Abstract
The variation of the proton chemical shifts due to the formation intermolecular hydrogen bonds is computed for a number of complexes which can be formed between the bases of the nucleic acids. The shifts expected for the isolated base pairs, in particular for the G-N1 H, T(or U)-N3H protons and the protons of the amino groups of A, G c, when combined with previous computations on the shifts to be expected upon base stacking, may enable a refined analysis of the high resolution NMR spectra of self complementary polynucleotides or tRNAs. Two examples are presented of a direct computation of proton shits associated with helix-coil transitions, helpful for deducing the helical structure in solution.
Full text
PDF


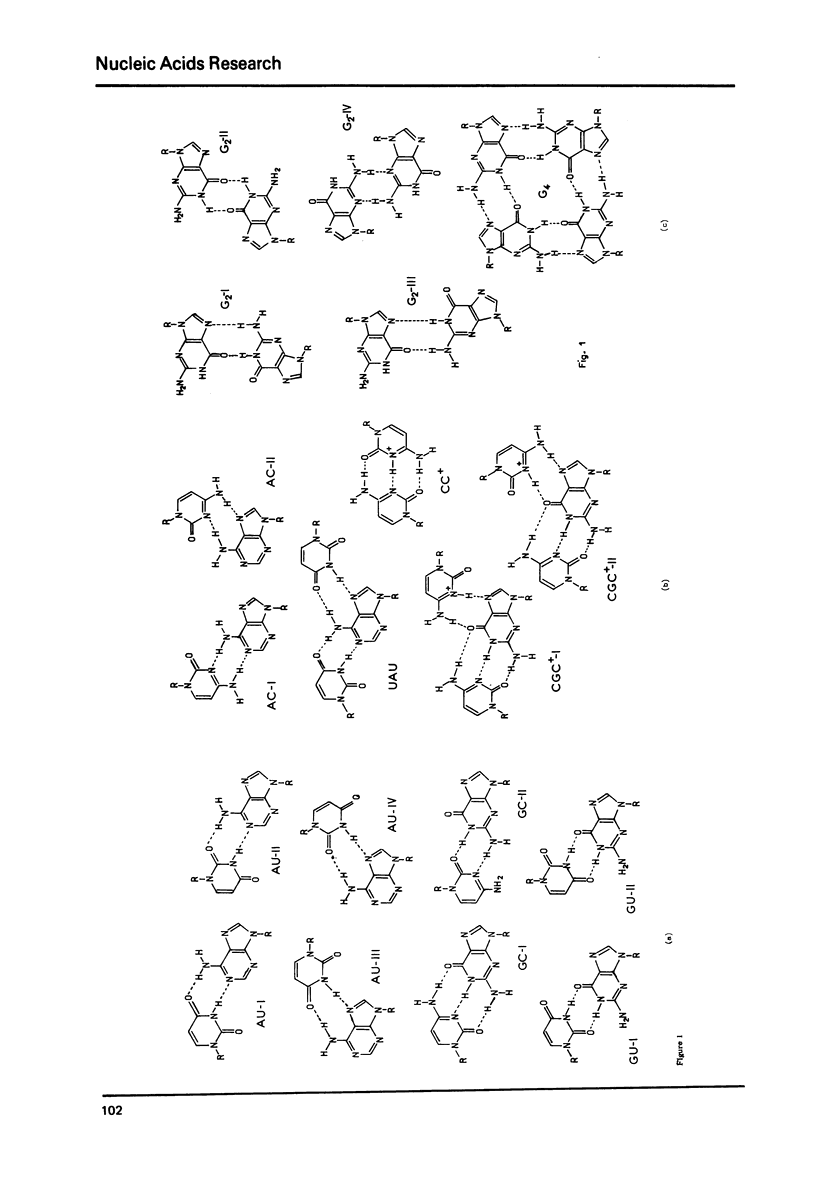
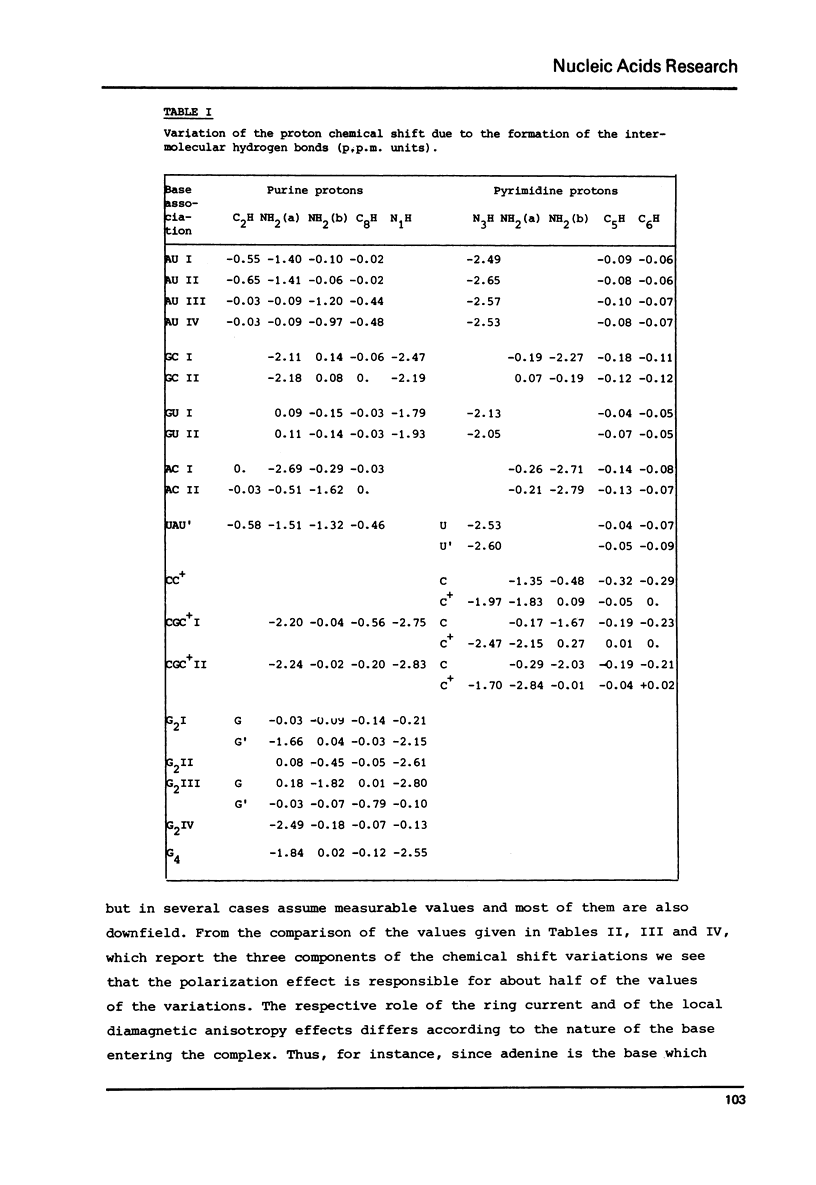
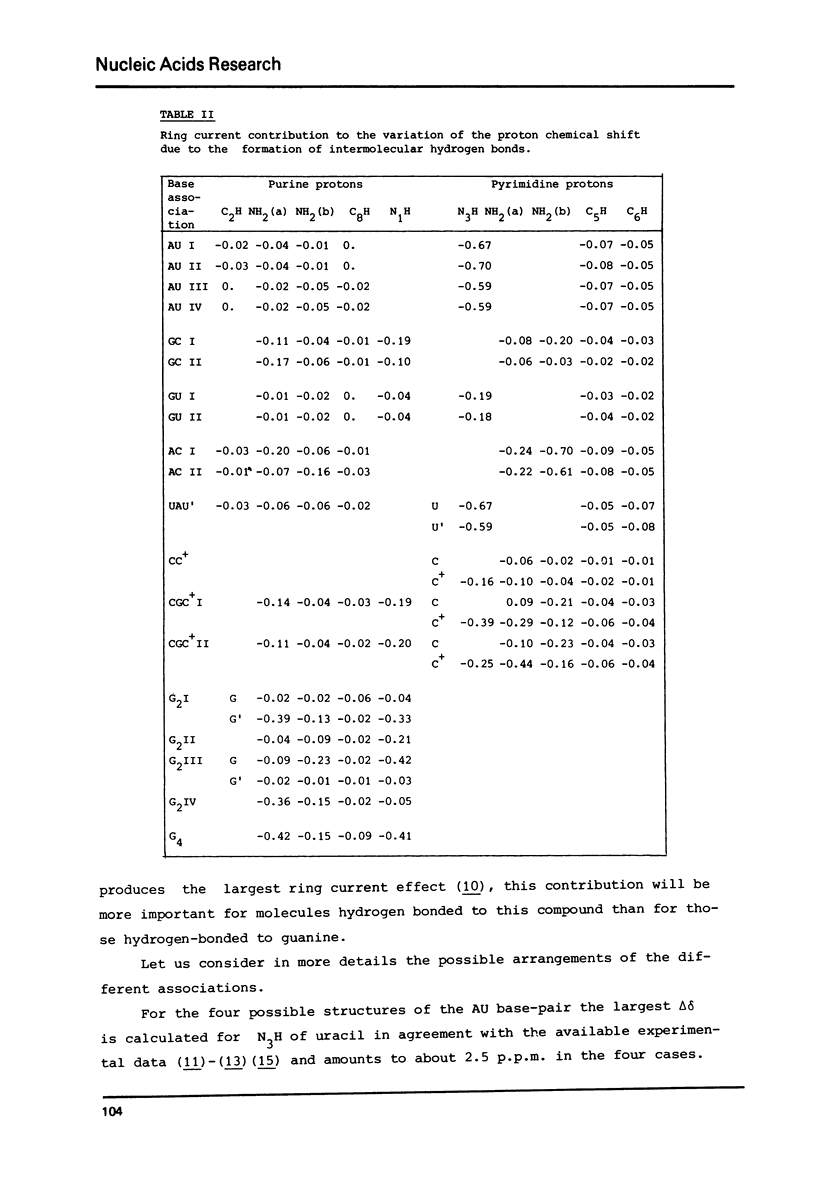
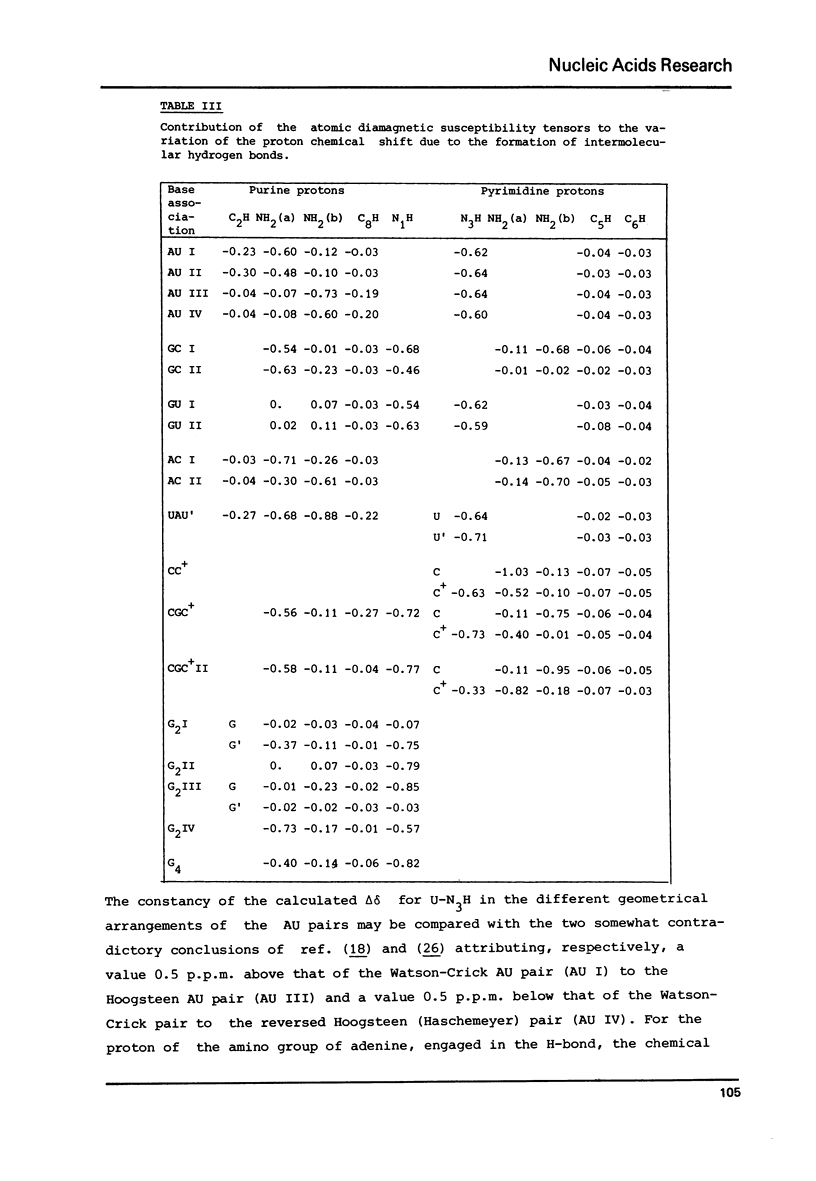







Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Arnott S., Chandrasekaran R., Hukins D. W., Smith P. J., Watts L. Structural details of double-helix observed for DNAs containing alternating purine and pyrimidine sequences. J Mol Biol. 1974 Sep 15;88(2):523–533. doi: 10.1016/0022-2836(74)90499-9. [DOI] [PubMed] [Google Scholar]
- Arnott S., Hukins D. W., Dover S. D. Optimised parameters for RNA double-helices. Biochem Biophys Res Commun. 1972 Sep 26;48(6):1392–1399. doi: 10.1016/0006-291x(72)90867-4. [DOI] [PubMed] [Google Scholar]
- Arnott S., Hukins D. W. Optimised parameters for A-DNA and B-DNA. Biochem Biophys Res Commun. 1972 Jun 28;47(6):1504–1509. doi: 10.1016/0006-291x(72)90243-4. [DOI] [PubMed] [Google Scholar]
