Abstract
The subclassification of glioblastoma (GBM) into clinically relevant subtypes using microRNA (miRNA)– and messenger RNA (mRNA)–based integrated analysis has been attempted. Because miRNAs regulate multiple gene-signaling pathways, understanding miRNA-mRNA interactions is a prerequisite for understanding glioma biology. However, such associations have not been thoroughly examined using high-throughput integrated analysis. To identify significant miRNA-mRNA correlations, we selected and quantified signature miRNAs and mRNAs in 82 gliomas (grade II: 14, III: 16, IV: 52) using real-time reverse-transcriptase polymerase chain reaction. Quantitative expression data were integrated into a single analysis platform that evaluated the expression relationship between miRNAs and mRNAs. The 21 miRNAs include miR-15b, -21, -34a, -105, -124a, -128a, -135b, -184, -196a-b, -200a-c, -203, -302a-d, -363, -367, and -504. In addition, we examined 23 genes, including proneural markers (DLL3, BCAN, and OLIG2), mesenchymal markers (YKL-40, CD44, and Vimentin), cancer stem cell-related markers, and receptor tyrosine kinase genes. Primary GBM was characterized exclusively by upregulation of mesenchymal markers, whereas secondary GBM was characterized by significant downregulation of mesenchymal markers, miR-21, and -34a, and by upregulation of proneural markers and miR-504. Statistical analysis showed that expression of miR-128a, -504, -124a, and -184 each negatively correlated with the expression of mesenchymal markers in GBM. Our functional analysis of miR-128a and -504 as inhibitors demonstrated that suppression of miR-128a and -504 increased the expression of mesenchymal markers in glioblastoma cell lines. Mesenchymal signaling in GBM may be negatively regulated by miR-128a and -504.
Keywords: glioma, mesenchymal, microRNA, miR-128a, miR-504
Recent technological advancements in molecular genotyping and expression profiling have shown that the molecular stratification of glioblastoma (GBM) provides better insight into tumor biology than does traditional histopathological classification.1–3 The profiling of GMB with regard to genomic alterations, transcripts, and the proteome has identified prognostic biomarkers. MicroRNAs (miRNAs), which are small noncoding RNA molecules, regulate the expression of a wide variety of genes at the posttranscriptional level. Recent evidence demonstrates that miRNAs can function as both negative gene regulators in normal tissues and tumor suppressors and oncogenes in various cancers.4–8 We previously reported that miR-196 may play an important role in the malignant progression of GBM.9 Because miRNAs have the potential to regulate the expression of a large number of genes, identifying the targets of miRNAs is critical for understanding glioma biology. Various algorithms have been used to make computational predictions about associations between miRNAs and mRNAs;10,11 nevertheless, these associations need to be validated, and statistical variability is a particular concern.
Using global gene expression profiling, several groups have categorized GBM into several subgroups using different methodologies. Representative classification schemes have been reported by Phillips et al. and Verhaak et al.12,13 Phillips et al. categorized high-grade gliomas into 3 subtypes: proneural, proliferative, and mesenchymal. The proneural subtype shows high expression of genes implicated in neurogenesis and is associated with better clinical outcomes.12 By contrast, the proliferative and mesenchymal subtypes are characterized by the high expression of genes correlated with cell proliferation or angiogenesis, respectively, and both subtypes are associated with poor clinical outcomes. Verhaak et al. classified GBM into proneural, neural, mesenchymal, and classic subtypes, and some subtypes show strong associations with specific genomic alterations.13 Although the classification schemes of Phillips et al. and Verhaak et al. used different sample sets and methodologies, Huse et al. used cross-validation analysis to show that the proneural and mesenchymal signature is concordant between the 2 studies; this analysis indicated that the classification of transcriptional subtypes into 3 groups—proneural, mesenchymal, and others—can be considered to be a general consensus.14
The Cancer Genome Atlas (TCGA) project indicated that receptor tyrosine kinase (RTK) signaling pathways, such as those involving the epidermal growth factor receptor (EGFR) or the platelet-derived growth factor receptor (PDGFR), are frequently altered and activated in GBM; therefore, RTK gene expression may provide diagnostic biomarkers.15 Moreover, the expression of cancer stem cell markers, such as CD133, Nestin, BMI-1, and MELK, has prognostic significance in GBM.16 Therefore, these mRNAs can be designated as signature GBM mRNAs and may provide insights into GBM biology. In this study, in an effort to identify associations between the expression of miRNAs that are aberrantly expressed in GBM and the signature mRNAs that play important roles in glioma biology, we used real-time reverse-transcriptase polymerase chain reaction (RT-PCR) to quantify the expression of selected miRNAs and mRNAs in 82 independent gliomas. Expression profiles were analyzed on the basis of the histological type and the grade of each tumor, and correlations were statistically evaluated.
Materials and Methods
Patients with Glioma and Tumor Specimens
We collected samples of 82 glioma cases from the Kyushu University Brain Tumor Bank; each sample was obtained from a patient during surgery and with the approval of the university ethics committee. For each tumor, a histological diagnosis of GBM was determined on the basis of WHO criteria by board-certified neuropathologists. The tumors consisted of 7 diffuse astrocytoma (DA), 7 oligodendroglioma (O), 7 anaplastic astrocytoma (AA), 9 anaplastic oligodendroglioma (AO), and 52 GBM, of which 43 tumors were primary GBM (pGBM) and 9 were secondary GBM (sGBM). Cases of recurrent GBM were excluded from this study. Normal brain reference RNA (NBRR) and normal brain RNA (NBR) were used as internal controls; NBRR was purchased from Ambion, and NBR was extracted from brain tissues resected during epilepsy surgery.
