Abstract
The monovalent cation (MVC) site of the tryptophan synthase from Salmonella typhimurium plays essential roles in catalysis and in the regulation of substrate channeling. In vitro, MVCs affect the equilibrium distribution of intermediates formed in the reaction of L-Ser with the α2β2 complex; the MVC-free, Cs+-, and NH4+-bound enzymes stabilize the α-aminoacrylate species, E(A-A), while Na+-binding stabilizes the L-Ser external aldimine species, E(Aex1). Two probes of β-site reactivity and conformation were used herein, the reactive indole analogue, indoline, and the L-Trp analogue, L-His. MVC-bound E(A-A) reacts rapidly with indoline to give the indoline quinonoid species, E(Q)indoline, which slowly converts to dihydroiso-L-tryptophan. MVC-free E(A-A) gives very little E(Q)indoline and turnover is strongly impaired; the fraction of E(Q)indoline formed is < 3.5% of that given by the Na+-bound form. The reaction of L-Ser with the MVC-free internal aldimine species, E(Ain), initially gives small amounts of an active E(A-A) which converts to an inactive species on a slower, conformational, time scale. This inactivation is abolished by the binding of MVCs. The inactive E(A-A), appears to have a closed β-subunit conformation with an altered substrate binding site that is different from the known conformations of tryptophan synthase. Reaction of L-His with E(Ain) gives an equilibrating mixture of external aldimine and quinonoid species, E(Aex)his and E(Q)his. The MVC-free and Na+ forms of the enzyme gave trace amounts of E(Q)his (~1% of the β-sites). The Cs+ and NH4+ forms gave, respectively, ~17% and ~14%. The reactivity of the MVC-free E(Ain) was restored by the binding of an α-site ligand. These studies show MVCs and α-site ligands act synergistically to modulate the switching of the β-subunit from the open to the closed conformation, and this switching is crucial to the regulation of β-site catalytic activity. Comparison of the structures of Na+ and Cs+ forms of the enzyme show Cs+ favors complexes with open indole binding sites poised for the conformational transition to the closed state, whereas the Na+ form does not. The β-subunits of Cs+ complexes exhibit preformed indole sub-sites, the indole sub-sites of the open Na+ complexes are collapsed, distorted and too small to accommodate indole.
The tryptophan synthase from Salmonella typhimurium is a substrate-channeling bienzyme complex that catalyzes the last two steps in the biosynthesis of L-Trp (Scheme 1) (1–5). The β-active site of this enzyme complex is activated by monovalent cation (MVC)1 binding to an allosteric site (6–10) located near the PLP cofactor (viz., the Cs+ site shown in Figure 1). In vitro, the enzyme can bind Na+, K+, Cs+, Li+, Rb+, NH4+, or guanidinium ion (6–11) at this site. These MVC complexes and the MVC-free form each exert different structural effects on the enzyme that alter the kinetic and thermodynamic properties of the β-subunit catalytic cycle, and the allosteric interactions linking the α- and β-sites (5, 7–10).
Scheme 1.
Organic Chemistry of the α- and β-Reactions
Figure 1.
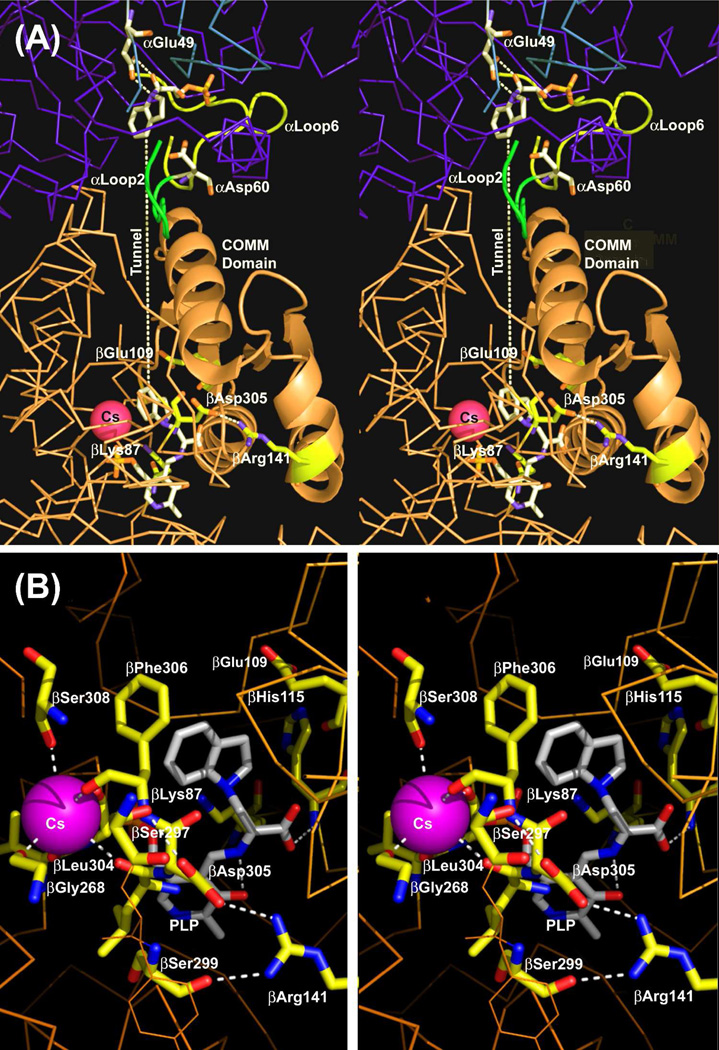
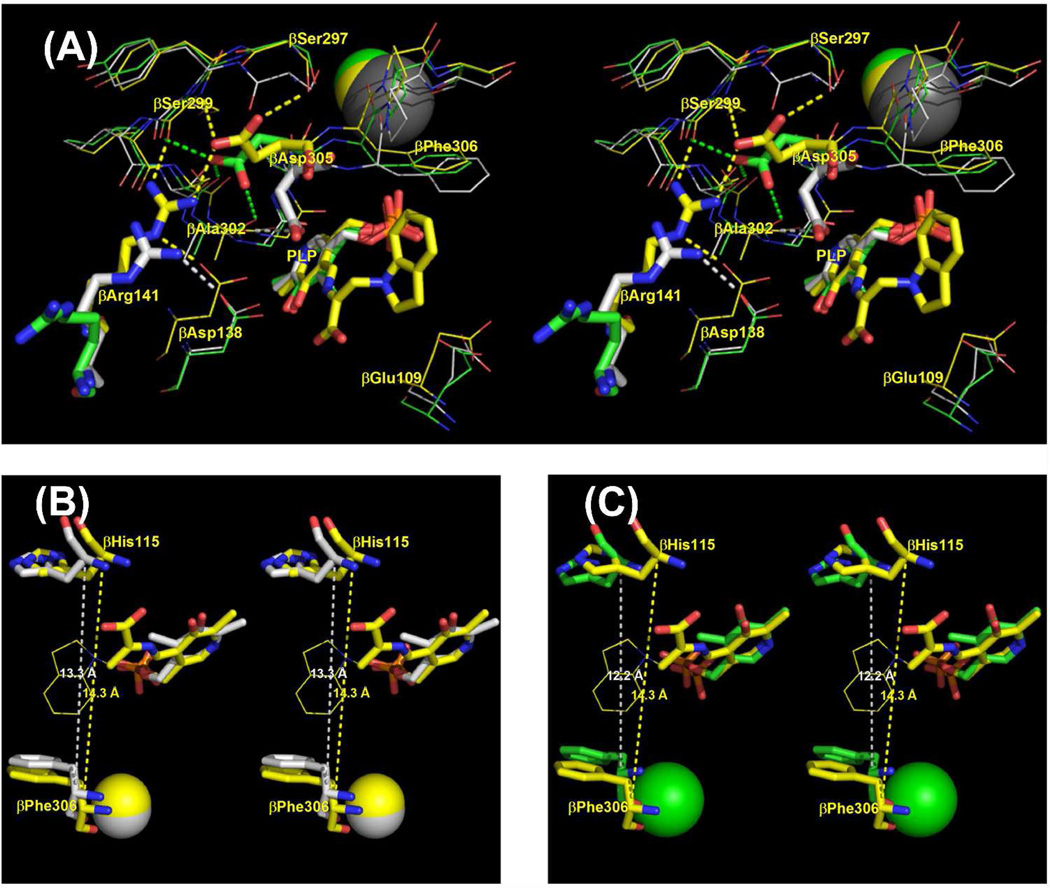
Stereo over view of the structure of an αβ-dimeric unit of the E(Q)indoline, and details of the β-catalytic site. (A) Stereo view of the catalytic sites of the α- and β-subunits, the MVC site and interconnecting tunnel. The dashed white line indicates the approximate position of the tunnel. The transition state analogue formed by the reaction of indoline with G3P at the α-site and the quinonoid intermediate at the β-site are shown in sticks. Cs+ is shown as a magenta ball. The α-subunit is represented by a backbone structure colored blue, the β-subunit is colored tan. αLoop L2 and αLoop L6 are shown, respectively, as yellow and blue ribbons. The COMM domain is shown in cartoon ribbon, the remainder of the β-subunit in backbone. At the α-site, catalytic residues αGlu49 and αAsp60 and the adduct formed between indoline and G3P are shown in CPK-colored sticks. At the β-site, catalytic residues βLys87 and βGlu109 and the salt bridging residues βArg141 and βAsp305 are shown as sticks with C yellow, N blue and O red. The structure of the indoline quinonoid intermediate is shown in CPK-colored sticks. (B) Stereo view of the β-catalytic site with the PLP-bound indoline quinonoid intermediate shown in CPK-colored sticks. The catalytic residues βLys87 and βGlu109, the βAsp305-βArg141 salt bridge, H-bonding residues βSer299 and βSer297, indole sub-site residues βHis115 and βPhe306 and Cs+ ligands βGly268, βLeu304, βPhe306 and βSer308 are shown in sticks with C yellow, N blue, and O red. Cs+ is shown as a purple ball. H-bonding interactions and coordination bonds to Cs+ are shown as white dashed lines. The β-subunit is represented as orange backbone. Structures are drawn from PDB code 3CEP with PyMOL 1.1r1.