- Arter D. B., Schmidt P. G. Ring current shielding effects in nucleic acid double helices. Nucleic Acids Res. 1976 Jun;3(6):1437–1447. doi: 10.1093/nar/3.6.1437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borer P. N., Kan L. S., Ts'o P. O. Conformation and interaction of short nucleic acid double-stranded helices. I. Proton magnetic resonance studies on the nonexchangeable protons of ribosyl ApApGpCpUpU. Biochemistry. 1975 Nov 4;14(22):4847–4863. doi: 10.1021/bi00693a012. [DOI] [PubMed] [Google Scholar]
- Cross A. D., Crothers D. M. A proton magnetic resonance study of single-stranded and double-helical deoxyribooligonucleotides. Biochemistry. 1971 Oct 26;10(22):4015–4023. doi: 10.1021/bi00798a002. [DOI] [PubMed] [Google Scholar]
- Engel J. D., von Hippel P. H. Effects of methylation on the stability of nucleic acid conformations: studies at the monomer level. Biochemistry. 1974 Sep 24;13(20):4143–4158. doi: 10.1021/bi00717a013. [DOI] [PubMed] [Google Scholar]
- Giessner-Prettre C., Pullman B. Intermolecular nuclear shielding values for protons of purines and flavins. J Theor Biol. 1970 Apr;27(1):87–95. doi: 10.1016/0022-5193(70)90130-x. [DOI] [PubMed] [Google Scholar]
- Giessner-Prettre C., Pullman B. On the atomic or "local" contributions to proton chemical shifts due to the anisotropy of the diamagnetic susceptibility of the nucleic acid base. Biochem Biophys Res Commun. 1976 May 17;70(2):578–581. doi: 10.1016/0006-291x(76)91086-x. [DOI] [PubMed] [Google Scholar]
- Giessner-Prettre C., Pullman B. Variation des déplacements chimiques des protons et formation des paires purines-pyrimidines complémentaires en solution non aqueuse. C R Acad Sci Hebd Seances Acad Sci D. 1970 Feb 9;270(6):866–868. [PubMed] [Google Scholar]