Target Genes and miRNAs
To quantify the relative expression of proneural and mesenchymal genes, we selected DLL3, BCAN (brevican), and OLIG2 as representative proneural genes and YKL40 (CHI3L1), CD44, and Vimentin (VIM) as representative mesenchymal genes based on the studies by Phillips et al. and Verhaak et al.12,13 Primer pairs for amplification were designed for nonredundant regions in the relevant National Center for Biotechnology Information (NCBI) Reference Sequence (RefSeq) using Primer 3 software. Primer sequences for each gene are provided in Supplementary Table 1. For miRNA quantification, we chose to analyze 21 miRNAs that were shown to be differentially expressed in GBM in a previous study.9 The list of miRNAs examined is composed of miR-196a, -196b, -128a, -200a, -200b, -200c, -302a, -302b, -302c, -302d, -184, -105, -203, -504, -367, -34a, -363, -124a, -135b, -15b, and -21. To evaluate stem cell marker and RTK gene expression, we quantified 5 cancer stem cell–related markers—CD133, Nestin, BMI-1, MELK, and Notch 1-4—and 6 RTK genes—EGFR, VEGFR1-3, FGFR1, FGFR2, PDGFRA, and PDGFRB—as described in a previous study.17
RNA Extraction, Reverse Transcription, and Real-Time RT-PCR
Total RNA was extracted from frozen samples and from cell lines using the commercial mirVana miRNA isolation kit (Life Technologies). Reverse transcription was performed using random hexamers and a High Capacity cDNA Reverse Transcription kit (Life Technologies). Reverse-transcribed products were amplified using the SYBR green method and the ABI PRISM 7500 Fast Real-time PCR System (Life Technologies). In brief, 2 µL of cDNA product was used as a template in a 20 µL PCR containing 10 µL Power SYBR Green Master Mix (Life Technologies), 200 mM of each primer, 1 µL DMSO, and 6.2 µL distilled water. Hypoxanthine phosphoribosyltransferase 1 (HPRT1) was used as a reference gene. Amplification protocols were as follows: 95°C for 10 min, 40 cycles of 95°C for 15 s, and 40 cycles of 60°C for 60 s, with melting curve analysis. The threshold cycle number (CT) was automatically determined by the ABI 7500 Fast System SDS software. All reactions were performed in duplicate. Quantification of miRNAs was performed using the TaqMan miRNA probes described in the previous study. Some of the pGBM data were derived from a previous study.9
Quantitative RT-PCR Data Analysis and Statistical Analysis
The mean CT value of duplicate runs was determined for each gene, and ΔCT was calculated by subtracting the CT value of the HPRT1 reference gene from that of each gene. For miRNA expression, RNU44 and RNU48 were used as endogenous controls. The relative expression of each gene was quantified using the ΔΔCT method. In this method, ΔΔCT was calculated by subtracting the ΔCT of NBRR from that of the target gene and then calculating the relative quantity (RQ). RQ was further normalized using the mean number of NBRR and NBR. For GBM cases, the Z score of BCAN, DLL3, CD44, and YKL-40 expression was calculated and plotted. Statistical analyses were performed using JMP , version 9 (SAS Institute).
Loss of Heterozygosity Analysis and Detection of IDH1/2 Mutations in Glioma Tissues
Loss of heterozygosity on chromosome 10, 1p, and 19q and detection of IDH1 and IDH2 mutations were performed for all 82 gliomas as described in the Supplementary Methods section.
Transfection of miRNA Inhibitors
MirVana miRNA inhibitors of miR-128a and -504 were purchased from Life Technologies. As a negative control for experiments using these inhibitors, Negative Control #1 (Life Technologies, cat. 4464076) of mirVana miRNA inhibitor was also purchased. These miRNA inhibitors and the negative control were transected into the conventional glioma cell line, U87, and the glioma initiating cell, KNS1295, using Lipofectamine RNAiMAX (Life Technologies) at the final concentration of 50 μM according to the forward transfection protocol. Transfection procedures were performed in duplicate. RNA was extracted and gene expression was evaluated using quantitative RT-PCR with in 48 h after transfection as described above.
Results
Differentially Regulated miRNAs and Genes Correlated with WHO Histological Grading
Relative expression values calibrated by normal brain tissue expression were obtained for all of the miRNAs and genes examined in this study. Raw relative expression values are shown in Supplementary Table 2. Statistical analysis using the Mann-Whitney U test indicated that the expressions of 10 miRNAs and 15 genes showed a positive association with tumor grading, whereas expressions of 4 miRNAs and 3 genes was negatively correlated with tumor grade (Fig. 1, Table 1).
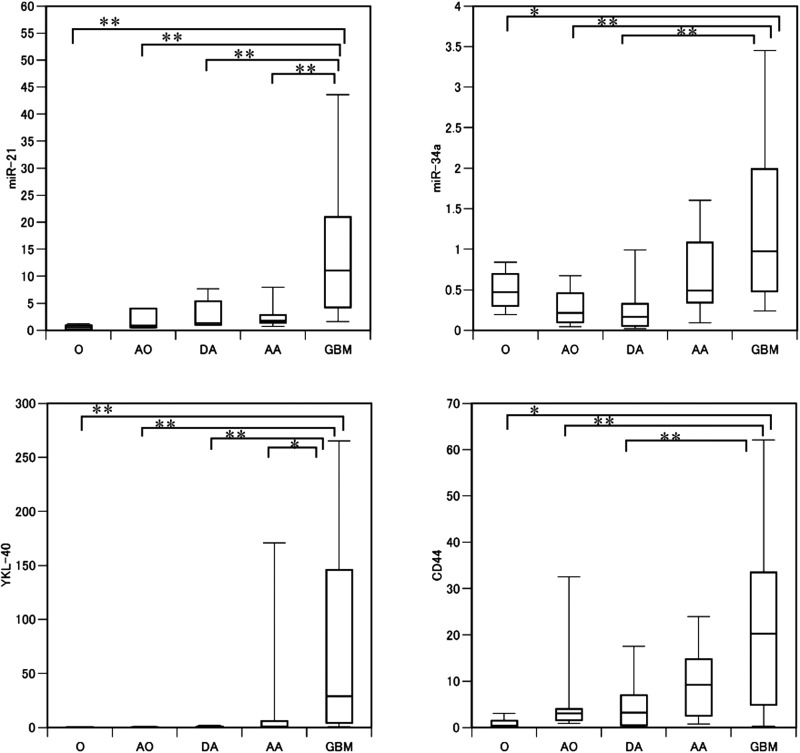
Fig. 1.