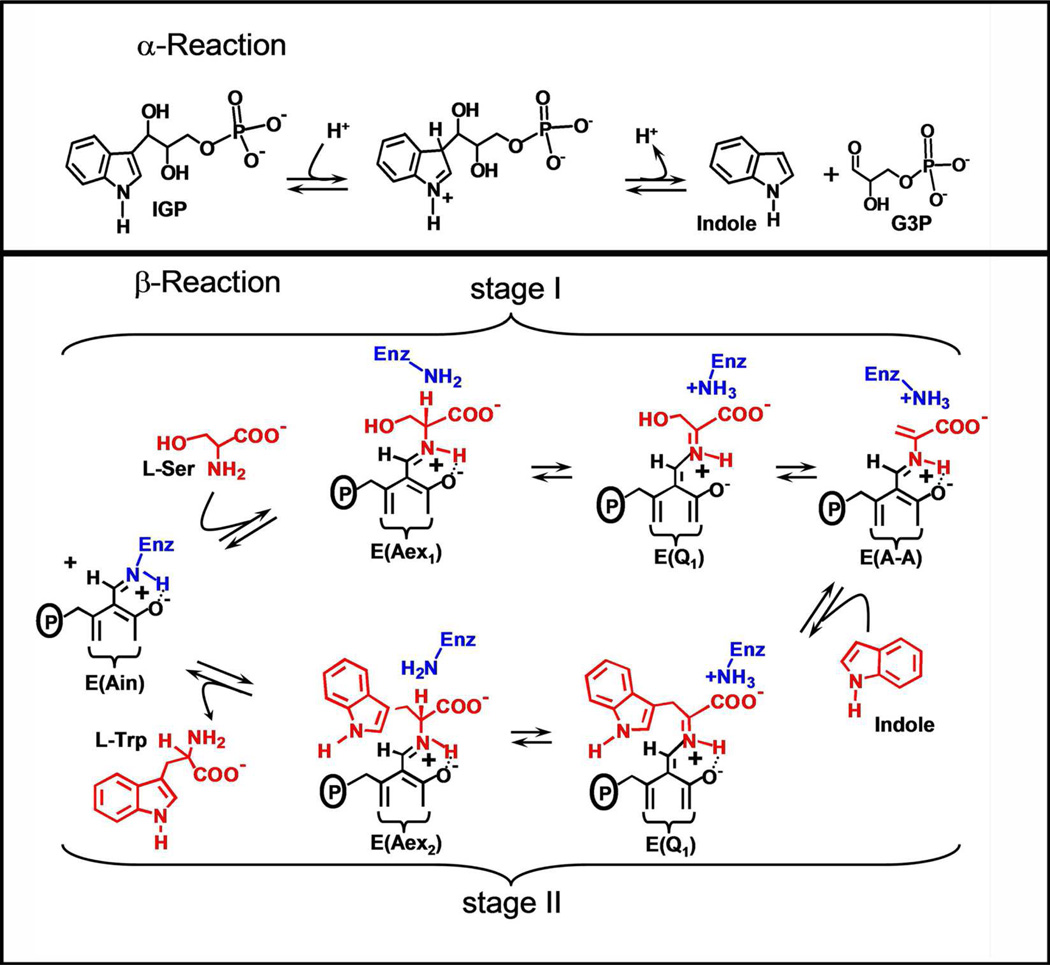
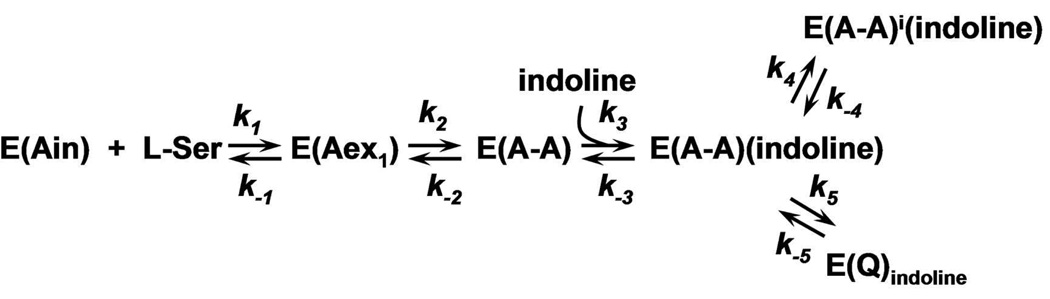
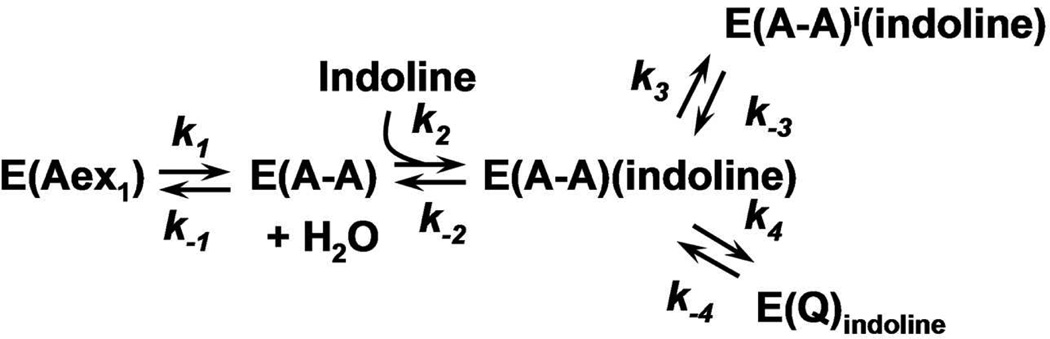
The α- and β-reactions catalyzed by tryptophan synthase are depicted in Scheme 1. The top panel shows the α-subunit-catalyzed conversion of 3-indole-D-glycerol 3’-phosphate (IGP)1 to indole and D-glyceraldehyde 3-phosphate (G3P) via an intermediate wherein the indole ring of IGP is protonated at C-3 (3, 5). The β-reaction (Scheme 1, lower panel) occurs in two stages. In stage I, L-Ser reacts with the internal aldimine form of the enzyme, E(Ain), to give the external aldimine species, E(Aex1), the L-Ser quinonoid species, E(Q1), and then the α-aminoacrylate species, E(A-A). As stage I approaches equilibrium, two intermediates dominate, E(Aex1) (λmax = 424 nm) and E(A-A) (λmax = 350 and 460 nm) (12–16). The ratio of E(Aex1) to E(A-A) is sensitive to the type of MVC bound, eg., Na+ favors E(Aex1) while Cs+ favors E(A-A) (9, 11).
The α-reaction is switched on by the conversion of E(Aex1) to E(A-A) at the β-site during stage I of the β-reaction. The α-reaction is switched off when E(Q3) is converted to E(Aex2) in stage II (5). To initiate stage II of the β-reaction, indole formed in the α-reaction is transferred (channeled) from the α-catalytic site to the β-catalytic site via the ~25 A long interconnecting tunnel (dashed white line in Figure 1A) where it reacts with E(A-A). MVC-bound forms of E(A-A) react quickly with indole and indole analogues such as indoline, to form quinonoid species which then undergo further transformation to give the corresponding new amino acids (Scheme 1, stage II) (2–5, 12, 13, 17–20). The binding of α-site ligands activate the rate limiting step in stage I of the β-reaction, the conversion of E(Aex1) to E(A-A) (4, 5, 21, 22). The switching between low and high states of catalytic activity is coupled to the switching of αβ-dimeric units of the enzyme complex between open and closed conformations (5, 18, 19, 22).
The indole analogue, indoline, reacts rapidly and reversibly with E(A-A) to give the indoline quinonoid species, E(Q)indoline, a quasi-stable intermediate with an intense absorption band at 466 nm (ε > 40,000 M−1cm−1) (17–20) (eq 1).
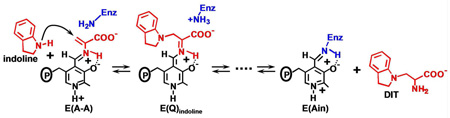 |
eq 1 |
The x-ray structure of the E(Q)indoline complex presented in Figure 1AB shows the closed conformation of the αβ dimeric unit of the α2β2 tetrameric complex (5, 20). At the closed β-site (Figure 1B), the indoline moiety, in the form of the indoline quinonoid species, E(Q)indoline, is bound to the indole ring sub-site and is poised for conversion to the corresponding external aldimine (20). Figure 1B also shows the β-site catalytic groups βLys87 and βGlu109, along with the salt bridge formed between βArg141 and βAsp305 and H-bonds involving βSer297 and βSer299, interactions that are crucially important for stabilization of the closed conformation (5, 20, 22).
Figure 1A shows a transition-state analogue bound to the α-site. This analogue consists of an adduct formed between indoline and G3P. The acid-base catalytic residue, αGlu49, is H-bonded to this adduct (5, 20). The residue which plays a charge-stabilization role in catalysis, αAsp60, is also shown (5, 20).
The activities of many enzymes, including tryptophan synthase, are known to be modulated by MVC effectors (6–11, 23–26). MVC activation/deactivation of enzyme catalysis has evolved to modulate biological function in a surprisingly large number of enzymes. In recent years, x-ray structure studies have provided detailed information about the nature of MVC binding sites, however, there is still relatively little known about the detailed mechanisms by which MVC binding brings about activation/deactivation of catalytic activity and biological function. Like conservative site-directed mutations, MVC substitution provides a means for creating subtle structural perturbations in MVC-activated enzymes that have the potential for revealing important structure-function relationships (25). The expected structural changes due to substitution of one MVC for another are relatively small due to the small change in ionic radius (for example replacement of Na+ by Cs+ increases ionic radius by ~ 0.77Å). Therefore, MVC substitutions often produce subtle structural changes with significant kinetic effects (25). In the tryptophan synthase system, as measured by the ratio kcat/Km, Peracchi et al. reported an ~ 45-fold activation due the binding of Na+ to the MVC-free enzyme (7). Woehl et al. (8) discovered that the binding of a monovalent cation to tryptophan synthase is essential both to β-site catalysis and to intersubunit communication. They found that in comparison to the Na+ form of the enzyme, the activity of the MVC-free form is greatly altered both in stages I and II of the β-reaction (Scheme 1). The MVC-free enzyme exhibits a turnover rate for the β-reaction that is decreased 5-fold and 6.4-fold relative to the Na+ and K+ substituted enzymes, respectively (8), and the αβ reaction is similarly affected (27). The most interesting effects involve the allosteric communication between the α- and β-sites. Reaction of L-Ser at the β-site with the Na+, K+ and NH4+ forms of the enzyme activate the α-site by ~30-fold, 26-fold, and 40-fold, respectively (27). When all monovalent metal ions are removed, reaction of L-Ser at the β-site does not activate the α-reaction (7–10, 27). Rhee et al. (6) showed that the MVC site is located in the β-subunit near the β-catalytic site (see Figure 1). The fraction of β-sites in the form of E(Q)indoline is dependent on the particular MVC bound, while the MVC-free species gives almost no E(Q)indoline. The amount of E(Q)indoline formed in the MVC-free system is less than 10% of that formed with the Na+-substituted enzyme (27). Kinetic, spectroscopic, and x-ray structural studies (4, 5, 8–10, 15, 16, 18–22) have shown that E(Ain), and E(Aex) species favor the open conformation of the β-subunit, while the E(Q) and E(A-A) species favor the closed conformation. Previous investigators (5, 8–10, 22) have proposed that the chemistry catalyzed by the β-site preferentially occurs via intermediates with the closed conformation, and that the switch from the open to the closed conformation is mediated by ligand binding to the α-subunit, MVC binding to the β-subunit and by the conversion of E(Aex1) to E(Q) and E(A-A).
