Abstract
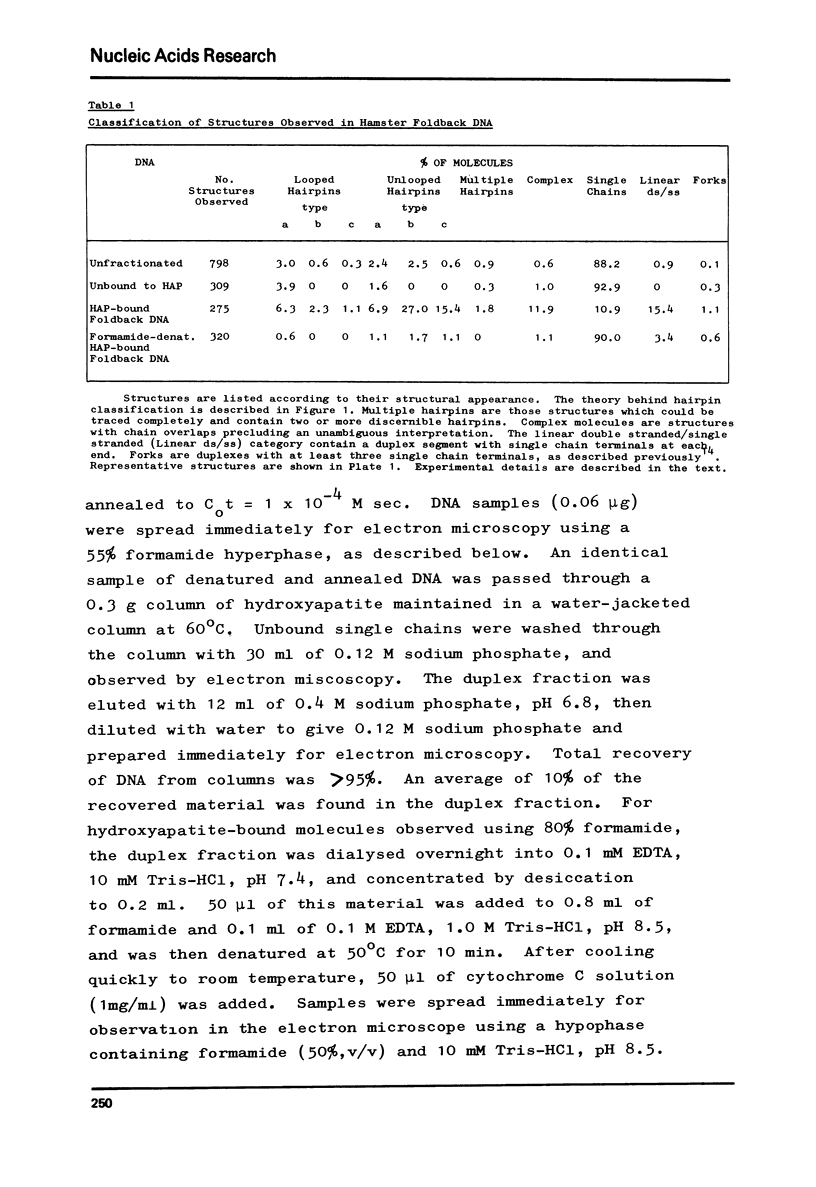
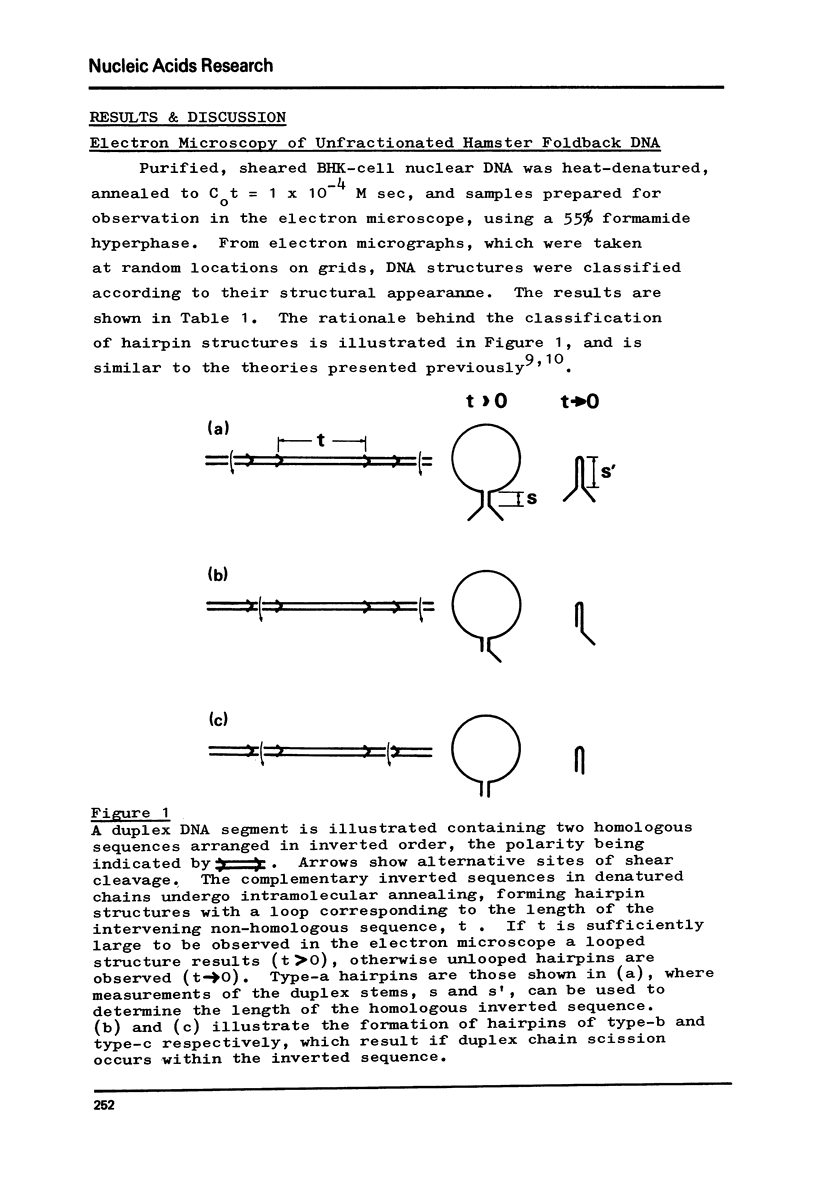
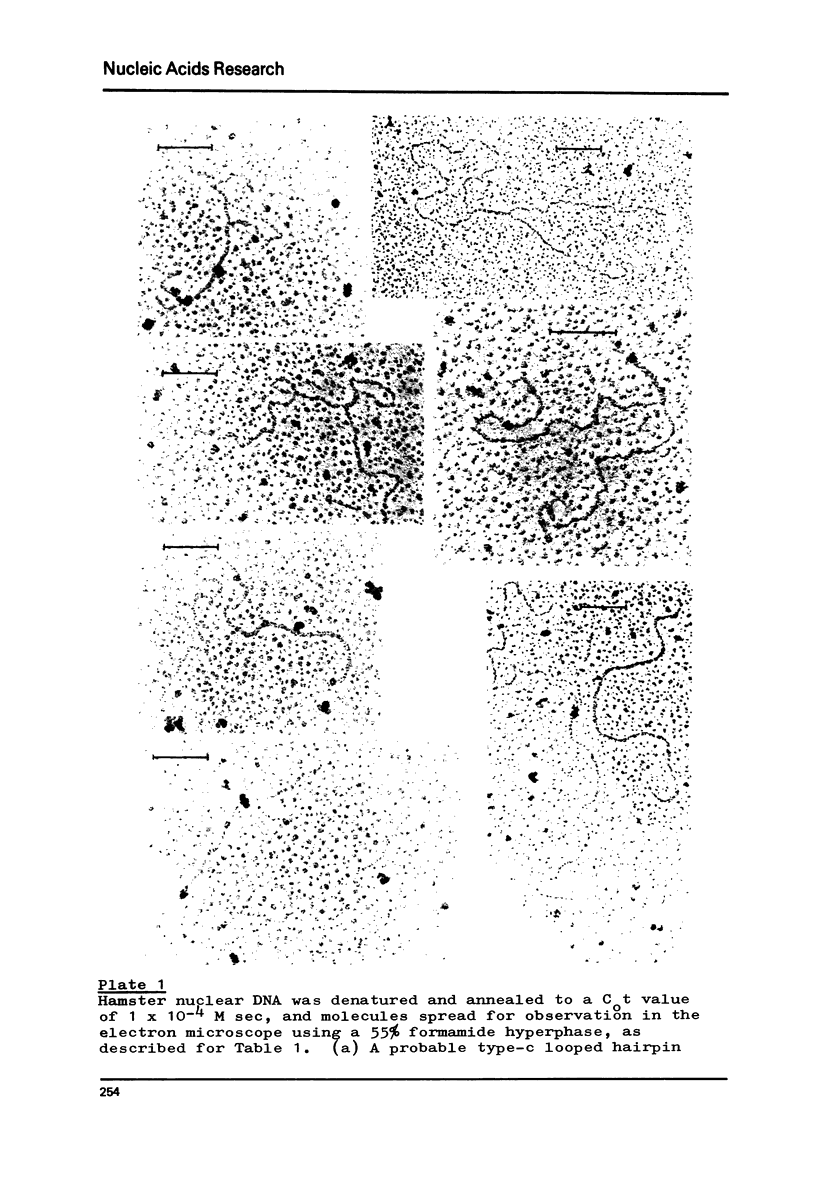
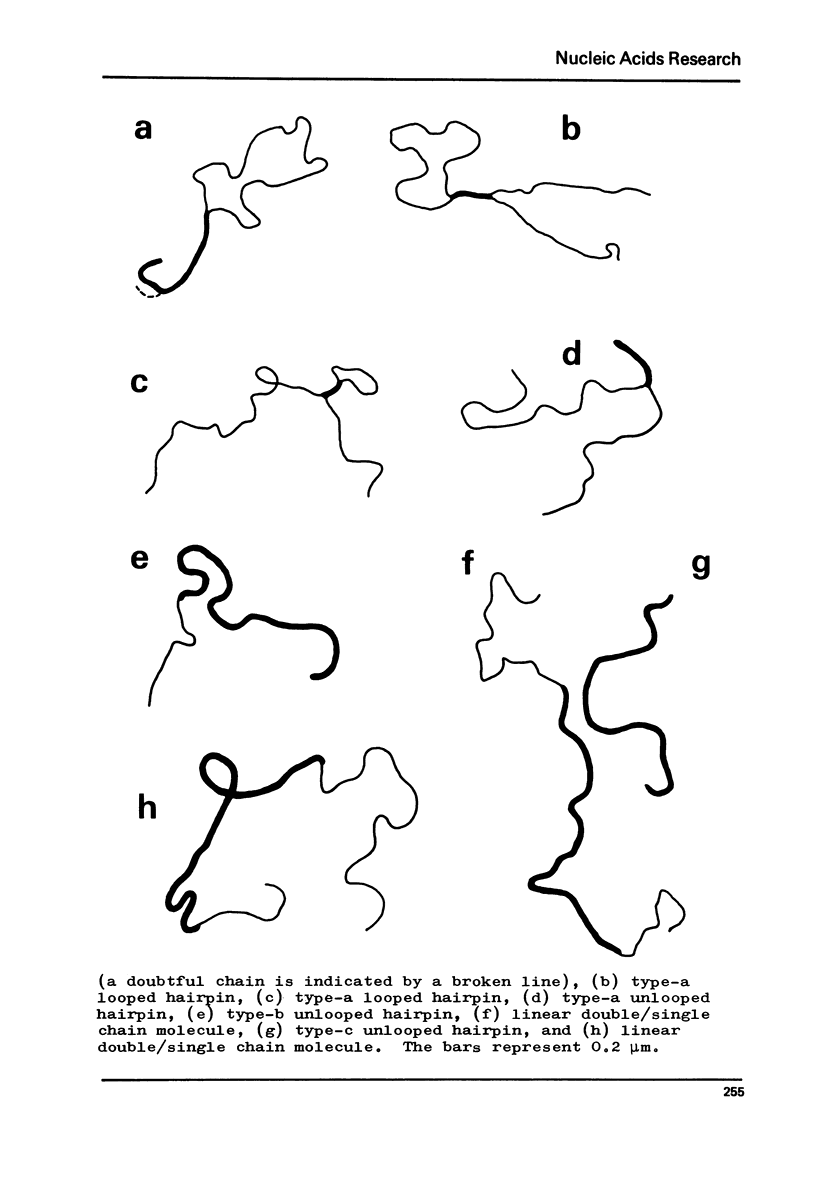
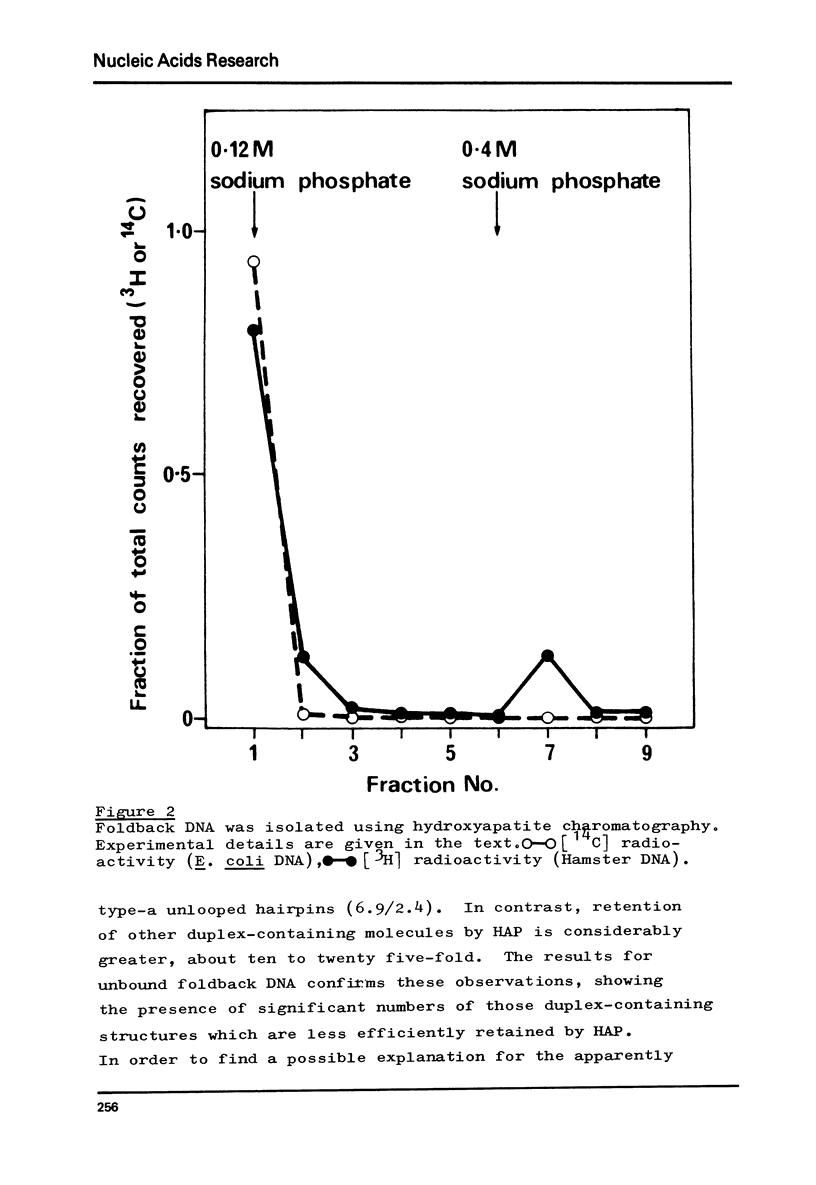
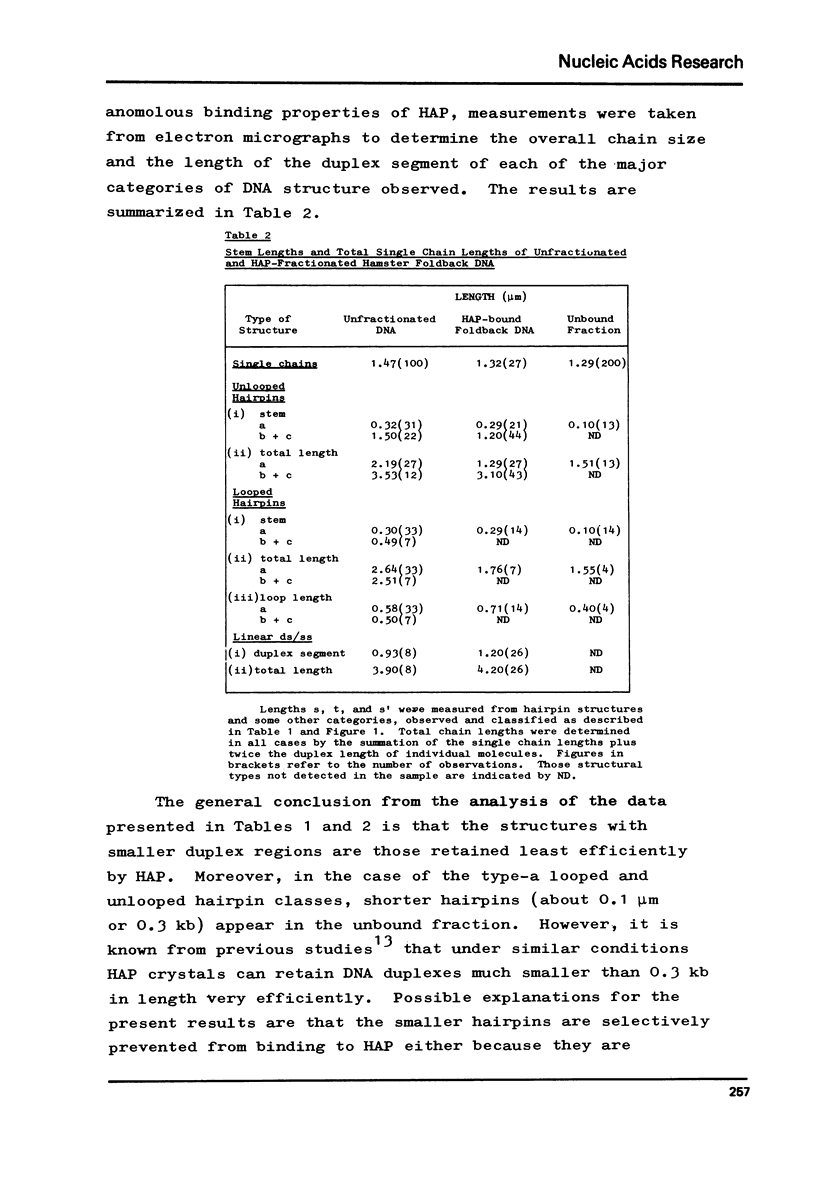
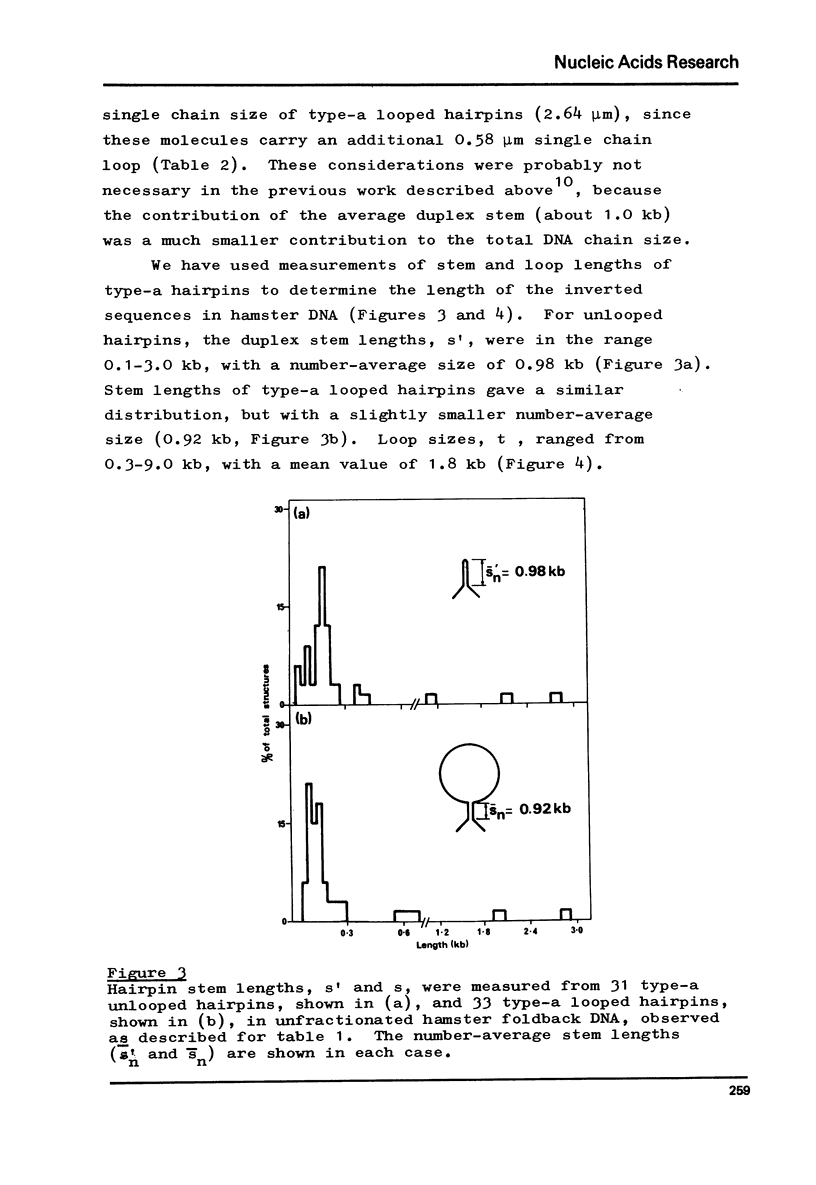
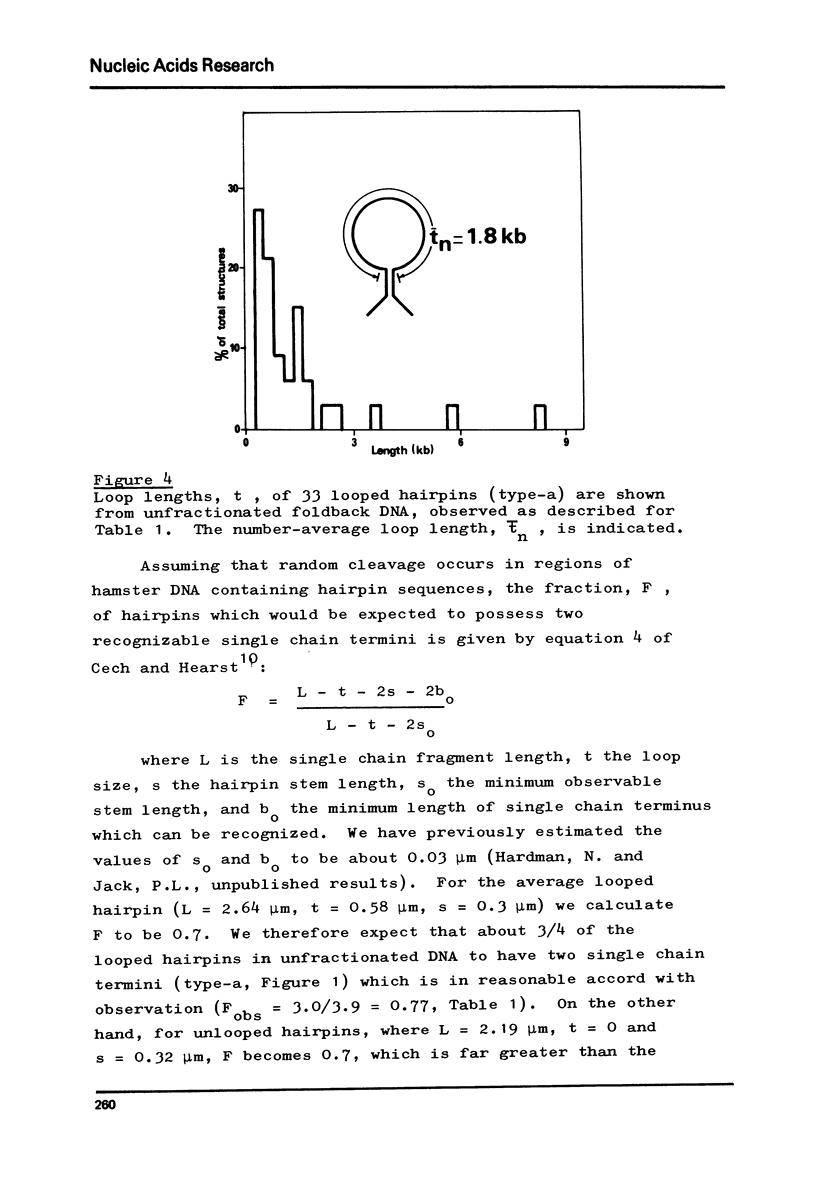
Foldback sequences in nuclear DNA from cultured Hamster fibroblasts (BHK-21/C13 cells) have been characterized by electron microscopy. One half of the structures observed when denatured hamster DNA is allowed to anneal in the range O less than Cot1 less than 1 x 10(-4) M sec result from the annealing of inverted sequences forming foldback DNA. The remainder