Relative expression of miR-21, miR-34a, YKL-40, and CD44 in glioma tissues by histology The expression of 2 microRNAs and of 2 mRNAs in tumor samples was normalized relative to expression of their respective expression in normal brain tissues; each of these microRNAs and mRNAs was significantly upregulated in high-grade glioma samples, particularly glioblastoma (Mann-Whitney U test). The relative expression value is shown on the ordinate. O, oligodendroglioma (WHO grade II); AO, anaplastic oligodendroglioma (III); DA, diffuse astrocytoma (II); AA, anaplastic astrocytoma (III); GBM, glioblastoma (IV). *P < .05, **P < .01.
Table 1.
List of microRNAs and genes that showed a statistical correlation with WHO glioma grade
| Mean expression value |
P-value |
|||||
|---|---|---|---|---|---|---|
| II | III | IV | II vs III | III vs IV | II vs IV | |
| mirR-196a | 0.455 | 17.306 | 224.882 | 0.3941 | <0.0001 | <0.0001 |
| mirR-196b | 1.777 | 18.609 | 215.098 | 0.5747 | <0.0001 | <0.0001 |
| mirR-15b | 1.801 | 2.261 | 6.193 | 0.3496 | <0.0001 | <0.0001 |
| mirR-21 | 1.807 | 2.266 | 16.811 | 0.3496 | <0.0001 | <0.0001 |
| mirR-200c | 0.97154 | 1.55 | 3.403 | 0.6929 | 0.0554 | 0.0092 |
| mirR-105 | 1.093 | 1.034 | 1.066 | 0.8679 | 0.0132 | 0.0211 |
| mirR-34a | 0.398 | 0.473 | 1.393 | 0.8192 | 0.0008 | 0.0004 |
| mirR-135b | 0.347 | 0.386 | 2.099 | 0.1515 | 0.0012 | 0.0129 |
| mirR-200a | 0.57 | 0.631 | 2.019 | 0.787 | 0.0033 | 0.0033 |
| mirR-203 | 1.109 | 0.301 | 0.655 | 0.0323 | 0.8283 | 0.0508 |
| YKL-40 | 0.434 | 14.118 | 111.683 | 0.4669 | <0.0001 | <0.0001 |
| CD44 | 3.1175 | 8.703 | 25.449 | 0.0396 | 0.008 | 0.0001 |
| VIM | 0.929 | 7.543 | 52.291 | 0.0168 | <0.0001 | <0.0001 |
| Nestin | 74.472 | 97.35 | 236.29 | 0.2797 | 0.0063 | 0.0016 |
| MELK | 21.19 | 159.75 | 2145.9 | 0.015 | <0.0001 | <0.0001 |
| BMI-1 | 0.293 | 3.453 | 3.997 | 0.0011 | 0.0302 | <0.0001 |
| CD133 | 5.063 | 8.804 | 51.248 | 0.0772 | 0.0949 | 0.0019 |
| Notch1 | 15.038 | 21.99 | 25.037 | 0.0613 | 0.734 | 0.037 |
| Notch2 | 7.434 | 10.125 | 13.761 | 0.2443 | 0.1883 | 0.0071 |
| Notch3 | 4.516 | 5.385 | 13.783 | 0.2706 | 0.0058 | 0.0033 |
| Notch4 | 1.137 | 3.122 | 12.531 | 0.0094 | 0.0014 | <0.0001 |
| VEGFR1 | 1.238 | 1.565 | 3.282 | 0.2444 | 0.0622 | 0.0077 |
| VEGFR2 | 7.222 | 10.771 | 25.187 | 0.3085 | 0.129 | 0.0094 |
| VEGFR3 | 5.805 | 11.153 | 28.015 | 0.0532 | 116 | <0.0001 |
| FGFR1 | 5.891 | 5.26 | 12.114 | 0.787 | 0.0028 | 0.0054 |
| mirR-504 | 0.553 | 0.368 | 0.221 | 0.0845 | 0.0036 | 0.0002 |
| mirR-184 | 1.643 | 0.867 | 0.775 | 0.14 | 0.0006 | 0.0001 |
| mirR-128a | 0.533 | 0.303 | 0.198 | 0.0586 | 0.0085 | <0.0001 |
| mirR-124a | 0.575 | 0.409 | 0.407 | 0.4928 | 0.2058 | 0.0323 |
| DLL3 | 441.958 | 406.483 | 211.165 | 0.5194 | 0.027 | 0.185 |
| BCAN | 70.332 | 102.253 | 57.354 | 0.0773 | 0.0091 | 0.8569 |
| OLIG2 | 26.225 | 51.581 | 76 | 0.0358 | 0.0051 | 0.8323 |
Abbreviations: II, grade II (diffuse astrocytoma and oligodendroglioma); III, grade III (anaplastic astrocytoma and anaplastic oligodendroglioma); IV, grade IV (glioblastoma).