Figure 1B shows the relationship of the MVC site to the β-catalytic site. The MVC site cavity is formed by the carbonyl oxygens of six amino acid residues, βVal231, βGly232, βGly268, βLeu304 βPhe306, and βSer308. Cs+ fills this cavity and makes coordination bonds with all six of these carbonyl oxygens (Figure 1B). Due to the smaller ionic radius of Na+, this ion bonds to only three of the six carbonyls (βGly232, βPhe306, and βSer308), and two waters are incorporated into the coordination sphere (4–6, 20–22). The coordination of K+ is similar to Na+ except that only a single water is included in the coordination sphere (6). Inspection of Figure 1B shows that the MVC site is linked to two residues with important roles in β-site function, βPhe306 and βAsp305 (5, 22). The aromatic ring of βPhe306 forms part of the binding site for substrate, indole. βAsp305, situated between MVC ligands βLeu304 and βPhe306, plays an important role in the switching between open and closed conformation states. In the open, inactive conformation, the βAsp305 carboxylate is H-bonded to the Aex1 hydroxyl. This interaction stabilizes E(Aex1) by holding the Aex1 hydroxyl away from the acid-base catalytic group, βGlu109. The switch to the closed conformation ruptures this H-bond, and repositions the carboxylate of βAsp305 to form the salt bridge with βArg141, allowing the Aex1 hydroxyl to rotate into position for the βGlu109 catalyzed elimination (5, 22). Consequently, the MVC site is directly involved in indole binding, the conformational switch, and β-site catalysis.
This work undertakes detailed studies of the MVC-free, Na+ and Cs+ forms of tryptophan synthase to further investigate the effects of MVC binding on the catalytic activity and conformational states of the enzyme. The 466 nm spectroscopic signature of the E(Q)indoline is used to quantify the reactivities of the MVC-free and MVC bound E(A-A) species (27). The L-Trp analogue, L-His is used to investigate stage II of the β-reaction. L-His reacts at the β-site to give a distribution of intermediates dominated by the L-His external aldimine, E(Aex)his, and the L-His quinonoid species, E(Q)his. It will be shown that the distribution of these two species is modulated by MVC binding to the β-site and by ligand binding to the α-site. We also examine the effects of α-site ligand binding on the switching between open and closed conformational states using the IGP analogue, N-(4'-trifluoromethoxybenzenesulfonyl)-2-aminoethyl phosphate (F9) (21, 22). F9 is chemically stable, binds tightly to the α-site and mimics the allosteric effects of IGP and G3P (22). The results presented herein reveal that (i) despite the observable formation of a closed conformation, the MVC-free E(A-A) is unreactive towards indoline. (ii) The slow turnover of L-Ser to pyruvate and NH4+ when L-Ser is incubated with the MVC-free enzyme (the pyruvate side reaction, eq 2)
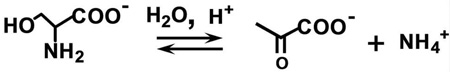 |
eq 2 |
generates traces of NH4+ which activate the enzyme by binding to the MVC site. This activation obscures the near zero reactivity of the MVC-free E(A-A) in the reaction with indoline. (iii) Reaction of L-Ser with E(Ain) initially gives two forms of E(A-A), one reactive the other inactive; the reactive form converts to the inactive form on a slower time scale. (iv) The binding of MVCs influence the distribution of the two forms of E(A-A), strongly favoring the reactive form. The likely conformational origins of the inactive MVC-free E(A-A) are discussed within the context of known β-site structures and the switching between active and inactive conformation states. (v) The Cs+ forms of the open β-subunit conformation are set up for conversion to the closed state with preformed indole binding sub-sites, while the corresponding Na+ forms are not set up for conversion to the closed state and the indole sub-sites are distorted with dimensions too small to accommodate indole. (vi) MVC-bound forms of the enzyme balance the relative energies of the open and closed conformations of the β-subunit during catalysis thereby modulating the switching between open and closed states which synchronizes the α- and β-catalytic cycles.
MATERIALS AND METHODS
Materials
L-Ser, L-His, indoline, CsCl, NaCl, and NH4Cl were purchased from Sigma-Aldrich. Indoline was purified as previously described (17). N-(4-trifluoromethyoxylbenzenesulfonyl)-2-amino-1-ethylphosphate (F9) was synthesized as described (22). 5,5'-Dithio-bis(2-nitrobenzoic acid) (DTNB) was purchased from Calbiochem. All solutions were prepared at 25 ± 2° C and maintained at pH 7.8 in 50 mM triethanolamine (TEA) buffer and (except for H+) maintained MVC-free. F9 was prepared free of monovalent metal ions by exchange of Na+ with cyclohexylammonium chloride. Salmonella tryphimurium α2β2 tryptophan synthase was purified as previously described (28).
UV-Visible Static and Stopped-Flow Spectral Measurements
Static UV/Vis absorbance spectra were collected on a Hewlett-Packard 8452A diode array spectrophotometer at 25°C ± 2° in 50 mM TEA buffer at pH 7.8. Titration curves were fitted to eq 3 using Sigma Plot 9.0, where A is the observed absorbance, Ain is the absorbance at time zero, and Afin is absorbance extrapolated to infinite titrant concentration.
| eq 3 |
| eq 4 |
Measurement of the rate of pyruvate formation was accomplished through monitoring the absorption maximum for pyruvate at 316 nm (ε316 = 22.7 M−1cm−1). The turnover rate for conversion of L-Ser and β-me to S-hydroxyethyl-L-cysteine was assayed by trapping unreacted β-me with DTNB and measuring production of 2-nitro-5-thiobenzoic acid (TNB), at 410 nm (ε410 = 14,150 M−1cm−1) (29). Single-wavelength and rapid-scanning stopped flow measurements were preformed as previously described (9, 10). Single wavelength time courses were fitted using the Levenberg–Marquardt algorithm shown in eq 4, where A and A0 are absorbance values at time t and time infinity, and An is the amplitude of the nth relaxation, τn.
Kinetic Simulations
Kinetic simulations were preformed using the program Kinetic 3.11 (30) and are described in the Supporting Information.
Structure Alignments
Structures were overlayed several different ways. It was found that overlaying the Cα atoms of residues 10 to 100 of the β-subunit using the software, PyMOL 1.1r1, gave alignments very similar to those reported by Rhee et al. (6) for comparison of the E(Ain) structures (PDB codes 1BKS and 1TTP) and the E(Q)indoline structure (PDB code 3CEP). This region of the β-subunit appears particularly insensitive to the conformational differences between open and closed states. Thus all of the subunit alignments discussed in this work were performed using this selection of atoms. Alignments to compare β-active sites of different MVC complexes and different conformation states were accomplished with PyMOL using paired selections consisting of atoms in the PLP cofactor and the MVC.
RESULTS
The Reactions of L-Ser, and L-Ser and Indoline, with MVC-Bound and -Free Forms of α2β2
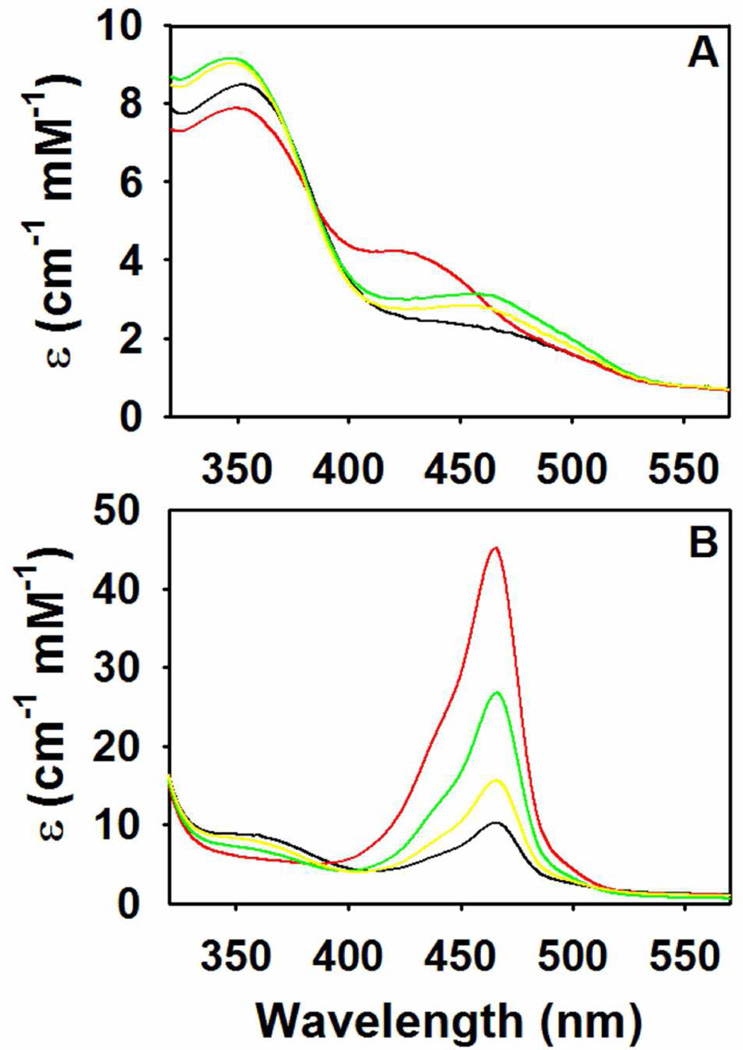
The UV/vis spectroscopic properties of the PLP-intermediates bound to tryptophan synthase provide sensitive probes of the conformational states of tryptophan synthase. Solution studies and x-ray structures have shown that the MVC-bound internal and external aldimine intermediates have the open conformation, while the α-aminoacrylate and quinonoid intermediates have the closed conformation (3, 5, 15, 16, 18–22). The reaction of L-Ser with the MVC-bound and -free forms of α2β2 results in the formation of equilibrating mixtures dominated by E(Aex1), λmax = 424 nm, and E(A-A), λmax = 350 and 460 nm (9, 10, 11, 13, 14, 31–33). Figure 2A compares the static spectra for the reaction of L-Ser in the presence or absence of Na+, Cs+, or NH4+. These spectra show that in the Cs+, NH4+, or MVC-free forms, the equilibrium distribution favors E(A-A), while the Na+ form favors E(Aex1). The relaxation rates for formation of E(Aex1) with the Na+, K+, NH4+, and Cs+ forms are 900 s−1, 660 s−1, 420 s−1, and 250 s−1 respectively, whereas the rates for E(Aex1) conversion to E(A-A) are 7 s−1, 20 s−1, 90 s−1, and 70 s−1, respectively. The MVC-free form gives rates of 200 s−1 and 21 s−1, respectively, for formation and decay of E(Aex1) (27, 34).
