.1093/jn/133.6.1806. [DOI] [PubMed] [Google Scholar]
- 7.Rice-Evans CA, Miller NJ, Paganga G. Structure-antioxidant activity relationships of flavonoids and phenolic acids. Free Radical Biology and Medicine. 1996;20(7):933–956. doi: 10.1016/0891-5849(95)02227-9. [DOI] [PubMed] [Google Scholar]
- 8.Kono Y, Shibata H, Kodama Y, Sawa Y. The suppression of the N-nitrosating reaction by chlorogenic acid. The Biochemical Journal. 1995;312, part 3:947–953. doi: 10.1042/bj3120947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Shibata H, Sakamoto Y, Oka M, Kono Y. Natural antioxidant, chlorogenic acid, protects against DNA breakage caused by monochloramine. Bioscience, Biotechnology and Biochemistry. 1999;63(7):1295–1297. doi: 10.1271/bbb.63.1295. [DOI] [PubMed] [Google Scholar]
- 10.Kasai H, Fukada S, Yamaizumi Z, Sugie S, Mori H. Action of chlorogenic acid in vegetables and fruits as an inhibitor of 8-hydroxydeoxyguanosine formation in vitro and in a rat carcinogenesis model. Food and Chemical Toxicology. 2000;38(5):467–471. doi: 10.1016/s0278-6915(00)00014-4. [DOI] [PubMed] [Google Scholar]
- 11.Feng R, Lu Y, Bowman LL, Qian Y, Castranova V, Ding M. Inhibition of activator protein-1, NF-κB, and MAPKs and induction of phase 2 detoxifying enzyme activity by chlorogenic acid. The Journal of Biological Chemistry. 2005;280(30):27888–27895. doi: 10.1074/jbc.M503347200. [DOI] [PubMed] [Google Scholar]
- 12.Larsson SC, Wolk A. Coffee consumption and risk of liver cancer: a meta-analysis. Gastroenterology. 2007;132(5):1740–1745. doi: 10.1053/j.gastro.2007.03.044. [DOI] [PubMed] [Google Scholar]
- 13.Bravi F, Bosetti C, Tavani A, et al. Coffee drinking and hepatocellular carcinoma risk: a meta-analysis. Hepatology. 2007;46(2):430–435. doi: 10.1002/hep.21708. [DOI] [PubMed] [Google Scholar]
- 14.Je Y, Liu W, Giovannucci E. Coffee consumption and risk of colorectal cancer: a systematic review and meta-analysis of prospective cohort studies. International Journal of Cancer. 2009;124(7):1662–1668. doi: 10.1002/ijc.24124. [DOI] [PubMed] [Google Scholar]
- 15.Tang N, Zhou B, Wang B, Yu R. Coffee consumption and risk of breast cancer: a metaanalysis. American Journal of Obstetrics and Gynecology. 2009;200(3):290.e1–290.e9. doi: 10.1016/j.ajog.2008.10.019. [DOI] [PubMed] [Google Scholar]
- 16.Naganuma T, Kuriyama S, Kakizaki M, et al. Coffee consumption and the risk of oral, pharyngeal, and esophageal cancers in Japan: the Miyagi Cohort Study. American Journal of Epidemiology. 2008;168(12):1425–1432. doi: 10.1093/aje/kwn282. [DOI] [PubMed] [Google Scholar]
- 17.Wilson KM, Kasperzyk JL, Rider JR, et al. Coffee consumption and prostate cancer risk and progression in the health professionals follow-up study. Journal of the National Cancer Institute. 2011;103(11):876–884. doi: 10.1093/jnci/djr151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nkondjock A. Coffee consumption and the risk of cancer: an overview. Cancer Letters. 2009;277(2):121–125. doi: 10.1016/j.canlet.2008.08.022. [DOI] [PubMed] [Google Scholar]
- 19.Ganmaa D, Willett WC, Li TY, et al. Coffee, tea, caffeine and risk of breast cancer: a 22-year follow-up. International Journal of Cancer. 2008;122(9):2071–2076. doi: 10.1002/ijc.23336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Li J, Seibold P, Chang-Claude J, et al. Coffee consumption modifies risk of estrogen-receptor negative breast cancer. Breast Cancer Research. 2011;13(3, article R49) doi: 10.1186/bcr2879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mosmann T. Rapid colorimetric assay for cellular growth and survival: application to proliferation and cytotoxicity assays. Journal of Immunological Methods. 1983;65(1-2):55–63. doi: 10.1016/0022-1759(83)90303-4. [DOI] [PubMed] [Google Scholar]