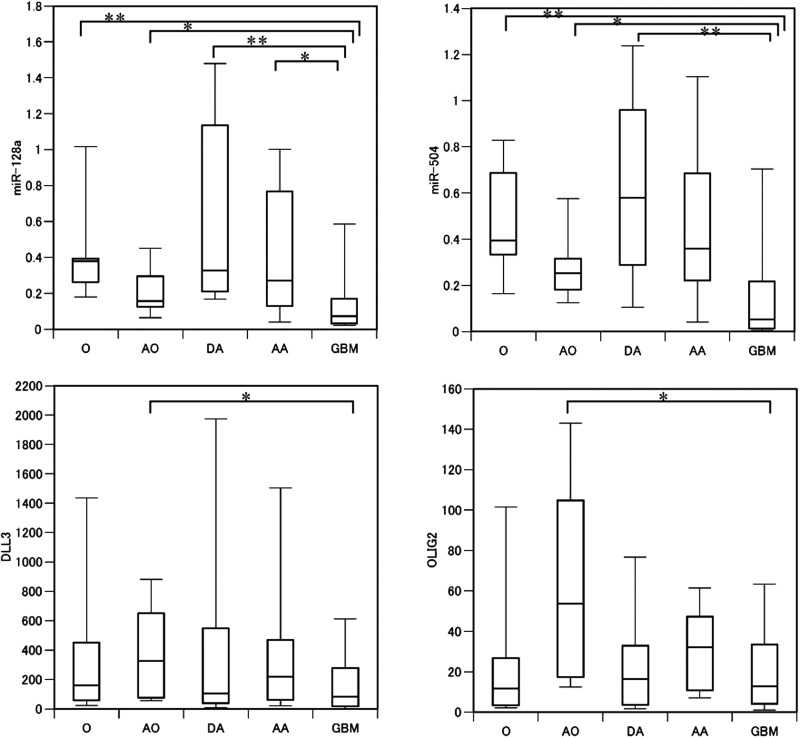
The expressions of 4 miRNAs (miR-196a, -196b, -15b, and -21) showed strong associations with tumor grade; moreover, expressions of 3 genes (mesenchymal markers YKL-40 and VIM and stem cell marker MELK) showed a significant correlation with tumor malignancy (Table 1). Conversely, expressions of 4 miRNAs and 3 mRNAs (miR-184, -504, -128a, and -124; DLL3, BCAN, and OLIG2, all proneural genes) was negatively correlated with tumor grade. However, proneural gene expression was not significantly different between grade II and IV. Thus, we reanalyzed the expression data from the proneural genes and from miR-184, -504, -128a, and -124 using both histology and tumor grade. A statistically significant difference was detected between AO and GBM for all of the 3 proneural genes (Mann-Whitney U test, P < .05), possibly explaining why no statistically significant difference was observed between grade II and IV gliomas (Fig. 2).
Fig. 2.
Relative expression of miR-128a, miR-504, DLL3, and OLIG2 in glioma. Expression of miR-128a and -504 were inversely associated with glioma malignancy. Expression of DLL3 and OLIG2 each showed a similar trend, although the differences were not statistically significant. *P < .05, **P < .01.
Differentially Regulated miRNAs and Genes in Primary and Secondary GBM
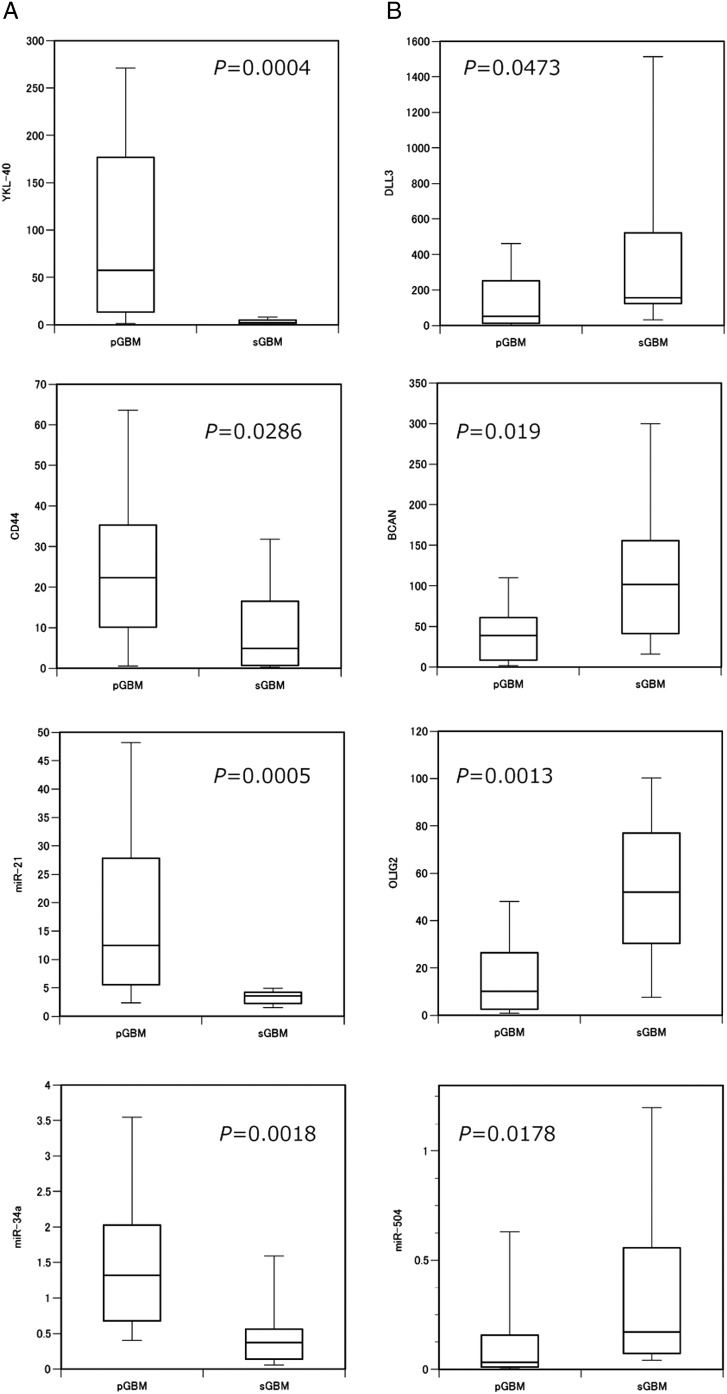
Primary GBM and sGBM differ in clinical and genetic characteristics; to better understand the molecular genetic stratification between these tumor grades, we compared pGBMs and sGBMs with regard to the expression of the 21 miRNAs and 18 mRNAs selected for this study. Our genetic analysis indicated that, in pGBM, the IDH1/2 mutation and total loss of chromosome 10 were detected in 1 of 43 (2%) and 30 of 43 (70%) samples, respectively; however, in sGBM, the frequency of IDH1/2 mutation (8 of 9; 89%) was high, and total chromosome 10 loss was not detected (Supplementary Table 3). Expressions of 2 genes, YKL-40 and CD44, was significantly lower in sGBM than in pGBM (Mann-Whitney U test, P = .0004 and P = .0286, respectively). Similarly, expressions of 2 miRNAs, miR-21 and -34a, was also significantly lower in sGBM than in pGBM (P = .0005 and P = .0018, respectively). In contrast, DLL3, BCAN, and OLIG2 were each significantly upregulated in pGBM relative to sGBM (P = .0473, P = .019, and P = .0013, respectively). The expression of miR-504 was higher in sGBM than in pGBM (P = .0178); further analyses are necessary to determine whether expression of miR-184, -128a, or -124 is elevated in sGBM relative to pGBM. No statistically significant difference in expression of any stem cell marker was detected between pGBM and sGBM. Of the TKR genes investigated, expression of PDGFRA was significantly upregulated, whereas PDGFRB, FGFR1, VEGFR1, and VEGFR3 indicated a significant decrease in expression of sGBM relative to pGBM. In summary, DLL3, BCAN, OLIG2 (proneural markers), miR-504, and PDGFRA each showed higher expression in sGBM than in pGBM; in contrast, YKL-40, CD44 (mesenchymal markers), miR-21, miR-34a, PDGFRB, FGFR1, VEGFR1, and VEGFR3 each showed lower expression in sGBM than in pGBM (Fig. 3, Supplementary Fig. 2).