- Howard F. B., Frazier J., Singer M. F., Miles H. T. Helix formation between polyribonucleotides and purines, purine nucleosides and nucleotides. II. J Mol Biol. 1966 Apr;16(2):415–439. doi: 10.1016/s0022-2836(66)80183-3. [DOI] [PubMed] [Google Scholar]
- Kallenbach N. R., Daniel W. E., Jr, Kaminker M. A. Nuclear magnetic resonance study of hydrogen-bonded ring protons in oligonucleotide helices involving classical and nonclassical base pairs. Biochemistry. 1976 Mar 23;15(6):1218–1224. doi: 10.1021/bi00651a007. [DOI] [PubMed] [Google Scholar]
- Katz L., Penman S. Association by hydrogen bonding of free nucleosides in non-aqueous solution. J Mol Biol. 1966 Jan;15(1):220–231. doi: 10.1016/s0022-2836(66)80222-x. [DOI] [PubMed] [Google Scholar]
- Katz L. Proton magnetic resonance investigation of the association of adenine and uracil derivatives in chloroform solution. J Mol Biol. 1969 Sep 14;44(2):279–296. doi: 10.1016/0022-2836(69)90175-2. [DOI] [PubMed] [Google Scholar]
- Kim S. H. Three-dimensional structure of transfer RNA. Prog Nucleic Acid Res Mol Biol. 1976;17:181–216. doi: 10.1016/s0079-6603(08)60070-7. [DOI] [PubMed] [Google Scholar]
- Kroon P. A., Kreishman G. P., Nelson J. H., Chan S. I. The effects of chain length on the secondary structure of oligoadenylates. Biopolymers. 1974 Dec;13(12):2571–2592. doi: 10.1002/bip.1974.360131214. [DOI] [PubMed] [Google Scholar]