Figure 2.
Comparison of MVC induced effects on the static UV/Vis spectra of tryptophan synthase. (A) The L-Ser reaction, and (B) the indoline reaction. (A) Reaction of MVC-bound and free α2β2, with L-Ser (stage I, β-reaction). (B) Reaction of α2β2 with L-Ser and indoline (the indoline reaction). Color code: Na+ form (red), NH4+ form (yellow), Cs+ form (green), and the MVC-free form (black). Final concentrations: 20 µM α2β2, 50 mM L-Ser, 5 mM indoline, and 50 mM MVC.
Spectra for the reaction of E(A-A) with indoline (the indoline reaction) are presented in Figure 2B. The MVC-bound forms of E(A-A) react with indoline to generate E(Q)indoline (λmax = 466 nm, εmax > 60,000 M−1cm−1), however, as indicated by the small amount of E(Q)indoline generated, the MVC-free form is severely impaired (8–10). Figure 2B shows that Na+ is more effective than NH4+ or Cs+ in stabilizing E(Q)indoline, and that under similar conditions very little E(Q)indoline is formed with the MVC-free system. The relative order of effectiveness is Na+ > Cs+ > NH4+ >> MVC-free. The lower yields obtained with NH4+ and with Cs+ reflect larger apparent dissociation constants for indoline in the reaction with E(A-A); the 5.0 mM indoline used in Figure 2B is sub-saturating for these MVCs. These spectral data indicate that the affinity of the MVC-free E(A-A) for indoline is at least 50 to 100-fold weaker than the affinities of the NH4+ or Cs+ bound E(A-A) species.
The appearance of the 466 nm spectral band in the indoline reaction (Figure 2B) occurs in two kinetic phases. Most of the amplitude change occurs with a relatively fast relaxation rate (1/τ1 ≈ 700 s−1). The amplitude of the slower phase is very small, and, therefore, this phase was not considered further in the effort to characterize the reactivity of the MVC-free enzyme.
Pyruvate and NH4+ Formation in the L-Ser Reaction
In previous work, the impaired catalytic properties of the MVC-free enzyme were not sufficiently quantified to determine to what extent the MVC-free E(A-A) is unreactive toward nucleophiles. Because both MVC-bound and MVC-free forms of E(A-A) undergo a slow side reaction with water to give pyruvate and NH4+ with regeneration of E(Ain) (eq. 2), experiments where the enzyme and excess L-Ser are pre-incubated result in the steady-state production of NH4+, a contaminating MVC. The rate of the pyruvate side reaction depends on the ligation state of the α-site, and the particular MVC bound to the MVC site (34). To test the MVC effect for the conditions employed in these studies, the rate of pyruvate formation from L-Ser catalyzed by MVC-free, NH4+, Cs+, or Na+ bound α2β2 was measured by recording the absorbance change due to appearance of pyruvate at 316 nm (Δε316= 22.7 M−1cm−1), at varying L-Ser incubation times. The MVC-free, NH4+, and Cs+ forms of the enzyme all gave similar turnover rates, kcat = 0.008 ±0.002 s−1, while the turnover rate for the Na+ form, kcat = 0.09 ±0.002 s−1, was ~ 11-fold greater.
Influence of L-Ser Incubation Time and NH4+ Production on the Indoline Reaction
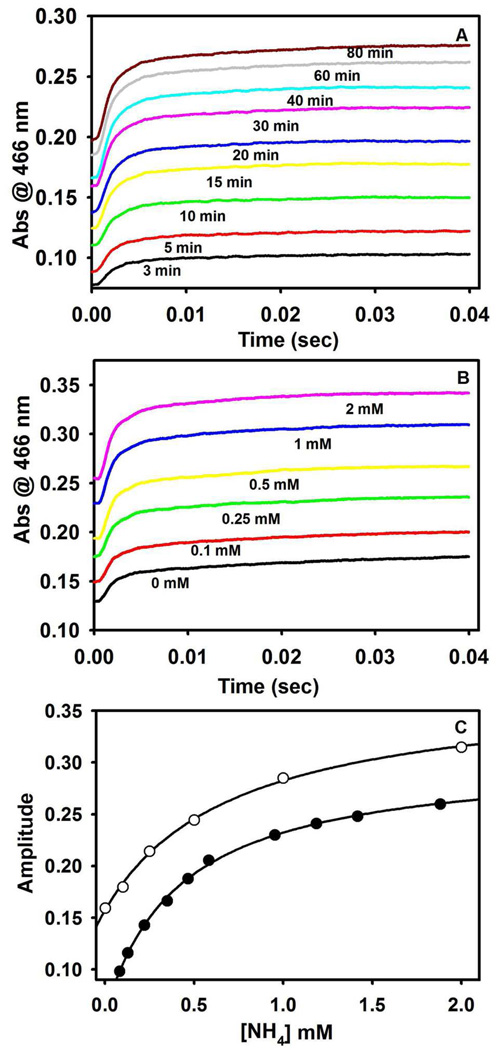
To quantify the effects of NH4+ on the activity of the MVC-free enzyme, experiments were performed to determine the significance of NH4+ contamination from the pyruvate side reaction on preparations of the MVC-free enzyme. Figure 3A compares the time courses for the indoline reaction obtained under conditions where α2β2 (prepared MVC-free) and L-Ser were pre-incubated prior to reaction for time periods ranging from 3 min to 80 min. At short incubation times, the traces show a strong dependence of the total amplitude on the incubation time; at longer incubation times this dependence attenuates. For comparison, Figure 3B shows the SWSF time courses obtained for the indoline reaction when E(A-A) is pre-mixed with various concentrations of NH4+. The dependence of amplitude on NH4+ concentration increases and then becomes independent as the NH4+ concentration increases (Figure 3B).
Figure 3.
Ammonium ion contamination effects from deamination of L-Ser in the tryptophan synthase-catalyzed pyruvate side reaction. (A) Effects of L-Ser - α2β2 incubation on the indoline reaction (incubation times are indicated). Final concentrations: α2β2, 10 µM; L-Ser 50 mM, indoline 5 mM. (B) Effects of NH4+ preincubation with α2β2. Final concentrations: α2β2, 10 µM; L-Ser 50 mM, indoline 5 mM, and NH4+ as indicated. Time courses in (A) and (B) were measured using SWSF instrumentation. (C) Comparison of the effect of L-Ser incubation time (black circles) on amplitude with the effects of added [NH4+] (white circles) on the fast phase of the indoline reaction (α2β2 preincubated with NH4+). Amplitudes taken from (A) and (B). To correct for fast phase amplitude loss in the instrument dead-time, the total amplitude of the fast phase was estimated as the sum of the observed fast phase and the y-intercept value for the reaction of α2β2 and L-Ser, with indoline. Traces were fit to the sum of two exponentials. The NH4+ concentrations for time resolved data points (black circles) were estimated using kcat = 0.098 s−1 for pyruvate formation at each incubation time in (A).
Amplitudes measured at 466 nm provide a quantifiable measure of the fraction of E(Q)indoline formed. Due to the magnitude of the fast phase, some amplitude is lost in the mixing dead-time (viz., Figures 3A and 3B). To adjust for this loss, theoretical y-intercept values were calculated by extrapolation to the time of mixing. The NH4+ concentrations generated during each incubation period were estimated from the kcat for pyruvate formation (kcat = 0.097 s−1), the incubation time, and the concentration of enzyme. Corrected amplitudes then were plotted as a function of the NH4+ concentration (Figure 3C, black dots). For comparison, the amplitudes measured as a function of NH4+ concentration in Figure 3B were also plotted in Figure 3C (open circles). Figure 3C establishes that the amplitudes of the time courses taken from both Figures 3A and 3B depend upon the concentration of NH4+. The apparent hyperbolic dependence of amplitude on NH4+ concentration indicates that these curves measure the apparent dissociation constant for NH4+ binding. When fitted to the positive lobe of a rectangular parabola, both data sets presented in Figure 3C yield a value of the apparent dissociation constant for NH4+ binding to the MVC site of ~ 0.67 mM.