Fig. 3.
The expressions of miR-21, miR-34a, miR-504, mesenchymal markers, and proneural markers were significantly different between pGBM and sGBM (A) Mesenchymal markers (YKL-40 and CD44), miR-21, and miR-34a were each downregulated in primary GBM; (B) In contrast, the proneural markers (DLL3, BCAN, and OLIG2) and miR-504 were each upregulated. The relative expression value is shown on the ordinate.
Statistical Association of miRNA and Gene Expression in GBM
To evaluate correlations between miRNA expression and gene expression, the nonparametric Spearman's rank test was used. Each miRNA that showed a significant positive correlation with a gene is listed in Table 2. Expression of miR-21 positively correlated with expression of each of the mesenchymal marker genes, and miR-34a expression positively correlated with YKL-40 expression. Moreover, expression of miR-21 positively correlated with miR-34a expression (Table 3). Expression miR-196a positively correlated with expression of each of PDGFRB, VEGFR1, and Notch3. Of interest, miR-128a, -504, -124a, and -184 each showed a negative correlation with each mesenchymal signature gene. Although miR-128a was weakly correlated with DLL3, no correlation between miR-124a, -184, or -504 expression and any proneural gene was detected (data not shown). Expression of miR-124a, -128a, -184, or -504 correlated significantly with expression of each other miRNA (Table 3). To investigate the correlations between proneural gene expression and mesenchymal signature gene expression in GBM tissues, we plotted the Z score of the relative expression of each gene (Supplementary Fig. 3). This figure shows a trend indicating that high expression of proneural and high expression of mesenchymal signature genes are mutually exclusive. These findings are consistent with findings from previous studies.12 Expression of YKL-40, CD44, or VIM correlated significantly with expression of each of the other 2 genes; similarly, expression of DLL3, BCAN, or OLIG2 correlated significantly with expression of each of the other 2 genes (Table 3). Taken together, these findings indicate that these 2 groups of genes (YKL-40, CD44, and VIM and DLL3, BCAN, and OLIG2) are reliable signature genes of mesenchymal and proneural GBM subtypes, respectively.
Table 2.
Statistical correlation between expression of individual miRNAs and individual mRNAs
| miRNA | Gene | Correlation coefficient | P-value | |
|---|---|---|---|---|
| Positive correlation | mirR-21 | YKL-40 | 0.5781 | <0.0001 |
| CD44 | 0.4431 | 0.001 | ||
| VIM | 0.2935 | 0.0347 | ||
| PDGFRB | 0.2891 | 0.0377 | ||
| miR-196a | PDGFRB | 0.3971 | 0.0036 | |
| VEGFR1 | 0.3617 | 0.0084 | ||
| Notch3 | 0.324 | 0.0191 | ||
| miR-128a | DLL3 | 0.2781 | 0.0459 | |
| miR-34a | YKL-40 | 0.3009 | 0.0302 | |
| Inverse correlation | mirR-128a | VIM | −0.7073 | <0.0001 |
| YKL-40 | −0.5544 | <0.0001 | ||
| Notch3 | −0.5332 | <0.0001 | ||
| Nestin | −0.5224 | <0.0001 | ||
| PDGFRB | −5225 | <0.0001 | ||
| VEGFR1 | −0.5209 | <0.0001 | ||
| mirR-504 | VIM | −0.6048 | <0.0001 | |
| YKL-40 | −0.5398 | <0.0001 | ||
| mirR-124a | VIM | −0.7236 | <0.0001 | |
| YKL-40 | −0.5599 | <0.0001 | ||
| Nestin | −0.5372 | <0.0001 | ||
| Notch2 | −0.5278 | <0.0001 | ||
| mirR-184 | VIMs | −0.5736 | <0.0001 | |
| mirR-105 | VIM | −0.5599 | <0.0001 | |
| Notch3 | −0.5232 | <0.0001 | ||
| miR-203 | VIM | −0.5232 | <0.0001 |
Table 3.