- 22.Selga E, Oleaga C, Ramírez S, de Almagro MC, Noé V, Ciudad CJ. Networking of differentially expressed genes in human cancer cells resistant to methotrexate. Genome Medicine. 2009;1(9, article 83) doi: 10.1186/gm83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Selga E, Morales C, Noe V, Peinado MA. Role of caveolin 1, E-cadherin, Enolase 2 and PKCalpha on resistance to methotrexate in human HT29 colon cancer cells. BMC Medical Genomics. 2008;1, article 35 doi: 10.1186/1755-8794-1-35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gillett CE, Lee AH, Millis RR, Barnes DM. Cyclin D1 and associated proteins in mammary ductal carcinoma in situ and atypical ductal hyperplasia. The Journal of Pathology. 1998;184:396–400. doi: 10.1002/(SICI)1096-9896(199804)184:4<396::AID-PATH1259>3.0.CO;2-G. [DOI] [PubMed] [Google Scholar]
- 25.Buckley MF, Sweeney KJ, Hamilton JA, et al. Expression and amplification of cyclin genes in human breast cancer. Oncogene. 1993;8(8):2127–2133. [PubMed] [Google Scholar]
- 26.Lim EJ, Joung YH, Jung SM, et al. Hemin inhibits cyclin D1 and IGF-1 expression via STAT5b under hypoxia in ERα-negative MDA-MB 231 breast cancer cells. International Journal of Oncology. 2010;36(5):1243–1251. doi: 10.3892/ijo_00000608. [DOI] [PubMed] [Google Scholar]
- 27.Joung YH, Lim EJ, Kim MS, et al. Enhancement of hypoxia-induced apoptosis of human breast cancer cells via STAT5b by momilactone B. International Journal of Oncology. 2008;33(3):477–484. [PubMed] [Google Scholar]
- 28.Fox EM, Bernaciak TM, Wen J, Weaver AM, Shupnik MA, Silva CM. Signal transducer and activator of transcription 5b, c-Src, and epidermal growth factor receptor signaling play integral roles in estrogen-stimulated proliferation of estrogen receptor-positive breast cancer cells. Molecular Endocrinology. 2008;22(8):1781–1796. doi: 10.1210/me.2007-0419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lange CA, Richer JK, Shen T, Horwitz KB. Convergence of progesterone and epidermal growth factor signaling in breast cancer: potentiation of mitogen-activated protein kinase pathways. The Journal of Biological Chemistry. 1998;273(47):31308–31316. doi: 10.1074/jbc.273.47.31308. [DOI] [PubMed] [Google Scholar]
- 30.Beier F, Lee RJ, Taylor AC, Pestell RG, Luvalle P. Identification of the cyclin D1 gene as a target of activating transcription factor 2 in chondrocytes. Proceedings of the National Academy of Sciences of the United States of America. 1999;96(4):1433–1438. doi: 10.1073/pnas.96.4.1433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sabbah M, Courilleau D, Mester J, Redeuilh G. Estrogen induction of the cyclin D1 promoter: involvement of a cAMP response-like element. Proceedings of the National Academy of Sciences of the United States of America. 1999;96(20):11217–11222. doi: 10.1073/pnas.96.20.11217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lewis JS, Thomas TJ, Pestell RG, Albanese C, Gallo MA, Thomas T. Differential effects of 16α-hydroxyestrone and 2-methoxyestradiol on cyclin D1 involving the transcription factor ATF-2 in MCF-7 breast cancer cells. Journal of Molecular Endocrinology. 2005;34(1):91–105. doi: 10.1677/jme.1.01599. [DOI] [PubMed] [Google Scholar]
- 33.Wakao H, Gouilleux F, Groner B. Mammary gland factor (MGF) is a novel member of the cytokine regulated transcription factor gene family and confers the prolactin response. EMBO Journal. 1994;13(9):2182–2191. doi: 10.1002/j.1460-2075.1994.tb06495.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hou J, Schindler U, Henzel WJ, Wong SC, McKnight SL. Identification and purification of human stat proteins activated in response to interleukin-2. Immunity. 1995;2(4):321–329. doi: 10.1016/1074-7613(95)90140-x. [DOI] [PubMed] [Google Scholar]