- Lightfoot D. R., Wong K. L., Kearns D. R., Reid B. R., Shulman R. G. Assignment of the low field proton nuclear magnetic resonance spectrum of yeast phenylalanine transfer RNA to specific base pairs. J Mol Biol. 1973 Jun 25;78(1):71–89. doi: 10.1016/0022-2836(73)90429-4. [DOI] [PubMed] [Google Scholar]
- MARVIN D. A., SPENCER M., WILKINS M. H., HAMILTON L. D. The molecular configuration of deoxyribonucleic acid. III. X-ray diffraction study of the C form of the lithium salt. J Mol Biol. 1961 Oct;3:547–565. doi: 10.1016/s0022-2836(61)80021-1. [DOI] [PubMed] [Google Scholar]
- Newmark R. A., Cantor C. R. Nuclear magnetic resonance study of the interactions of guanosine and cytidine in dimethyl sulfoxide. J Am Chem Soc. 1968 Aug 28;90(18):5010–5017. doi: 10.1021/ja01020a041. [DOI] [PubMed] [Google Scholar]
- Paetkau V., Coulter M. B., Flintoff W. F., Morgan A. R. Thymine-guanine base pairing during transcription of polydeoxypyrimidines in vitro. J Mol Biol. 1972 Nov 14;71(2):293–306. doi: 10.1016/0022-2836(72)90352-x. [DOI] [PubMed] [Google Scholar]
- Patel D. J., Canuel L. Nuclear magnetic resonance studies of the helix-coil transition of poly (dA-dT) in aqueous solution. Proc Natl Acad Sci U S A. 1976 Mar;73(3):674–678. doi: 10.1073/pnas.73.3.674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel D. J., Tonelli A. E. Assignment of the proton Nmr chemical shifts of the T-N3H and G-N1H proton resonances in isolated AT and GC Watson-Crick base pairs in double-stranded deoxy oligonucleotides in aqueous solution. Biopolymers. 1974;13(10):1943–1964. doi: 10.1002/bip.1974.360131003. [DOI] [PubMed] [Google Scholar]