Pairwise correlations among miRNAs and mRNAs
| Correlation coefficient | P-value | |
|---|---|---|
| miRNA | ||
| miR-21 vs miR-34a | 0.6942 | <0.001 |
| miR-124a vs miR-128a | 0.8951 | <0.001 |
| miR-124a vs miR-184 | 0.6379 | <0.001 |
| miR-124a vs miR-504 | 0.8053 | <0.001 |
| miR-128a vs miR-184 | 0.6903 | <0.001 |
| miR-128a vs miR-504 | 0.8355 | <0.001 |
| miR-184 vs miR-504 | 0.6642 | <0.001 |
| Gene | ||
| YKL-40 vs CD44 | 0.6966 | <0.0001 |
| YKL-40 vs VIM | 0.6733 | <0.001 |
| CD44 vs VIM | 0.7654 | <0.001 |
| DLL3 vs BCAN | 0.7594 | <0.001 |
| DLL3 vs OLIG2 | 0.6881 | <0.001 |
| BCAN vs OLIG2 | 0.7817 | <0.001 |
Inhibition of mir-128a and mir-504 Increase Mesenchymal Gene Expression and Decrease Proneural Gene Expression
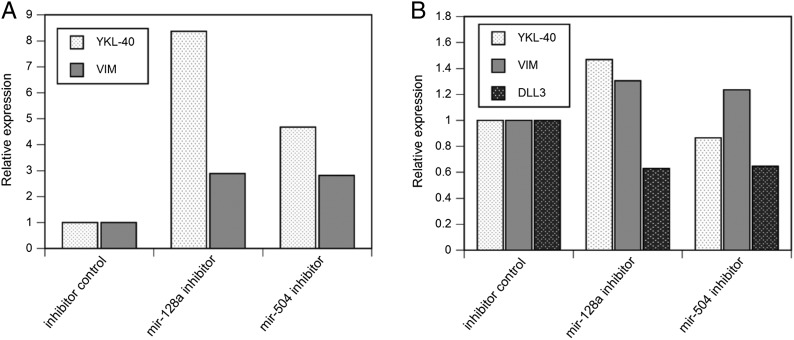
To determine whether there are any functional associations of mir-128a or mir-504 expression with mesenchymal or proneural gene expression, we transfected a mir-128a or mir-504 inhibitor into U87 or KNS1295 cells. We selected U87 and KNS1295 cells, because mesenchymal genes were most strongly expressed in U87 cells and proneural markers were most strongly expressed in KNS1295 cells (Supplementary Table 4). After transfection of mir-128a or mir-504 inhibitors, YKL-40 or VIM expression was increased in U87 and separately in KNS1295 (Fig. 4). Although the transfection of the different miRNA inhibitors had similar effects on VIM expression, transfection of mir-128a inhibitor resulted in stronger induction of YKL-40 expression than did transfection of the mir-504 inhibitor. In KNS1295 cells, DLL3 expression was suppressed by the transfection of either the mir-128a or the mir-504 inhibitor, compared to negative control.
Fig. 4.
Inhibition of mir-128a or mir-504 resulted in increased mesenchymal gene expression in U87 and KNS1295 cells and decreased proneural gene expression in KNS1295 cells (A) Transfection of a mir-128a or a mir-504 inhibitor, relative to that of a negative control miRNA, into U87 cells enhanced expression of YKL-40 and of VIM expression. (B) Similarly, transfection of a mir-128a or a mir-504 inhibitor into KNS1295 cells increased VIM expression, but only inhibition of mir-128a enhanced YKL-40 expression. Inhibition of mir-128a or mir-504 suppressed DLL3 expression.
Discussion
To stratify heterogeneous GBM into clinically relevant subtypes, global mRNA expression profiling has been used to establish novel classifications, and proneural and mesenchymal subtypes have been identified as representative classifiers.12–14 Although how many and which markers should be used in classifying GBM remains undetermined, we found that proneural and mesenchymal features can be efficiently assessed by monitoring 6 markers (YKL-40, CD44, VIM, DLL3, BCAN, and OLIG2) and that expression profiles comprising these markers reveal characteristic patterns that reflect glioma histology and grade. Specifically, sGBM could be differentiated from pGBM because proneural markers were upregulated and mesenchymal markers were downregulated in sGBMs relative to pGMBs. Although mRNA-based GBM subclassifications are reportedly not associated with significant survival differences,13 our results indicated that mesenchymal markers, stem cell markers, and some genes involved in RTK signaling were exclusively upregulated in pGBM. These results are consistent with previous findings, specifically that the mesenchymal phenotype is a molecular hallmark of GBM and is positively associated with tumor aggressiveness.18 Although Carro et al. have recently shown that 2 transcription factors, C/EBPb and STAT3, are master regulators of mesenchymal signaling, therapeutic targets have not be identified among mesenchymal signaling components.19
MicroRNAs have been shown to participate in the regulation of almost every cellular process; moreover, they contribute to tumorigenesis by modulating both oncogenic and tumor suppressor pathways; therefore, miRNA-mRNA regulation is likely to be crucial for glioma development and progression.4,8 Several reports have shown that profiles of miRNA expression can be used to classify human cancers, and these profiles are useful in diagnostic and prognostic assessments.9,20,21 However, the significance of the classifications based on these profiles needs to be further validated. Moreover, miRNA-mRNA regulation has not been thoroughly explored in the context of glioma biology. Here, we identified miRNAs and mRNAs that were differentially regulated between pGBM and sGBM. Expression of miR-504 was significantly higher in sGBM than in pGBM; however, expression of miR-21 and -34a were lower in sGBM. Proneural marker genes (DLL3, BCAN, and OLIG2) and PDGFRA showed significantly higher expression in sGBM than in pGBM; in contrast, mesenchymal marker genes (YKL-40 and CD44) including several other genes showed lower expression in sGBM, compared to pGBM. These results indicated that both gene signatures and miRNA signatures can differentiate the 2 types of glioma. Reportedly, miR-21 is overexpressed in various types of cancers, including glioblastoma, and has been designated as an oncomiR because of its oncogenic potential.22,23 Conversely, miR-34a functions as a tumor suppressor that regulates the p53 signaling pathway;24,25 moreover, Li et al. reported that miR-34a is inactivated in some gliomas.26 However, our results indicate that miR-34 was slightly upregulated in GBM, compared with normal brain tissues (mean relative quantification of 1.39). This discrepancy between the findings of Li et al. and our finding may be attributable to the difference in the number of GBM samples analyzed in the respective studies. Li et al. analyzed only 12 samples, whereas we examined 52 GBM samples, some of which showed lower expression than in normal brain (Supplementary Table 2). Recently, Silber et al. reported that miR-34a expression is significantly lower in proneural glioma; this finding is consistent with our results.27 Moreover, they demonstrated that PDGFRA is a direct target of miR-34a and that suppression of miR-34a inhibits proliferation only of proneural gliomas, but not of mesenchymal gliomas. Taken together, their findings indicate that the functional significance of miR-34a expression is dependent on cellar context.