- 35.Liu X, Robinson GW, Gouilleux F, Groner B, Hennighausen L. Cloning and expression of Stat5 and an additional homologue (Stat5b) involved in prolactin signal transduction in mouse mammary tissue. Proceedings of the National Academy of Sciences of the United States of America. 1995;92(19):8831–8835. doi: 10.1073/pnas.92.19.8831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lin JX, Mietz J, Modi WS, John S, Leonard WJ. Cloning of human Stat5B: reconstitution of interleukin-2-induced Stat5A and Stat5B DNA binding activity in COS-7 cells. The Journal of Biological Chemistry. 1996;271(18):10738–10744. [PubMed] [Google Scholar]
- 37.Mui AL, Wakao H, O’Farrell AM, Harade N, Miyajima A. Interleukin-3, granulocyte-macrophage colony stimulating factor and interleukin-5 transduce signals through two STAT5 homologs. EMBO Journal. 1995;14(6):1166–1175. doi: 10.1002/j.1460-2075.1995.tb07100.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Udy GB, Towers RP, Snell RG, et al. Requirement of STAT5b for sexual dimorphism of body growth rates and liver gene expression. Proceedings of the National Academy of Sciences of the United States of America. 1997;94(14):7239–7244. doi: 10.1073/pnas.94.14.7239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Teglund S, McKay C, Schuetz E, et al. Stat5a and Stat5b proteins have essential and nonessential, or redundant, roles in cytokine responses. Cell. 1998;93(5):841–850. doi: 10.1016/s0092-8674(00)81444-0. [DOI] [PubMed] [Google Scholar]
- 40.Kanda K, Nishi K, Kadota A, Nishimoto S, Liu MC, Sugahara T. Nobiletin suppresses adipocyte differentiation of 3T3-L1 cells by an insulin and IBMX mixture induction. Biochimica et Biophysica Acta. 2011;1820(4):461–468. doi: 10.1016/j.bbagen.2011.11.015. [DOI] [PubMed] [Google Scholar]
- 41.Chattopadhyay S, Bhattacharyya S, Saha B, et al. Tumor-shed PGE2 impairs IL2Rγc-signaling to inhibit CD4+ T cell survival: regulation by theaflavins. PLoS ONE. 2009;4(10) doi: 10.1371/journal.pone.0007382.e7382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Singh RP, Raina K, Deep G, Chan D, Agarwal R. Silibinin suppresses growth of human prostate carcinoma PC-3 orthotopic xenograft via activation of extracellular signal-regulated kinase 1/2 and inhibition of signal transducers and activators of transcription signaling. Clinical Cancer Research. 2009;15(2):613–621. doi: 10.1158/1078-0432.CCR-08-1846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Pandey MK, Sung B, Ahn KS, Aggarwal BB. Butein suppresses constitutive and inducible signal transducer and activator of transcription (stat) 3 activation and stat3-regulated gene products through the induction of a protein tyrosine phosphatase SHP-1. Molecular Pharmacology. 2009;75(3):525–533. doi: 10.1124/mol.108.052548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Nadeau K, Hwa V, Rosenfeld RG. STAT5b deficiency: an unsuspected cause of growth failure, immunodeficiency, and severe pulmonary disease. Journal of Pediatrics. 2011;158(5):701–708. doi: 10.1016/j.jpeds.2010.12.042. [DOI] [PubMed] [Google Scholar]
- 45.Scaglia PA, Martinez AS, Feigerlova E, et al. A novel missense mutation in the SH2 domain of the STAT5B gene results in a transcriptionally inactive STAT5b associated with severe IGF-I deficiency, immune dysfunction, and lack of pulmonary disease. The Journal of Clinical Endocrinology & Metabolism. 2012;97:E830–E839. doi: 10.1210/jc.2011-2554. [DOI] [PubMed] [Google Scholar]