Reaction of Pre-incubated MVC-free α 2β2 and Indoline with L-Ser
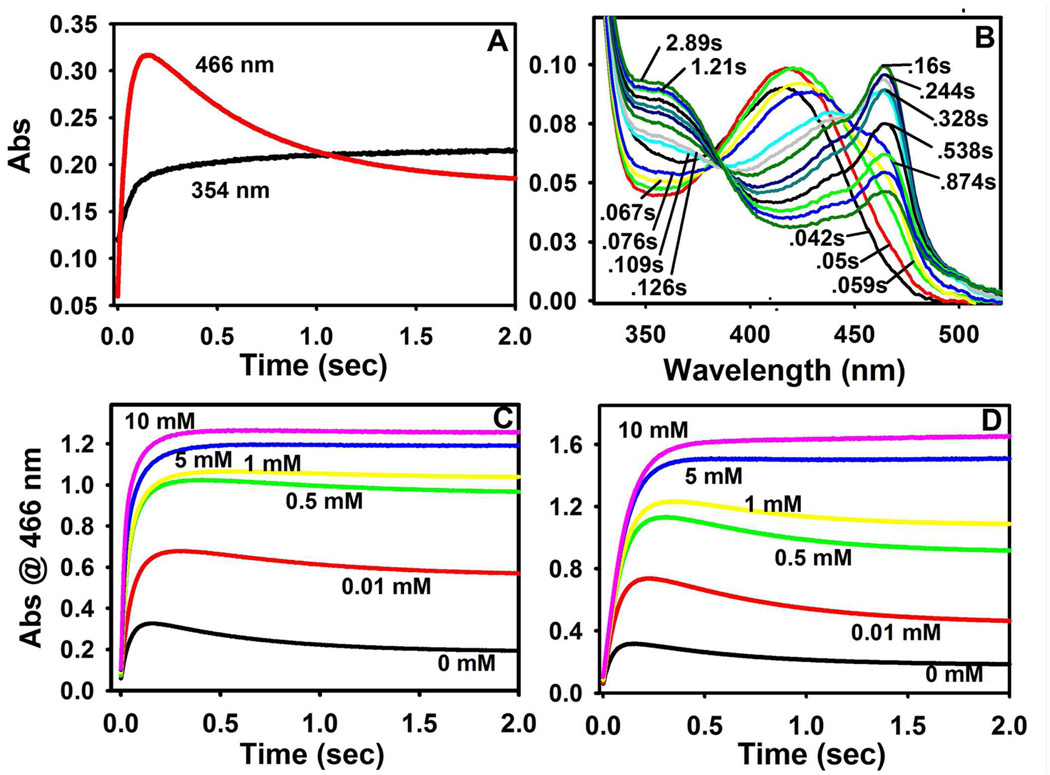
In order to further investigate the reactivity of the MVC-free enzyme, MVC-free α2β2 was pre-incubated with indoline, and mixed with L-Ser in the SWSF apparatus (Figure 4A). The reaction was monitored at both 466 nm and 354 nm. At 466 nm, three relaxations were detected in the trace, two with increasing phases (1/τ1 ≈ 23 s−1, and 1/τ2 ≈ 14 s−1) and one with a decreasing phase (1/τ3 ≈ 1.7 s−1) (Table 1). At 354 nm, two relaxation rates with increasing amplitudes were detected (1/τ1 ≈ 19 s−1, and 1/τ2 ≈ 2.2 s−1; Table 1), values in good agreement with the rates of the first two relaxations observed at 466 nm for the formation of E(Q)indoline.
Figure 4.
(A) Comparison of the indoline reaction time courses at 466 nm and 354 nm under MVC-free conditions where L-Ser is mixed with 20 µM α2β2 preincubated with indoline in the SWSF. (B) Rapid-scanning stopped-flow experiment showing time-resolved spectra (300 to 550 nm) for the formation of E(Q)indoline under MVC-free conditions. L-Ser was mixed with MVC-free 10 µM α2β2 preincubated with indoline. Spectra were acquired at 0.042, 0.0504, 0.0588, 0.0672, 0.0756, 0.1092, 0.126, 0.1596, 0.2436, 0.3276, 0.5376, 0.8736, 1.21, 2.89 s following mixing. (C) and (D) Time courses at 466 nm for the reaction of L-Ser mixed in the SWSF apparatus with α2β2, indoline and different concentrations of CsCl (C) or NaCl (D). 50 mM L-Ser and 5 mM indoline were used in (A) – (D), and the indicated MVC concentrations were used in (C) and (D).
Table 1.
Effects of Saturating and Sub-saturating Monovalent Cation Concentrations on the Indoline Reactiona
| Wavelength, 466 nm | ||||||
|---|---|---|---|---|---|---|
| MVC | A1b | 1/τ1 (s−1) | A2b | 1/τ2 (s−1) | A3c | 1/τ3 (s−1) |
| MVC free | 0.26 | 22.9 | 0.07 | 14.3 | 0.20 | 1.73 |
| CsCl, 0.5 mM | 0.66 | 40.3 | 0.39 | 9.2 | 0.13 | 1.20 |
| CsCl, 10 mM | 0.86 | 75.0 | 0.43 | 11.7 | 0.07 | 0.71 |
| NaCl, 0.5 mM | 1.38 | 10.7 | 0.50 | 1.98 | ||
| NaCl, 10 mM | 1.63 | 10.1 | 0.09 | 0.34 | ||
| Wavelength, 354 nm | ||||||
| MVC | A1b | 1/τ1 (s−1) | A2b | 1/τ2 (s−1) | A3b | 1/τ3 (s−1) |
| MVC free | 0.07 | 18.7 | 0.03 | 2.21 | ||
| CsCl, 0.5 mM | 0.07 | 34.7 | 0.02 | 1.83 | ||
| CsCl, 10 mM | 0.04 | 60.0 | 0.01 | 1.73 | ||
| NaCl, 0.5 mM | 0.04 | 13.7 | 0.05 | 1.63 | ||
| NaCl, 10 mM | 0.02 | 14.4 | 0.02 | 2.77 | ||
Enzyme and indoline in one syringe of the SWSF apparatus were mixed with indoline and L-Ser in the other syringe. The rates and amplitudes (absorbance changes) reported were derived from analysis of the time courses presented in Figure 4.
Increasing absorbance.
Decreasing absorbance.
RSSF time courses (Figure 4B) for the indoline reaction were measured to unambiguously identify the species formed during the three relaxations observed in Figure 4A. These time-resolved spectra establish that the absorbance changes at 466 nm accompanying 1/τ1, 1/τ2 and 1/τ3 are due to the formation and decay of E(Q)indoline. Figure 4B shows that after 0.16 s, the E(Q)indoline band at 466 nm reaches an absorbance maximum, and then decays, extrapolating to the absorbance values observed in MVC-free static spectra at the same enzyme concentration. Throughout the 2.89 s time interval for the RSSF experiment shown in Figure 4B, the band at 354 nm continues to increase in absorbance, and these changes are dominated by the formation of E(A-A). Kinetic slices from the RSSF data set measured at both 466 nm and 354 nm were fitted using PeakFit, and the relaxation rates were found to agree with those obtained from the SWSF time courses run under the same conditions (data not shown).
Panels C and D of Figure 4 compare the influence of Na+ and Cs+ on the SWSF time courses for the indoline reaction under conditions where α2β2 pre-incubated with indoline in one syringe was mixed with L-Ser pre-incubated with indoline in the other syringe. In panels C and D, the MVC was added to both syringes before mixing in the SWSF. In these 466 nm time courses, the decay of E(Q)indoline observed at low Na+ or Cs+ concentrations either disappears or becomes obscured by the slow phases of increasing amplitude that appear as the MVC concentration increases. Table 1 summarizes rates and amplitudes measured at 466 nm and 354 nm for the MVC-free enzyme, and compares these values with rates and amplitudes measured at sub-saturating (0.5 mM) and saturating (10 mM) concentrations of Cs+ or Na+. The Cs+ form of the enzyme exhibited 3 phases at 466 nm (two increasing, one decreasing), and two phases at 354 nm (both increasing). The Na+ form of the enzyme exhibited two phases at 466 nm (one increasing, one decreasing) and two phases at 354 nm (both increasing).
Characterization of the MVC-free E(A-A) Conformation
The data presented in Figures 2–4 establish that the reaction of MVC-free α2β2 with L-Ser gives a MVC-free E(A-A) that is essentially inactive towards indoline. One possibility is that the inactive form of MVC-free E(A-A) has an open β-subunit conformation (9). In 1996, a method was devised to probe the conformational state of tryptophan synthase, using the fluorescent ligand, 8-anilino-1-naphthalenesulfonate (ANS) (35). When bound, ANS exhibits a strongly enhanced fluorescence emission spectrum; free in aqueous solution, the fluorescence of ANS is strongly quenched. Pan and Dunn (35) demonstrated that ANS binds much more strongly to the open conformation than to the closed conformation of the tryptophan synthase αβ-dimeric unit. Consequently, when the tryptophan synthase subunits switch to the closed conformation, ANS is released and fluorescence due to bound ANS is strongly decreased (35). To determine the conformation state of inactive MVC-free E(A-A), ANS fluorescence was used to probe the conformation states of the Na+, Cs+, and MVC free forms of the enzyme. From the quenched ANS fluorescence it was estimated that in the absence or presence of F9, L-Ser reaction displaces 81% and 89%, respectively, of bound ANS from the Cs+-bound enzyme, and 85% and 96%, respectively, of bound ANS from the MVC-free enzyme. These findings indicate that the MVC-free E(A-A) adopts a closed conformation even though it is essentially inactive towards indoline.