Our most significant finding is that expression of miR-128a, -504, -124a, or -184 demonstrated a significant inverse correlation with expression of each of the mesenchymal markers. This set of findings prompted us to speculate that miR-128a, -504, -124a, or-184 may each function as a suppressor of the mesenchymal signaling pathway. This speculation was partly confirmed by functional analysis using miR-128a and -504 inhibitors. Our result showed that VIM expression was increased by inhibiting miR-128a or -504 expression and that YKL-40 expression was more strongly increased by inhibiting miR-128a than miR-504; these findings indicated that miR-128a was a stronger suppressor of the mesenchymal signaling pathway than miR-504. In KNS1295 cells, inhibition of miR-128a or mR-504 resulted in only minimal induction of YKL-40 expression; these small responses may occur because baseline expression of YKL-40 was low in this cell line. Of interest, inhibition of miR-128a resulted in suppression of DLL3 expression; these finding were consistent with our observation that expression of miR-128a was weakly correlated with DLL3 expression. Taken together, our finding indicated that mesenchymal signaling in GBM may be negatively regulated by miR-128a and -504.
Reportedly, miR-128 is a proneural glioma tumor suppressor that targets mitotic kinases; of interest, BMI-1 is also a direct target.28,29 However, our analysis did not show a correlation between miR-128 expression and BMI-1 expression in glioma (data not shown). In addition, Wuchty et al. recently reported that miR-128 confers tumor suppressive activity by downregulating the expression of WEE1, a tyrosine kinase that phosphorylates CDK1.10 They also identified an association between the expression of extracellular matrix proteins and expression of miR-124, which has been reported to exert tumor suppressive functions inhibiting stem cell activity and inducing tumor differentiation.30,31 These data support our finding that the expression of miR-128 and of miR-124 is associated with mesenchymal signaling. Although miR-184 reportedly plays an important role in the progression of malignancy in glioma, no functional target of miR-184 has yet been identified.21 miR-504 has oncogenic activity through its negative regulation of p53 protein levels.32 However, our results indicated that miR-504 expression was downregulated in GBM; this finding indicated that miR-504 may be tumor suppressive rather than oncogenic. Kim et al. recently reported an miRNA-based subclassification scheme in comparison with Verhaak's mRNA-based classification.33 According to this report, miR-128a, -504, and -124a are categorized into neural precursor clusters by their heterogeneous mRNA-based category. Our results indicated that the functional relevance of the expression of these microRNAs can be explained, in some part, by the suppression of mesenchymal signaling.
Although recent prediction algorithms have identified many miRNA and mRNA interactions,10 important interactions may be missed by these computational predictions. Here, we provided data indicating putative interactions between miRNAs and mesenchymal marker genes that have not been reported previously. Future experiments will be needed to validate these interactions in functional studies. Nonetheless, we believe that this study provides an important framework for identifying candidate targets of miRNAs and that identification of these candidates may lead to the development of new therapeutic targets.
Supplementary Material
Acknowledgments
We thank Ms. Fumie Doi for her technical assistance.
Conflict of interest statement. None declared.
Funding
This project was supported by the Ministry of Education, Culture, Sports, Science and Technology of Japan (22390279 to K.Y. and 22390280 to M. M.).
References
- 1.Freije WA, Castro-Vargas FE, Fang Z, et al. Gene expression profiling of gliomas strongly predicts survival. Cancer Res. 2004;64(18):6503–6510. doi: 10.1158/0008-5472.CAN-04-0452. [DOI] [PubMed] [Google Scholar]
- 2.Fuller GN, Hess KR, Rhee CH, et al. Molecular classification of human diffuse gliomas by multidimensional scaling analysis of gene expression profiles parallels morphology-based classification, correlates with survival, and reveals clinically-relevant novel glioma subsets. Brain Pathol. 2002;12(1):108–116. doi: 10.1111/j.1750-3639.2002.tb00427.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sulman EP, Aldape K. The use of global profiling in biomarker development for gliomas. Brain Pathol. 2011;21(1):88–95. doi: 10.1111/j.1750-3639.2010.00456.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ventura A, Jacks T. MicroRNAs and cancer: short RNAs go a long way. Cell. 2009;136(4):586–591. doi: 10.1016/j.cell.2009.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Esquela-Kerscher A, Slack FJ. Oncomirs - microRNAs with a role in cancer. Nat Rev Cancer. 2006;6(4):259–269. doi: 10.1038/nrc1840. [DOI] [PubMed] [Google Scholar]
- 6.Babashah S, Soleimani M. The oncogenic and tumour suppressive roles of microRNAs in cancer and apoptosis. Eur J Cancer. 2011;47(8):1127–1137. doi: 10.1016/j.ejca.2011.02.008. [DOI] [PubMed] [Google Scholar]
- 7.Chen CZ. MicroRNAs as oncogenes and tumor suppressors. N Engl J Med. 2005;353(17):1768–1771. doi: 10.1056/NEJMp058190. [DOI] [PubMed] [Google Scholar]
- 8.Calin GA, Croce CM. MicroRNA signatures in human cancers. Nat Rev Cancer. 2006;6(11):857–866. doi: 10.1038/nrc1997. [DOI] [PubMed] [Google Scholar]
- 9.Guan Y, Mizoguchi M, Yoshimoto K, et al. MiRNA-196 is upregulated in glioblastoma but not in anaplastic astrocytoma and has prognostic significance. Clin Cancer Res. 2010;16(16):4289–4297. doi: 10.1158/1078-0432.CCR-10-0207. [DOI] [PubMed] [Google Scholar]