- 46.Rotwein P. Mapping the growth hormone—Stat5b—IGF-I transcriptional circuit. Trends in Endocrinology & Metabolism. 2012;23:186–193. doi: 10.1016/j.tem.2012.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Han X, Osuntokun B, Benight N, Loesch K, Frank SJ, Denson LA. Signal transducer and activator of transcription 5b promotes mucosal tolerance in pediatric Crohn’s disease and murine colitis. American Journal of Pathology. 2006;169(6):1999–2013. doi: 10.2353/ajpath.2006.060186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Han X, Ren X, Jurickova I, et al. Regulation of intestinal barrier function by signal transducer and activator of transcription 5b. Gut. 2009;58(1):49–58. doi: 10.1136/gut.2007.145094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Du W, Wang YC, Hong J, et al. STAT5 isoforms regulate colorectal cancer cell apoptosis via reduction of mitochondrial membrane potential and generation of reactive oxygen species. Journal of Cellular Physiology. 2012;227:2421–2429. doi: 10.1002/jcp.22977. [DOI] [PubMed] [Google Scholar]
- 50.Garcia R, Bowman TL, Niu G, et al. Constitutive activation of Stat3 by the Src and JAK tyrosine kinases participates in growth regulation of human breast carcinoma cells. Oncogene. 2001;20(20):2499–2513. doi: 10.1038/sj.onc.1204349. [DOI] [PubMed] [Google Scholar]
- 51.Garcia R, Yu CL, Hudnall A, et al. Constitutive activation of Stat3 in fibroblasts transformed by diverse oncoproteins and in breast carcinoma cells. Cell Growth and Differentiation. 1997;8(12):1267–1276. [PubMed] [Google Scholar]
- 52.Yamashita H, Iwase H, Toyama T, Fujii Y. Naturally occurring dominant-negative Stat5 suppresses transcriptional activity of estrogen receptors and induces apoptosis in T47D breast cancer cells. Oncogene. 2003;22(11):1638–1652. doi: 10.1038/sj.onc.1206277. [DOI] [PubMed] [Google Scholar]
- 53.Tan SH, Nevalainen MT. Signal transducer and activator of transcription 5A/B in prostate and breast cancers. Endocrine-Related Cancer. 2008;15(2):367–390. doi: 10.1677/ERC-08-0013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Yamashita H, Iwase H. The role of Stat5 in estrogen receptor-positive breast cancer. Breast Cancer. 2002;9(4):312–318. doi: 10.1007/BF02967610. [DOI] [PubMed] [Google Scholar]
- 55.Frasor J, Barkai U, Zhong L, Fazleabas AT, Gibori G. PRL-induced ERα gene expression is mediated by Janus kinase 2 (Jak2) while signal transducer and activator of transcription 5b (Stat5b) phosphorylation involves Jak2 and a second tyrosine kinase. Molecular Endocrinology. 2001;15(11):1941–1952. doi: 10.1210/mend.15.11.0722. [DOI] [PubMed] [Google Scholar]
- 56.Frasor J, Gibori G. Prolactin regulation of estrogen receptor expression. Trends in Endocrinology & Metabolism. 2003;14(3):118–123. doi: 10.1016/s1043-2760(03)00030-4. [DOI] [PubMed] [Google Scholar]
- 57.Peck AR, Witkiewicz AK, Liu C, et al. Loss of nuclear localized and tyrosine phosphorylated Stat5 in breast cancer predicts poor clinical outcome and increased risk of antiestrogen therapy failure. Journal of Clinical Oncology. 2011;29(18):2448–2458. doi: 10.1200/JCO.2010.30.3552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Perotti C, Liu R, Parusel CT, et al. Heat shock protein-90-alpha, a prolactin-STAT5 target gene identified in breast cancer cells, is involved in apoptosis regulation. Breast Cancer Research. 2008;10(6, article R94) doi: 10.1186/bcr2193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Hai TW, Liu F, Coukos WJ, Green MR. Transcription factor ATF cDNA clones: an extensive family of leucine zipper proteins able to selectively form DNA-binding heterodimers. Genes and Development. 1989;3(12):2083–2090. doi: 10.1101/gad.3.12b.2083. [DOI] [PubMed] [Google Scholar]
- 60.Benbrook DM, Jones NC. Heterodimer formation between CREB and JUN proteins. Oncogene. 1990;5(3):295–302. [PubMed] [Google Scholar]