- Pinnavaia T. J., Miles H. T., Becker E. D. Letter: Self-assembled 5'-guanosine monophosphate. Nuclear magnetic resonance evidence for a regular, ordered structure and slow chemical exchange. J Am Chem Soc. 1975 Nov 26;97(24):7198–7200. doi: 10.1021/ja00857a059. [DOI] [PubMed] [Google Scholar]
- Quigley G. J., Seeman N. C., Wang A. H., Suddath F. L., Rich A. Yeast phenylalanine transfer RNA: atomic coordinates and torsion angles. Nucleic Acids Res. 1975 Dec;2(12):2329–2341. doi: 10.1093/nar/2.12.2329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raszka M., Kaplan N. O. Association by hydrogen bonding of mononucleotides in aqueous solution. Proc Natl Acad Sci U S A. 1972 Aug;69(8):2025–2029. doi: 10.1073/pnas.69.8.2025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robillard G. T., Hilbers C. W., Reid B. R., Gangloff J., Dirheimer G., Shulman R. G. A study of secondary and tertiary solution structure of yeast tRNA(Asp) by nuclear magnetic resonance. Assignment of G.U ring NH and hydrogen-bonded base pair proton resonances. Biochemistry. 1976 May 4;15(9):1883–1888. doi: 10.1021/bi00654a014. [DOI] [PubMed] [Google Scholar]
- Robillard G. T., Tarr C. E., Vosman F., Berendsen H. J. Similarity of the crystal and solution structure of yeast tRNAPhe. Nature. 1976 Jul 29;262(5567):363–369. doi: 10.1038/262363a0. [DOI] [PubMed] [Google Scholar]
- Rordorf B. F., Kearns D. R., Hawkins E., Chang S. H. High-resolution NMR study of yeast tRNA Leu CUA and the native and denatured conformers of yeast tRNA Leu UUG. Biopolymers. 1976 Feb;15(2):325–336. doi: 10.1002/bip.1976.360150210. [DOI] [PubMed] [Google Scholar]