Effects of F9 on the Formation and Decay of the E(Q)indoline
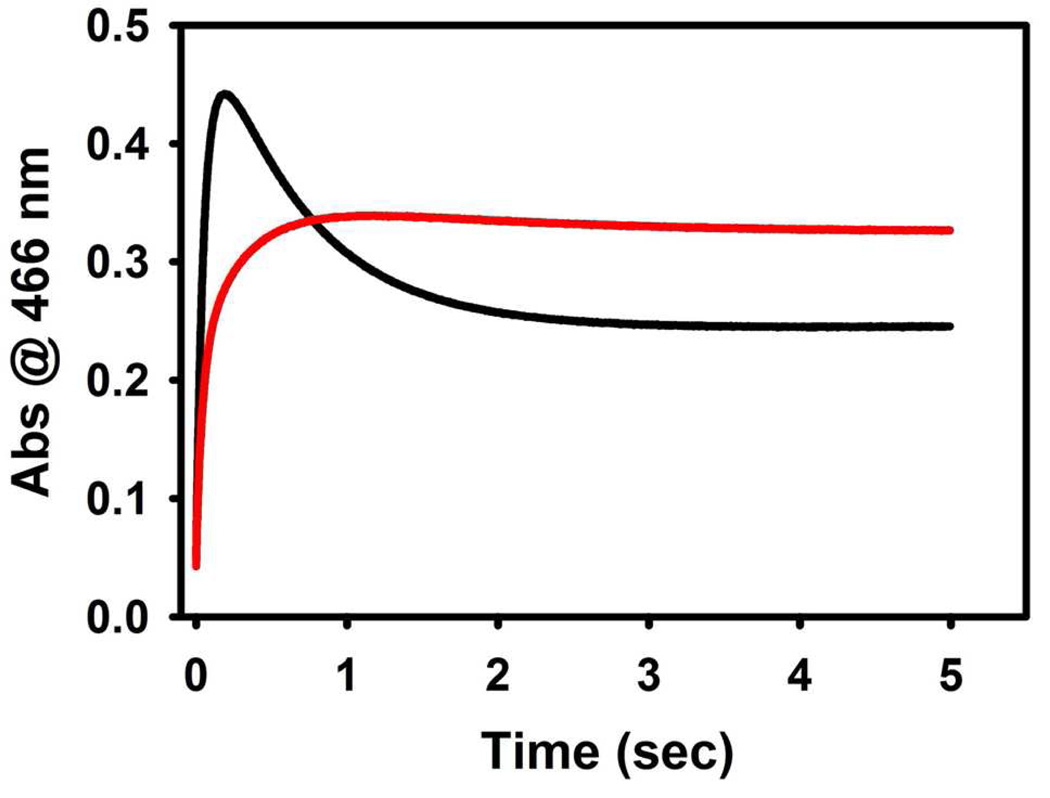
Ngo et al. (21, 22) demonstrated that the IGP analogue, F9, is a relatively specific, high-affinity ligand for the α-site, and that F9 mimics the allosteric effects of product G3P. Like other IGP analogues, the binding of F9 stabilizes the closed conformation of the α-subunit, preventing the entry of indole and indole analogues such as indoline into the β-site from solution by blocking access to the interconnecting tunnel at the α-site (19, 21, 22, 33). Therefore the use of F9 in combination with indoline provides a sensitive kinetic probe of the conformational switch to the closed conformation. When MVC-free α2β2, pre-incubated with indoline, is mixed with L-Ser in the SWSF apparatus, there are two increasing phases (1/τ1 ≈ 20 s−1, and 1/τ2≈10 s−1) and a decay phase (1/τ3 ≈ 2 s−1) (Figure 5, black trace). The time course also was examined under conditions where the α-site is occupied by F9 (Figure 5, red trace). For this experiment, the F9 used was prepared monovalent metal ion-free. These data show that when F9 is bound to the α-site, the time course of the indoline reaction is changed; the F9 complex gives a slow increasing phase with 1/τ ~ 2 s−1, followed by a slow decreasing phase of very small amplitude with a rate that is reduced by a factor of 4 (Figure 5, red trace).
Figure 5.
Comparison of time courses for the formation and decay of E(Q)indoline in the presence and absence of α-site ligand, F9. L-Ser and indoline in one syringe were mixed with α2β2 and indoline in the other syringe of the SWSF apparatus in the presence (red) or absence (black) of F9. Final concentrations: 20 µM α2β2, 50 mM L-Ser, 5 mM indoline, and 200 µM F9.
Influence of MVCs and F9 on the reaction of L-His with E(Ain)
L-His reacts with E(Ain) to form a distribution of covalent adducts at the β-active site dominated by the corresponding external aldimine and quinonoid species (36) (eq 5).
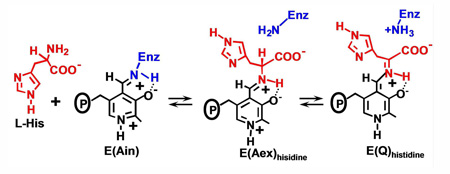 |
eq 5 |
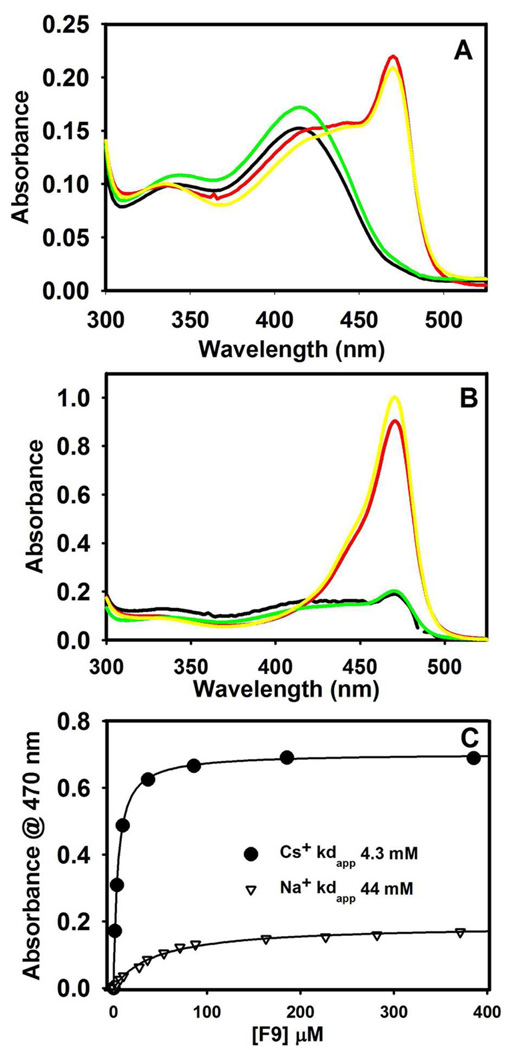
UV/vis absorption spectra of the L-His reaction provide a means for quantifying the dependence of the distribution of E(Aex)his and E(Q)his species on the binding of MVCs and α-site ligands. Spectra were collected with Cs+, NH4+, Na+, and MVC-free forms of the enzyme, both in the absence (Figure 6A) and in the presence of F9 (Figure 6B). For the Cs+ and NH4+ forms, these spectra exhibit contributions from two species, E(Aex)his (λmax = 414 nm) and E(Q)his (λmax = 470 nm) (37). The spectra of the MVC-free and Na+ forms are dominated by E(Aex)his and show only trace amounts of E(Q)his (~ 1.5% and ~ 2%, respectively). For each of the MVC-bound complexes, comparison of Figures 6A and 6B, reveals a significant increase in the fraction of enzyme sites occupied by E(Q)his when F9 is bound to the α-site (from ~ 2% to ~ 18.2 % for Na+, from ~ 14 % to 100 % for NH4+, and from ~ 17.3 % to ~ 89 % for Cs+). The percentages of E(Q)his and E(Aex)his formed were estimated from the static spectra and are summarized in Table 2. The simplifying assumption (eq 5) was made that the observed spectral bands in the static spectra arise from E(Aex)his and E(Q)his, and that contributions from E(GD)his, or E(Ain) are minor and can be neglected (16).
Figure 6.
Static spectra for the reactions of various MVC forms of α2β2 with L-His (eq 5) in the absence (A) and presence (B) of F9. MVC forms: Na+ (black), Cs+ (red), MVC-free (green), and NH4+ (yellow). (C) Binding isotherms measured at 470 nm for the titration of F9 into a premixed solution of α2β2, L-His and the Cs+ (black circles) or Na+ (open triangles). Each line is the least squares best fit of the data. Final Concentrations: 100 mM L-His, 50 mM MVC, and when present 200 µM F9; 10 mM α2β2 in (A) and (C), and 20 µM α2β2 in (B).
Table 2.
Dependence of E(Q)his Yield on the Binding of Monovalent Cations and on the α-Site Ligand, F9a,.
| Estimated Yield of E(Q)his (%)a | ||
|---|---|---|
| MVC | −F9 | +F9 |
| MVC-free | ~1.5 | ~16.0 |
| Na+ | ~2.0 | ~18.2 |
| NH4+ | ~14.0 | 100b |
| Cs+ | ~17.3 | ~89 |
Percentages estimated from spectra in Figure 6.
The yield obtained with the NH4+ form of the F9 complex was assumed to be 100% of the β-sites.
The effects of Cs+ and Na+ on the formation of E(Q)his when α2β2 is titrated with L-His in the presence of F9 were determined. L-His has a slightly higher affinity for the Cs+ form of the enzyme than for the Na+ form, giving KD values of 6 mM and 11 mM, respectively (data not shown). Binding of F9, increases the affinity of the Cs+ and Na+ forms of the enzyme for L-His ~ 3-fold. Figure 6C shows that the affinity of F9 for the L-His complex is enhanced ~ 10-fold by the substitution of Cs+ for Na+.
Analysis of the Effects of Cs+ Substitution for Na+ on Open and Closed Structures
The comparison of β-subunit structures was accomplished using alignment by sequence of the Cα atom positions for residues 10 to 100 with the program, PyMOL. Comparison of indole sub-sites was accomplished using paired selections consisting of the MVC and atoms in the PLP cofactor.
The superposition of the Na+ and Cs+ forms of the β-subunit in the closed conformation show very small structural differences. The most significant changes include the following: (a) Cs+ is displaced relative to the position of Na+ by ~ 1 A. (b) Na+ is coordinated by the carbonyl oxygens of βGly232, βPhe306, and βSer308 and two waters, while Cs+ is coordinated by the carbonyl oxygens of βGly232, βPhe306, βSer308, βVal231, βGly268, βLeu304, and one water molecule. (c) Those residues involved in coordination show a slight perturbation when Na+ is replaced by Cs+ with the largest effect on βGly232 (positional shift ~ 0.4 A). (d) In all of the β-subunit closed structures, the side chain of βAsp305 takes up a single orientation, a swing out position pointing away from the active site, and makes a salt bridge to βArg141. (e) The side chains of βTyr279 and βPhe280 take up positions against the tunnel wall leaving the tunnel unhindered. In some structures, the side chain of βPhe280 is disordered.
In the open conformation, substitution of Cs+ for Na+ has a larger effect on structure than in the closed conformation. Rhee et al. (6) made very careful comparisons of the effects of Na+, K+ and Cs+ on the structure of the open conformation. Their comparison of the Na+ and Cs+ complexes of E(Ain) used PDB codes 1BKS and 1TTP, respectively2. We re-examined the comparison of the Na+ and Cs+ forms of E(Ain) using a different structure for the Na+ complex, PDB code 1KFK. We found that this Na+ structure differs significantly from PDB code 1BKS only in the position of the βAsp305 side chain. The likely origins of this difference stem from the different temperatures used in the x-ray diffraction experiments (300°K for PDB code 1BKS vs. 100 K for PDB code 1KFK). In PDB code 1BKS, the side chain takes up two positions, a swing in position toward the active site, and a swing out position pointed away. The more favored, swing-in, position makes a salt bridge to βLys167. In the PDB code 1KFK structure, the βAsp305 side chain takes up a somewhat different swing out position.