- 61.Bhoumik A, Ronai Z. ATF2: a transcription factor that elicits oncogenic or tumor suppressor activities. Cell Cycle. 2008;7(15):2341–2345. doi: 10.4161/cc.6388. [DOI] [PubMed] [Google Scholar]
- 62.Recio JA, Merlino G. Hepatocyte growth factor/scatter factor activates proliferation in melanoma cells through p38 MAPK, ATF-2 and cyclin D1. Oncogene. 2002;21(7):1000–1008. doi: 10.1038/sj.onc.1205150. [DOI] [PubMed] [Google Scholar]
- 63.Takasawa S, Ikeda T, Akiyama T, et al. Cyclin D1 activation through ATF-2 in Reg-induced pancreatic β-cell regeneration. FEBS Letters. 2006;580(2):585–591. doi: 10.1016/j.febslet.2005.12.070. [DOI] [PubMed] [Google Scholar]
- 64.Rubach M, Lang R, Seebach E, Somoza MM, Hofmann T, Somoza V. Multi-parametric approach to identify coffee components that regulate mechanisms of gastric acid secretion. Molecular Nutrition & Food Research. 2012;56:325–335. doi: 10.1002/mnfr.201100453. [DOI] [PubMed] [Google Scholar]
- 65.Mujtaba T, Dou QP. Black tea polyphenols inhibit tumor proteasome activity. In Vivo. 2012;26:197–202. [PMC free article] [PubMed] [Google Scholar]
- 66.Shen M, Chan TH, Dou QP. Targeting tumor ubiquitin-proteasome pathway with polyphenols for chemosensitization. doi: 10.2174/187152012802649978. Anti-Cancer Agents in Medicinal Chemistry. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Xue L, Firestone GL, Bjeldanes LF. DIM stimulates IFNγ gene expression in human breast cancer cells via the specific activation of JNK and p38 pathways. Oncogene. 2005;24(14):2343–2353. doi: 10.1038/sj.onc.1208434. [DOI] [PubMed] [Google Scholar]
- 68.Kole L, Giri B, Manna SK, Pal B, Ghosh S. Biochanin-A, an isoflavon, showed anti-proliferative and anti-inflammatory activities through the inhibition of iNOS expression, p38-MAPK and ATF-2 phosphorylation and blocking NFκB nuclear translocation. European Journal of Pharmacology. 2011;653(1–3):8–15. doi: 10.1016/j.ejphar.2010.11.026. [DOI] [PubMed] [Google Scholar]
- 69.Ip YT, Davis RJ. Signal transduction by the c-Jun N-terminal kinase (JNK)—from inflammation to development. Current Opinion in Cell Biology. 1998;10(2):205–219. doi: 10.1016/s0955-0674(98)80143-9. [DOI] [PubMed] [Google Scholar]
- 70.Romier B, Schneider YJ, Larondelle Y, During A. Dietary polyphenols can modulate the intestinal inflammatory response. Nutrition Reviews. 2009;67(7):363–378. doi: 10.1111/j.1753-4887.2009.00210.x. [DOI] [PubMed] [Google Scholar]
- 71.Hollenbach E, Vieth M, Roessner A, Neumann M, Malfertheiner P, Naumann M. Inhibition of RICK/nuclear factor-κB and p38 signaling attenuates the inflammatory response in a murine model of Crohn disease. The Journal of Biological Chemistry. 2005;280(15):14981–14988. doi: 10.1074/jbc.M500966200. [DOI] [PubMed] [Google Scholar]
- 72.Derer S, Waetzig GH, Seegert D, Nikolaus S, Schreiber S, Rosenstiel P. A possible link between TIMP-1 induction and response to infliximab. Gut. 2009;58(6):888–889. [PubMed] [Google Scholar]
- 73.Mao Y, Li Z, Lou C, Zhang Y. Expression of phosphorylated Stat5 predicts expression of cyclin D1 and correlates with poor prognosis of colonic adenocarcinoma. International Journal of Colorectal Disease. 2011;26(1):29–35. doi: 10.1007/s00384-010-1090-7. [DOI] [PubMed] [Google Scholar]
- 74.Belt EJ, Brosens RP, Delis-van Diemen PM, et al. Cell cycle proteins predict recurrence in stage II and III colon cancer. doi: 10.1245/s10434-012-2216-7. Annals of Surgical Oncology. In press. [DOI] [PubMed] [Google Scholar]