have a probable bimolecular origin. arising from rapidly-annealing sequences of satellite-like complexity. The average length of the inverted sequences in the foldback molecules is about 0.9 kilobases. There is estimated to be about 42,000 such sequences (21,000 pairs) in the hamster genome, approximately 45% of which form looped structures with a mean loop length of 1.74 kilobases. Contrary to previous reports, binding of the renatured duplex molecules to hydroxyapatite results in a poor recovery of structures containing identifiable foldback sequences, due to preferential enrichment of the bound fraction with duplexes formed by intermolecular annealing.
Full text
PDF






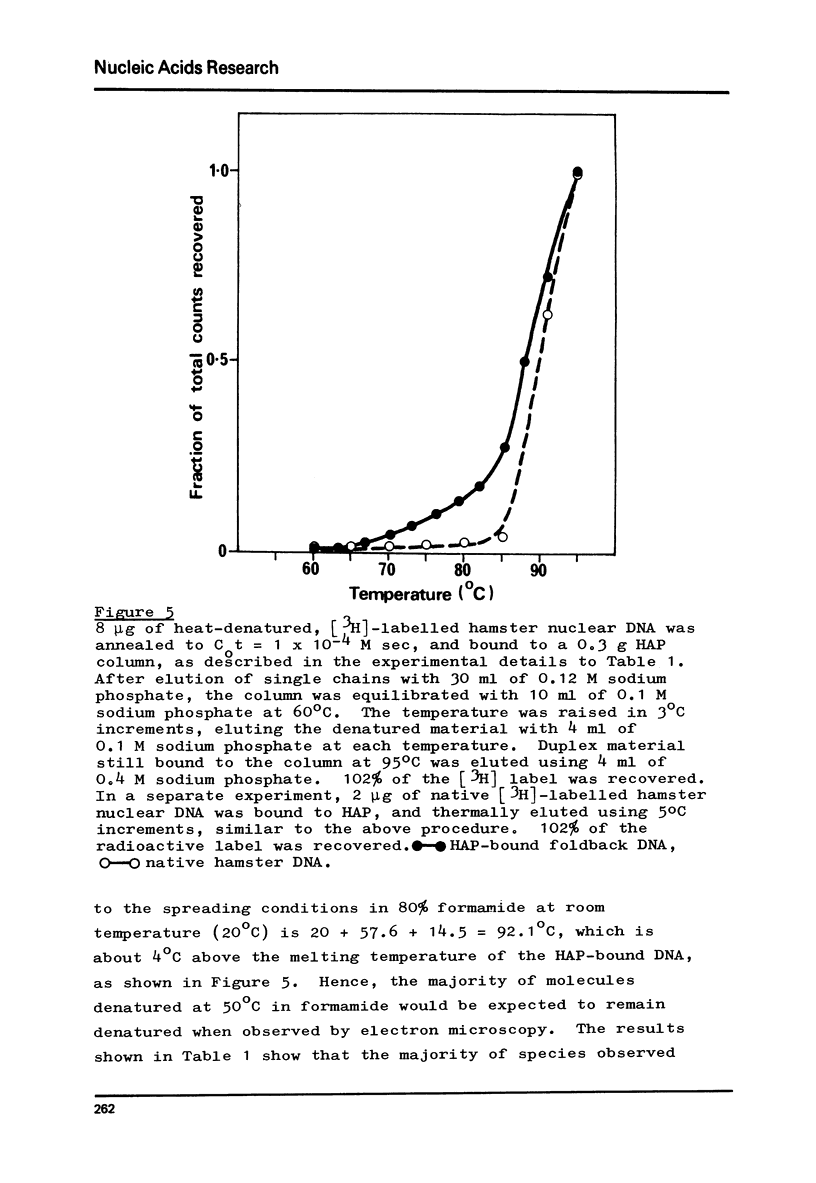

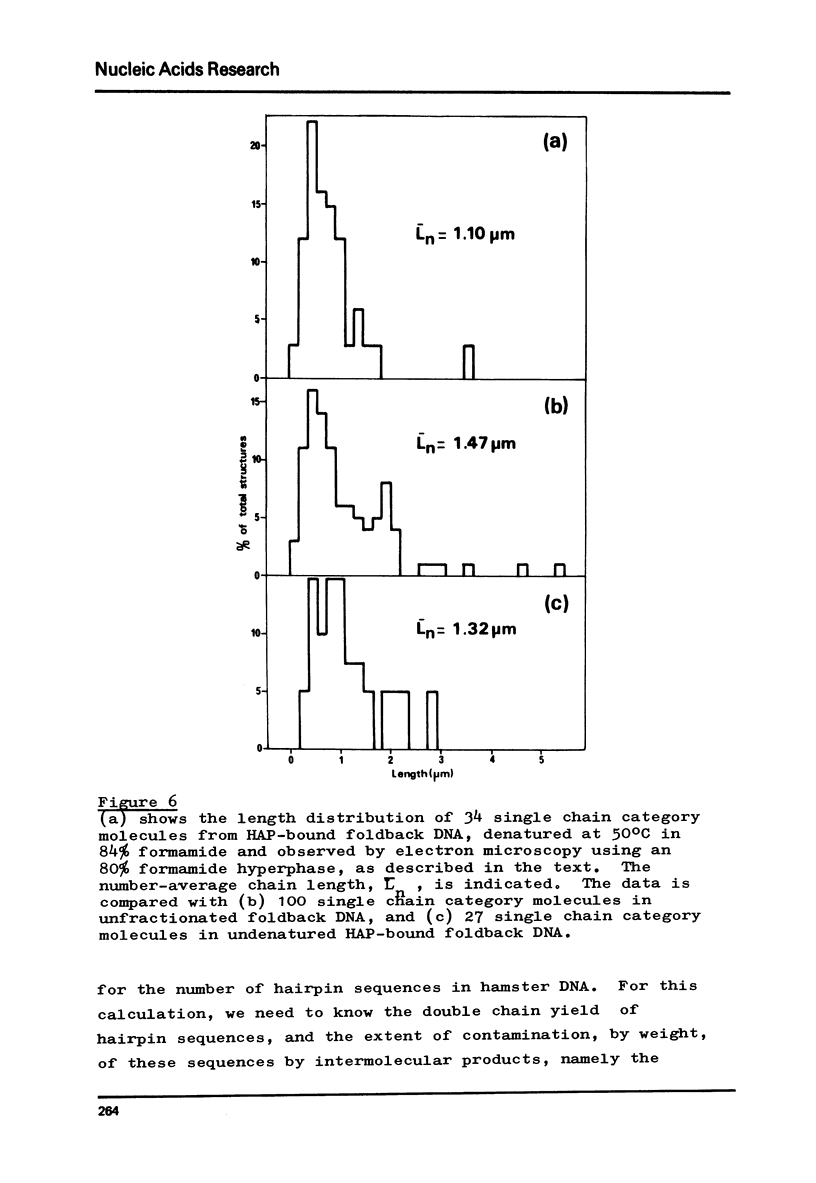




Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Cech T. R., Hearst J. E. An electron microscopic study of mouse foldback DNA. Cell. 1975 Aug;5(4):429–446. doi: 10.1016/0092-8674(75)90062-8. [DOI] [PubMed] [Google Scholar]
- Cech T. R., Hearst J. E. Organization of highly repeated sequences in mouse main-band DNA. J Mol Biol. 1976 Jan 25;100(3):227–256. doi: 10.1016/s0022-2836(76)80061-7. [DOI] [PubMed] [Google Scholar]
- Cech T. R., Rosenfeld A., Hearst J. E. Characterization of the most rapidly renaturing sequences in mouse main-band DNA. J Mol Biol. 1973 Dec 15;81(3):299–325. doi: 10.1016/0022-2836(73)90143-5. [DOI] [PubMed] [Google Scholar]