Kinetic Simulation of the Transient Formation and Decay of the MVC-free E(Q)indoline in the Indoline Reaction
The formation of a closed, inactive E(A-A) species is consistent with Scheme 2, and is supported by the kinetic and spectroscopic results shown in Figures 4 and 5. Kinetic simulation was performed to further test the validity of Scheme 2 (for full details see the Supporting Information, Figure S1 and Tables S1 and S2). Because the binding of L-Ser and the conversion of E(Ain)(L-Ser) to E(Aex1) are relatively rapid processes, and because there is a strong forward commitment to give E(Aex1) (Scheme 2), these steps are decoupled from subsequent steps and do not perturb the rates of subsequent relaxations. Consequently for the simulations, Scheme 2 was simplified and rewritten as Scheme 3. Comparison of the simulated time courses with the experimental time courses obtained at both 466 nm and 350 nm show good agreement (see Figure S1 of the Supporting Information). Furthermore, it was found that the effects of Cs+ substitution for Na+ and the presence or absence of an MVC are well-accommodated by Scheme 3 (see Supporting Information, Figure S1 and Tables S1 and S2). For example, the experimental data and the corresponding simulations show that when Cs+ is substituted for Na+, the fast relaxation is increased by greater than 7-fold, whereas the MVC-free and Na+ forms of the enzyme show similar relaxation rates (22.9 s−1 and 10.1 s−1, respectively) (see Supporting Information, Table S2). The greater than 6-fold difference in amplitudes of the MVC-free and Na+ enzymes for this relaxation (0.26 A vs. 1.63 A) are consistent with the conclusion that most of the MVC-free E(A-A) is present in an unreactive state while the reactive state predominates in the MVC-bound forms.
Scheme 2.
Formation of E(A-A) and Reaction with Indoline via the Branched Path for the Preincubation of E(Ain) with Indoline
Scheme 3.
Simulation Scheme for Formation and Reaction of E(A-A) via the Simplified Branched Path
DISCUSSION
As is true for allosteric effectors in general, the action of MVCs in the regulation of enzyme activity and protein function has its origins in the modulation of protein conformation between states of altered activity. This modulation involves at least two levels of conformation hierarchy; global changes in subunit structure such as the switching between T- and R-states, and local changes such as the perturbation of the dimensions and/or properties of a ligand binding site (25, 26). As discussed below, the results presented in this study demonstrate that the binding of an MVC is essential for switching between open (inactive) and closed (active) states of tryptophan synthase, and for maintenance of the correct conformation of the indole sub-site at the β-subunit.
The influence of Na+ binding on the activation energies for steps in the β-reaction have been examined by Woehl and Dunn (10). They concluded that in stage I of the β-reaction Na+ binding (a) slightly stabilizes the binding of L-Ser to E(Ain), (b) strongly stabilizes E(Aex1), (c) lowers the activation energy for E(Aex1) formation, and (d) increases the activation energy for E(A-A) formation. In stage II, both the complex of indole with E(A-A) and E(Q) are stabilized by Na+ binding, while the activation energies for E(Q) formation and decay are unaffected by Na+ binding.
Analysis of MVC Effects on β-Subunit Structure and Function
The ionic radii of Na+, K+ and Cs+ are 0.97 A, 1.33 A and 1.74 A, respectively. This size variation dictates MVC-dependent differences in coordination number and geometry for inner-sphere ligand complexes in small molecules and in proteins (25) The MVC effects described in this work and in previous studies no doubt have their origins in the propagation of these differences from the MVC site to the surrounding protein (6–10, 27). Alignment of β-subunit structures taken from the protein data base show only two global conformations of the β-subunit, the open conformation typically found for E(Ain) and E(Aex1), and the closed conformation recently determined for an E(A-A) complex (21, 22) and for a quinonoid complex (20). Closed β-subunit structures have also been reported for two different tryptophan synthase mutants (38, 39). There are no published structures for MVC-free forms of the enzyme. Because the open conformation is easily crystallized, most structural studies have focused on the Na+ form of the enzyme with the β-subunit in this conformation. However, there are six crystal structures of the enzyme with the β-subunit in the closed conformation, four of the Na+ form (β-subunit mutants with βLys87 replaced by Thr; PDB codes, 2TRS, 2TRY, 2TYS, and the mutant with βGln114 replaced with Asn; PDB code 2J9Y) and two of the Cs+ form (wild-type enzyme, PDB codes 3CEP and 2J9X). These structures make possible comparisons of the effects of Cs+ substitution for Na+ on the closed conformation of the β-subunit.
Rhee et al. (6) described and compared the structures of the Na+, K+ and Cs+ forms of E(Ain) where the β-subunit has the open conformation. The salient findings of Rhee et al. (6) are summarized as follows: (a) When Cs+ (PDB code 1TTP) is substituted for Na+ in E(Ain), significant changes are found in the locations of certain key β-site residues2. (b) Substitution of Cs+ for Na+ shifts Cs+ ~1.1 Å away from the Na+ position. (c) Na+ coordinates the carbonyl oxygens of βGly232, βPhe306, and βSer308 and two waters, while the Cs+ coordination sphere expands to also include βVal231, βGly268, and βLeu304, but no waters. (d) The MVC binding loop (residues β259–β310) is slightly displaced by Cs+ substitution with the largest effects on βPhe306 and βAsp305 (positional shifts ~ 0.8 Å). (e) In the Na+ structure, the βTyr279 and βPhe280 side chains partially block the tunnel connecting the α- and β-sites. In the Cs+ structure, these side chains take up positions against the tunnel wall and may cause a repositioning of αAsp56.
Figure 7A presents a composite overlay that compares details of the β-sites and MVC sites of wild-type enzyme for the open β-subunits of the Na+ (PDB code 1KFK)2,3 and Cs+ (PDB code 1TTP) structures of E(Ain) with the closed β-subunit of the Cs+ (PDB code 3CEP) structure of E(Q)indoline. The open structures of the Na+ E(Aex1) complexes (PDB codes 1KFJ and 1KFE, (40); and PDB codes 2CLL, 2CLM, 2CLO, (22) all show placements of the βAsp305 side chain that are essentially identical to the position found in the Na+ E(Ain) complex (not shown). Figure 7A shows that the replacement of Na+ by Cs+ gives an open E(Ain) structure where the side chain of βAsp305 is shifted toward the position found in the closed Cs+ conformation (the swing out position). Previous structural and kinetic studies (4, 5, 11, 20, 22, 34, 38, 39) strongly indicate that formation of the H-bonded salt bridge between βAsp305 and βArg141 with the associated H-bonding network involving βSer197 and βSer199 (Figures 1B and 8A) is essential both for conversion of the β-subunit from the open to the closed conformation, and for activation of the β-site for the conversion of E(Aex1) to E(A-A) and E(Q) (Scheme 1). The K+ substituted E(Ain) complex (PDB code 1TTQ) shows the side chain of βAsp305 in a position nearly identical to that found in the Cs+ E(Ain) complex, the swing out position (6). Thus, increasing MVC size favors the swing out position for the βAsp305 side chain, a conformation that is poised to form the salt bridge with βArg141 when the β-subunit is switched to the closed conformation. Since, the MVC site of tryptophan synthase in vivo likely is occupied by K+, these comparisons (Figure 7A) suggest that the Cs+ structures are better models for the physiologically relevant form of E(Ain) than are the Na+ structures. Accordingly, we hypothesize that the MVC effects reported in this work result from shifts between the open and closed states brought about by the preferential stabilization of one state relative to the other by the particular MVC.
Figure 7.
(A) Stereoscopic diagram comparing details of the β-sites and MVC sites of the Na+-and Cs+-E(Ain) complexes (respectively, PDB codes 1KFK and 1TTP) with the Cs+ E(Q)indoline complex (PDB code 3CEP). The E(Ain) complexes have the open β-subunit conformation, the E(Q)indoline complex has the closed conformation. Structures were aligned using the Cα atom coordinates of β-subunit residues 10 to 100. Color scheme: PDB code 1KFK, carbons, Na+ and dashes are gray; PDB code 1TTP, carbons, Cs+ and dashes are green; PDB code 3CEP, carbons, Cs+ and dashes are yellow. All other atoms are shown in CPK colors. βAsp305, βArg141, and the PLP moieties are shown as sticks, other residues are shown as wireframe. Dashed lines indicate H-bonds and/or van der Waals contacts between the side chain of βAsp305, βArg141 and nearby residues. (B) and (C): Stereo views comparing the indole sub-sites of the open Cs+ and Na+ E(Ain) complexes, respectively, with the closed Cs+ E(Q)indoline complex. Coloring schemes: in (B) and (C), the C and Cs+ atoms are yellow in the Cs+ E(Q)indoline complex. In (B), the C and Cs+ atoms of the Cs+ E(Ain) complex are gray. In (C) the C and Na+ atoms of the Na+ E(Ain) complex are green. All other atoms are shown with CPK coloring. The indoline ring of the Cs+ E(Q)indoline complex is shown in wireframe. The dashed lines measure the distances between the Cα atoms of βHis115 and βPhe306. In (B) and (C), structures were superimposed using atoms of the PLP moieties and the MVC in the PyMOL paired selection mode. Structures rendered with PyMOL 1.1r1.
Detailed comparisons of the indole ring sub-site (Figure 7B) show that the open Cs+ E(Ain) and closed Cs+ E(Q)indoline complexes have very similar cavities for the indole ring (modeled by the indoline group), whereas the open Na+ E(Ain) complex exhibits a somewhat different cavity (Figure 7C). The rings of βPhe306 and βHis115 define the long dimension of the indole sub-site cavity (Figures 8B and 8C). (Figure 7B) shows that these residues are similarly aligned both in open and in closed Cs+ complexes, with the cavity diameter decreased by ~ 1.0 Å (measured as the distance between the βHis115 and βPhe306 Cα positions) in the open E(Ain) structure compared to the closed E(Q)indoline structure. However, in the open Na+ complex, βHis115 is shifted relative to the positions found in the Cs+ complexes and the diameter of the cavity is reduced by ~2 Å compared to the closed E(Q)indoline structure (Figure 7C). Comparisons of the closed Cs+ E(Q)indoline and the Cs+ E(A-A) complexes and the closed Na+ complexes of E(Aex) species (38) show that all the indole cavities in these closed conformations are very similar even though the cavity is only partially filled in some of these structures (data not shown). These comparisons establish that the indole cavities of the open Na+ E(Ain) and Na+ E(Aex1) complexes are collapsed/distorted relative to the cavities of the closed structures. As argued in the following paragraphs, this constriction/distortion of the cavity in the open conformation of Na+- bound enzyme likely accounts for the MVC-dependent differences reported herein.