- 75.Wangefjord S, Manjer J, Gaber A, Nodin B, Eberhard J, Jirstrom K. Cyclin D1 expression in colorectal cancer is a favorable prognostic factor in men but not in women in a prospective, population-based cohort study. Biology of Sex Differences. 2011;2, article 10 doi: 10.1186/2042-6410-2-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Hsu CP, Shih YT, Lin BR, Chiu CF, Lin CC. Inhibitory effect and mechanisms of an anthocyanins- and anthocyanidins-rich extract from purple-shoot tea on colorectal carcinoma cell proliferation. Journal of Agricultural and Food Chemistry. 2012;60:3686–3692. doi: 10.1021/jf204619n. [DOI] [PubMed] [Google Scholar]
- 77.Turktekin M, Konac E, Onen HI, Alp E, Yilmaz A, Menevse S. Evaluation of the effects of the flavonoid apigenin on apoptotic pathway gene expression on the colon cancer cell line (HT29) Journal of Medicinal Food. 2011;14:1107–1117. doi: 10.1089/jmf.2010.0208. [DOI] [PubMed] [Google Scholar]
- 78.Lim DY, Jeong Y, Tyner AL, Park JH. Induction of cell cycle arrest and apoptosis in HT-29 human colon cancer cells by the dietary compound luteolin. American Journal of Physiology. 2007;292(1):G66–G75. doi: 10.1152/ajpgi.00248.2006. [DOI] [PubMed] [Google Scholar]
- 79.Seelinger G, Merfort I, Wölfle U, Schempp CM. Anti-carcinogenic effects of the flavonoid luteolin. Molecules. 2008;13(10):2628–2651. doi: 10.3390/molecules13102628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Suh Y, Afaq F, Johnson JJ, Mukhtar H. A plant flavonoid fisetin induces apoptosis in colon cancer cells by inhibition of COX2 and Wnt/EGFR/NF-κB-signaling pathways. Carcinogenesis. 2009;30(2):300–307. doi: 10.1093/carcin/bgn269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Zukerberg LR, Yang WI, Gadd M, et al. Cyclin D1 (PRAD1) protein expression in breast cancer: approximately one-third of infiltrating mammary carcinomas show overexpression of the cyclin D1 oncogene. Modern Pathology. 1995;8(5):560–567. [PubMed] [Google Scholar]
- 82.Weinstat-Saslow D, Merino MJ, Manrow RE, et al. Overexpression of cyclin D mRNA distinguishes invasive and in situ breast carcinomas from non-malignant lesions. Nature Medicine. 1995;1(12):1257–1260. doi: 10.1038/nm1295-1257. [DOI] [PubMed] [Google Scholar]
- 83.Simpson JF, Quan DE, O’Malley F, Odom-Maryon T, Clarke PE. Amplification of CCND1 and expression of its protein product, cyclin D1, in ductal carcinoma in situ of the breast. American Journal of Pathology. 1997;151(1):161–168. [PMC free article] [PubMed] [Google Scholar]
- 84.Hsiang CH, Straus DS. Cyclopentenone causes cell cycle arrest and represses cyclin D1 promoter activity in MCF-7 breast cancer cells. Oncogene. 2002;21(14):2212–2226. doi: 10.1038/sj.onc.1205293. [DOI] [PubMed] [Google Scholar]
- 85.Sawatsri S, Samid D, Malkapuram S, Sidell N. Inhibition of estrogen-dependent breast cell responses with phenylacetate. International Journal of Cancer. 2001;93(5):687–692. doi: 10.1002/ijc.1399. [DOI] [PubMed] [Google Scholar]
- 86.Sauter ER, Nesbit M, Litwin S, Klein-Szanto AJ, Cheffetz S, Herlyn M. Antisense cyclin D1 induces apoptosis and tumor shrinkage in human squamous carcinomas. Cancer Research. 1999;59(19):4876–4881. [PubMed] [Google Scholar]
- 87.Arber N, Doki Y, Han EK, et al. Antisense to cyclin D1 inhibits the growth and tumorigenicity of human colon cancer cells. Cancer Research. 1997;57(8):1569–1574. [PubMed] [Google Scholar]
- 88.Kornmann M, Arber N, Korc M. Inhibition of basal and mitogen-stimulated pancreatic cancer cell growth by cyclin D1 antisense is associated with loss of tumorigenicity and potentiation of cytotoxicity to cisplatinum. The Journal of Clinical Investigation. 1998;101(2):344–352. doi: 10.1172/JCI1323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Carlson B, Lahusen T, Singh S, et al. Down-regulation of cyclin D1 by transcriptional repression in MCF-7 human breast carcinoma cells induced by flavopiridol. Cancer Research. 1999;59(18):4634–4641. [PubMed] [Google Scholar]