- Davidson E. H., Hough B. R., Amenson C. S., Britten R. J. General interspersion of repetitive with non-repetitive sequence elements in the DNA of Xenopus. J Mol Biol. 1973 Jun 15;77(1):1–23. doi: 10.1016/0022-2836(73)90359-8. [DOI] [PubMed] [Google Scholar]
- Davis R. W., Hyman R. W. A study in evolution: the DNA base sequence homology between coliphages T7 and T3. J Mol Biol. 1971 Dec 14;62(2):287–301. doi: 10.1016/0022-2836(71)90428-1. [DOI] [PubMed] [Google Scholar]
- HJERTEN S., LEVIN O., TISELIUS A. Protein chromatography on calcium phosphate columns. Arch Biochem Biophys. 1956 Nov;65(1):132–155. doi: 10.1016/0003-9861(56)90183-7. [DOI] [PubMed] [Google Scholar]
- Hardman N. Formation of rings from segments of HeLa-cell nuclear deoxyribonucleic acid. Biochem J. 1974 Dec;143(3):521–534. doi: 10.1042/bj1430521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jelinek W., Mollov G., Salditt M., Wall R., Sheiness D., Darnell J. E., Jr Origin of mRNA in HeLa cells and the implications for chromosome structure. Cold Spring Harb Symp Quant Biol. 1974;38:891–898. doi: 10.1101/sqb.1974.038.01.091. [DOI] [PubMed] [Google Scholar]
- Lewin B. Units of transcription and translation: sequence components of heterogeneous nuclear RNA and messenger RNA. Cell. 1975 Feb;4(2):77–93. doi: 10.1016/0092-8674(75)90113-0. [DOI] [PubMed] [Google Scholar]