- 10.Wuchty S, Arjona D, Li A, et al. Prediction of Associations between microRNAs and Gene Expression in Glioma Biology. PLoS One. 2011;6(2):e14681. doi: 10.1371/journal.pone.0014681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bandyopadhyay S, Mitra R, Maulik U, Zhang MQ. Development of the human cancer microRNA network. Silence. 2010;1(1):6. doi: 10.1186/1758-907X-1-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Phillips HS, Kharbanda S, Chen R, et al. Molecular subclasses of high-grade glioma predict prognosis, delineate a pattern of disease progression, and resemble stages in neurogenesis. Cancer Cell. 2006;9(3):157–173. doi: 10.1016/j.ccr.2006.02.019. [DOI] [PubMed] [Google Scholar]
- 13.Verhaak RG, Hoadley KA, Purdom E, et al. Integrated genomic analysis identifies clinically relevant subtypes of glioblastoma characterized by abnormalities in PDGFRA, IDH1, EGFR, and NF1. Cancer Cell. 2010;17(1):98–110. doi: 10.1016/j.ccr.2009.12.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Huse JT, Phillips HS, Brennan CW. Molecular subclassification of diffuse gliomas: seeing order in the chaos. Glia. 2011;59(8):1190–1199. doi: 10.1002/glia.21165. [DOI] [PubMed] [Google Scholar]
- 15.Cancer Genome Atlas Research Network. Comprehensive genomic characterization defines human glioblastoma genes and core pathways. Nature. 2008;455(7216):1061–1068. doi: 10.1038/nature07385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Natsume A, Kinjo S, Yuki K, et al. Glioma-initiating cells and molecular pathology: implications for therapy. Brain Tumor Pathol. 2011;28(1):1–12. doi: 10.1007/s10014-010-0011-3. [DOI] [PubMed] [Google Scholar]
- 17.Yoshimoto K, Ma X, Guan Y, et al. Expression of stem cell marker and receptor kinase genes in glioblastoma tissue quantified by real-time RT-PCR. Brain Tumor Pathol. 2011;28(4):291–296. doi: 10.1007/s10014-011-0046-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tso CL, Shintaku P, Chen J, et al. Primary glioblastomas express mesenchymal stem-like properties. Mol Cancer Res. 2006;4(9):607–619. doi: 10.1158/1541-7786.MCR-06-0005. [DOI] [PubMed] [Google Scholar]
- 19.Carro MS, Lim WK, Alvarez MJ, et al. The transcriptional network for mesenchymal transformation of brain tumours. Nature. 2010;463(7279):318–325. doi: 10.1038/nature08712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lawler S, Chiocca EA. Emerging functions of microRNAs in glioblastoma. J Neurooncol. 2009;92(3):297–306. doi: 10.1007/s11060-009-9843-2. [DOI] [PubMed] [Google Scholar]
- 21.Malzkorn B, Wolter M, Liesenberg F, et al. Identification and functional characterization of microRNAs involved in the malignant progression of gliomas. Brain Pathol. 2010;20(3):539–550. doi: 10.1111/j.1750-3639.2009.00328.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Krichevsky AM, Gabriely G. miR-21: a small multi-faceted RNA. J Cell Mol Med. 2009;13(1):39–53. doi: 10.1111/j.1582-4934.2008.00556.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Papagiannakopoulos T, Shapiro A, Kosik KS. MicroRNA-21 targets a network of key tumor-suppressive pathways in glioblastoma cells. Cancer Res. 2008;68(19):8164–8172. doi: 10.1158/0008-5472.CAN-08-1305. [DOI] [PubMed] [Google Scholar]
- 24.Hermeking H. p53 enters the microRNA world. Cancer Cell. 2007;12(5):414–418. doi: 10.1016/j.ccr.2007.10.028. [DOI] [PubMed] [Google Scholar]
- 25.Hermeking H. The miR-34 family in cancer and apoptosis. Cell Death Differ. 2010;17(2):193–199. doi: 10.1038/cdd.2009.56. [DOI] [PubMed] [Google Scholar]
- 26.Li Y, Guessous F, Zhang Y, et al. MicroRNA-34a inhibits glioblastoma growth by targeting multiple oncogenes. Cancer Res. 2009;69(19):7569–7576. doi: 10.1158/0008-5472.CAN-09-0529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Silber J, Jacobson A, Ozawa T, et al. miR-34a repression in proneural malignant gliomas upregulates expression of its target PDGFRA and promotes tumorigenesis. PLoS One. 2012;7(3):e33844. doi: 10.1371/journal.pone.0033844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Papagiannakopoulos T, Friedmann-Morvinski D, Neveu P, et al. Pro-neural miR-128 is a glioma tumor suppressor that targets mitogenic kinases. Oncogene. 2012;31(15):1884–1895. doi: 10.1038/onc.2011.380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Godlewski J, Nowicki MO, Bronisz A, et al. Targeting of the Bmi-1 oncogene/stem cell renewal factor by microRNA-128 inhibits glioma proliferation and self-renewal. Cancer Res. 2008;68(22):9125–9130. doi: 10.1158/0008-5472.CAN-08-2629. [DOI] [PubMed] [Google Scholar]
- 30.Xia H, Cheung WK, Ng SS, et al. Loss of brain-enriched miR-124 enhances the stem-like traits and invasiveness of glioma cells. J Biol Chem. 2012;287(13):9962–9971. doi: 10.1074/jbc.M111.332627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Silber J, Lim DA, Petritsch C, et al. miR-124 and miR-137 inhibit proliferation of glioblastoma multiforme cells and induce differentiation of brain tumor stem cells. BMC Med. 2008;6:14. doi: 10.1186/1741-7015-6-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hu W, Chan CS, Wu R, et al. Negative regulation of tumor suppressor p53 by microRNA miR-504. Mol Cell. 2010;38(5):689–699. doi: 10.1016/j.molcel.2010.05.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kim TM, Huang W, Park R, Park PJ, Johnson MD. A developmental taxonomy of glioblastoma defined and maintained by MicroRNAs. Cancer Res. 2011;71(9):3387–3399. doi: 10.1158/0008-5472.CAN-10-4117. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.