- Rosenberg J. M., Seeman N. C., Day R. O., Rich A. RNA double helices generated from crystal structures of double helical dinucleoside phosphates. Biochem Biophys Res Commun. 1976 Apr 19;69(4):979–987. doi: 10.1016/0006-291x(76)90469-1. [DOI] [PubMed] [Google Scholar]
- Scruggs R. L., Ross P. D. A calorimetric study of monomer-polymer complexes formed by polyribouridylic acid and some adenine derivatives. J Mol Biol. 1970 Jan 14;47(1):29–40. doi: 10.1016/0022-2836(70)90399-2. [DOI] [PubMed] [Google Scholar]
- Shoup R. R., Miles H. T., Becker E. D. NMR evidence of specific base-pairing between purines and pyrimidines. Biochem Biophys Res Commun. 1966 Apr 19;23(2):194–201. doi: 10.1016/0006-291x(66)90527-4. [DOI] [PubMed] [Google Scholar]
- Shulman R. G., Hilbers C. W. Ring-current shifts in the 300 MHz nuclear magnetic resonance spectra of six purified transfer RNA molecules. J Mol Biol. 1973 Jun 25;78(1):57–69. doi: 10.1016/0022-2836(73)90428-2. [DOI] [PubMed] [Google Scholar]
- Wong K. L., Wong Y. P., Kearns D. R. Investigation of the thermal unfolding of secondary and tertiary structure in E. coli tRNAfMet by high-resolution Nmr. Biopolymers. 1975 Apr;14(4):749–762. doi: 10.1002/bip.1975.360140407. [DOI] [PubMed] [Google Scholar]
- Young M. A., Krugh T. R. Proton magnetic resonance studies of double helical oligonucleotides. The effect of base sequence on the stability of deoxydinucleotide dimers. Biochemistry. 1975 Nov 4;14(22):4841–4847. doi: 10.1021/bi00693a011. [DOI] [PubMed] [Google Scholar]
- Zimmerman S. B., Cohen G. H., Davies D. R. X-ray fiber diffraction and model-building study of polyguanylic acid and polyinosinic acid. J Mol Biol. 1975 Feb 25;92(2):181–192. doi: 10.1016/0022-2836(75)90222-3. [DOI] [PubMed] [Google Scholar]