- Lewin B. Units of transcription and translation: the relationship between heterogeneous nuclear RNA and messenger RNA. Cell. 1975 Jan;4(1):11–20. doi: 10.1016/0092-8674(75)90128-2. [DOI] [PubMed] [Google Scholar]
- Manning J. E., Schmid C. W., Davidson N. Interspersion of repetitive and nonrepetitive DNA sequences in the Drosophila melanogaster genome. Cell. 1975 Feb;4(2):141–155. doi: 10.1016/0092-8674(75)90121-x. [DOI] [PubMed] [Google Scholar]
- McConaughy B. L., Laird C. D., McCarthy B. J. Nucleic acid reassociation in formamide. Biochemistry. 1969 Aug;8(8):3289–3295. doi: 10.1021/bi00836a024. [DOI] [PubMed] [Google Scholar]
- Perlman S., Phillips C., Bishop J. O. A study of foldback DNA. Cell. 1976 May;8(1):33–42. doi: 10.1016/0092-8674(76)90182-3. [DOI] [PubMed] [Google Scholar]
- Rommelaere J., Miller-Faurès A. Detection by density equilibrium centrifugation of recombinant-like DNA molecules in somatic mammalian cells. J Mol Biol. 1975 Oct 15;98(1):195–218. doi: 10.1016/s0022-2836(75)80109-4. [DOI] [PubMed] [Google Scholar]
- Ryskov A. P., Saunders G. F., Farashyan V. R., Georgiev G. P. Double-helical regions in nuclear precursor of mRNA (pre-mRNA). Biochim Biophys Acta. 1973 Jun 8;312(1):152–164. doi: 10.1016/0005-2787(73)90060-9. [DOI] [PubMed] [Google Scholar]
- Schmid C. W., Manning J. E., Davidson N. Inverted repeat sequences in the Drosophila genome. Cell. 1975 Jun;5(2):159–172. doi: 10.1016/0092-8674(75)90024-0. [DOI] [PubMed] [Google Scholar]
- Wensink P. C., Finnegan D. J., Donelson J. E., Hogness D. S. A system for mapping DNA sequences in the chromosomes of Drosophila melanogaster. Cell. 1974 Dec;3(4):315–325. doi: 10.1016/0092-8674(74)90045-2. [DOI] [PubMed] [Google Scholar]
- Wilson D. A., Thomas C. A., Jr Hydroxyapatite chromatography of short double-helical DNA. Biochim Biophys Acta. 1973 Dec 21;331(3):333–340. doi: 10.1016/0005-2787(73)90019-1. [DOI] [PubMed] [Google Scholar]
- Wilson D. A., Thomas C. A., Jr Palindromes in chromosomes. J Mol Biol. 1974 Mar 25;84(1):115–138. doi: 10.1016/0022-2836(74)90216-2. [DOI] [PubMed] [Google Scholar]