Ligand Binding Effects on Stage I of the β-Reaction
Kinetic and spectroscopic studies in aqueous solution have established that ligand binding to three different loci, the α- and β-catalytic sites and the MVC site (Figure 1), can alter the distribution of covalent intermediates formed in stages I and II of the β-reaction (Scheme 1). It is now clear that these ligand-induced changes in distribution result from the switching of the β-subunit between the open and closed conformation states (5, 22).
Figure 2A compares the effects of MVCs on the interconversion of the open E(Aex1) and the closed E(A-A) complexes. As shown in Figure 2A, stage I of the β-reaction gives a distribution of intermediates that is MVC-dependent. This distribution is determined by the relative stabilities of the MVC complexes of the open E(Aex1), λmax 424 nm, and the closed E(A-A) species, λmax 354 nm, shoulder 460 nm. The spectra show that for NH4+ and Cs+, conversion to E(A-A) is nearly complete, indicating that both ions have a significantly higher affinity for E(A-A) than for E(Aex1). For Na+ complexes, although E(A-A) is still the predominant species, there is a significant amount of E(Aex1) present at equilibrium. The open Na+ E(Aex1) complex (like the Na+ E(Ain) complex, (21, 22)) has an active site with the side chains of βAsp305 and βArg141 incorrectly positioned to form the salt bridge, and the indole sub-site has a collapsed/distorted structure (viz., Figure 7). Since the open Cs+ E(Ain) complex and all known closed structures with either Na+ or Cs+ bound have preformed indole binding sites in the β-subunit (viz., Figure 7B), it is reasonable to conclude that the reason Na+ is less effective than NH4+ and Cs+ in stabilizing E(A-A) is because the difference in Na+ affinity for E(Aex1) vs. E(AA) is smaller.
Effects of MVC and ASL Binding on the Reaction of L-His with E(Ain)
L-His reacts with MVC-bound forms of tryptophan synthase to give equilibrating mixtures of E(Aex)his and E(Q)his (eq 5) Figure 6 and Table 2 show that the MVC-free and Na+ forms of the enzyme react with L-His to give E(Aex)his with only small amounts of E(Q)his, respectively ~1.5% and ~2%; whereas the Cs+ and NH4+ forms give respectively ~17.3% and ~14%. Binding of the α-site ligand, F9, gives ~16%, ~18%, ~89%, and ~100% E(Q)his, respectively, for the MVC-free, Na+, Cs+, and NH4+ enzymes. These findings establish that Na+ is ineffective in stabilizing E(Q)his, whereas NH4+ and Cs+ binding cause a significant reordering of the relative stabilities of E(Aex)his and E(Q)his in favor of E(Q)his (Figure 6A). The binding of ASLs further stabilize the NH4+ and Cs+ forms of E(Q)his (Figure 6B) but exert only a small effect on the Na+ forms. By analogy to the L-Ser and indoline reactions, it is likely that E(Aex)his has the open conformation while E(Q)his has the closed conformation. Binding of L-His to give E(Aex)his is ineffective in displacing ANS, indicating the conformation of this species is open (35). Therefore, the stabilizing effects of NH4+ and Cs+ on E(Q)his likely reflect higher affinities of these MVCs for the closed conformation of E(Q)his than for the open conformation of E(Aex)his. Conversely, the difference in affinity of Na+ for E(Aex)his and E(Q)his must be relatively small. The further stabilization of E(Q)his by the ASL, F9 (Figure 6B), is consistent with a ligand-induced allosteric shift in favor of the closed state (4, 5, 11, 22, 33, 36, 37, 41). The trend toward tighter binding at the β-site (Figure 6C) is similar to that seen with L-Ser; for example, F9 binds 25-fold more tightly to the closed E(A-A) (with KD ~ 2 µM), than to the open E(Ain) (with KD ~ 50 µM) (22).
All of the reported ligands specific for the α-site appear to exert allosteric interactions that favor the closed β-subunit conformation (5, 22, 42) (this work, Figures 5 and 6). In stage I of the β-reaction, the binding of unreactive indole analogues (e.g., benzimidazole) to the β-subunit indole sub-site shifts the distribution of species completely to E(A-A) in the closed β-subunit conformation (33, 36, 37). The substitution of NH4+ or Cs+ for Na+ at the MVC site also shifts the distribution of species essentially completely to E(A-A) in the closed β-subunit conformation (4, 8–11, 20, 22) (Figure 2, this work).
The Reactivity of E(A-A) with Indoline
This work and previous studies show that indoline reacts rapidly and reversibly with MVC-bound forms of E(A-A) to give E(Q)indoline (Figures 2–5) (8–10, 17–19, 33). At concentrations of indoline approaching saturation of binding, there is little difference in yield for these MVC-bound forms. However because the apparent dissociation constant for indoline is MVC-dependent, at 5 mM concentrations of indoline the yields of E(Q)indoline shown in Figure 2B are substantially different among the MVC-free and MVC-bound forms of the enzyme (8). Consequently, under the conditions of Figure 2B, the Na+-bound enzyme is converted to E(Q)indoline in nearly quantitative yield, while the Cs+-bound and NH4+-bound enzymes give 60% to 70%, and 30% to 40%, respectively. The experiments presented in Figures 3 and 4 establish that the amount of E(Q)indoline formed (~3.5%) in the absence of added MVC is due either to contaminating MVC (likely NH4+ generated by the pyruvate side reaction, eq 2), or to a residual amount of active, MVC-free E(A-A).
When L-Ser is reacted with MVC-free α2β2 pre-incubated with indoline, three relaxations are observed at 466 nm, two with increasing phases and one with a decreasing phase (Figure 4). Comparison of time courses shows, as expected, that E(A-A) formation and E(Q)indoline formation are kinetically linked. These indoline reaction time courses indicate that the MVC-free enzyme reacts with L-Ser to give a mixture of active and inactive E(A-A) forms strongly weighted toward the inactive form (Figure 4). The time courses in Figure 4 show that when L-Ser reacts with MVC-free enzyme in the presence of indoline, the reactive MVC-free E(A-A) initially formed is quickly consumed by reaction with indoline to give MVC-free E(Q)indoline. In this experiment (Figure 4) the rate of formation of E(Q)indoline is determined by the rate of formation of the active MVC-free E(A-A) from E(Aex1) is limited by the rate of E(A-A) formation, a process occurring at ~20 s−1 (9). The slow decay (1/τ3 ≈ 2 s−1) at 466 nm, and the slow increase (1/τ2 ≈ 2 s−1) at 354 nm reflect the conversion of the active E(A-A) to the inactive E(A-A) (9, 10), and the results presented herein agree with this conclusion. Consequently, the slow decay of MVC-free E(Q)indoline occurs in response to the slow redistribution of enzyme forms in favor of the inactive E(A-A) (Figure 4). Thus, conversion to inactive E(A-A) occurs at the expense of E(Q)indoline. The bifurcated pathway in Scheme 2 describes the partitioning of active E(A-A) between E(Q)indoline and inactive E(A-A). Because formation of E(Q)indoline is rapid and reversible, and conversion to the inactive E(A-A) is slow, this scheme predicts that formation of E(Q)indoline will show a transient increase and decrease, just as is seen in Figure 4. The relaxation for the slow interconversion (~2 s−1), is present in the time courses both for the formation of E(A-A) and for the decay of E(Aex)1 (9). This slow process is most likely due to a conformational change which transforms the active E(A-A) into the inactive species.
ANS has been shown to be a sensitive probe of tryptophan synthase conformation states (35, 42). ANS is displaced when the subunits are switched from the open to the closed conformations. ANS is displaced when E(A-A) is formed regardless of whether or not an MVC is bound. This finding is consistent with the assignment of closed conformations for both the active and the inactive MVC-free E(A-A) species. Since the nucleophilic attack of indoline on E(A-A) is the step most effected in the indoline reaction by removal of the MVC, it seems likely that the switch from the active to the inactive MVC-free E(A-A), involves a structural rearrangement of the β-subunit indole binding sub-site that interferes with the binding of indoline. The switch from the open to the closed conformation of the β-subunit as E(Aex1) is converted to E(A-A) (Scheme 1) creates a relatively high affinity binding site capable of binding indole and indole analogues such as benzimidazole or indoline, and this conformation is stabilized by MVC binding. We propose that without a bound MVC, this sub-site slowly relaxes to a “collapsed” conformation, effectively decreasing the affinity of indoline and interfering with formation of E(Q)indoline. The binding of Na+ either prevents or makes negligible the conformational transition to the collapsed conformation, allowing accumulation of E(Q)indoline in nearly stoichiometric amounts (Figures 2 and 3).
Effect of α-Site Ligand Binding on the Formation of Inactive E(A-A)
In the absence of an ASL, reaction of indole or indole analogues with E(A-A) to give E(Q) is relatively rapid (200 s−1 to 900 s−1, depending on the MVC and the substrate) (31, 33, 43–46). ASL binding to the α-site with the β-site in the form of E(A-A) switches both the α- and the β-subunits to the closed conformation, thus greatly retarding the entry of ligands from solution into the sites and the interconnecting tunnel (5, 18, 19, 21, 22, 32, 33, 42, 47). Figure 5 shows the influence of the high affinity α-site ligand, F9 (21, 22), on formation and decay of E(Q)indoline under MVC-free conditions. When L-Ser is mixed with F9-bound α2β2 and indoline under MVC-free conditions, two increasing phases are seen at 466 nm with relaxation rates of 1/τ1 ≈ 20 s−1 and 1/τ2 ≈ 2 s−1, followed by a decay phase of very small amplitude (Figure 5, red trace). The presence of F9 does not effect the rate of E(A-A) formation at 354 nm (data not shown), yet the decay phase at 466 nm (compare the black and red traces), observed in the absence of F9, disappears almost entirely. Therefore, it appears that the binding of F9 to the α-site shifts the equilibrium shown in Scheme 2 towards the active branch of the E(A-A) system, thus increasing the yield of E(Q)indoline and reducing the fraction of sites in the inactive E(A-A) form. This result establishes that F9 binding to the α-site and MVC binding to the β-site have similar effects on the distribution of active and inactive forms of MVC-free E(A-A), and these effects are synergistic.
Kinetic Simulation of MVC-free E(Q)indoline Formation and Decay
Substitution of Cs+ for Na+ increases the rate of E(Q)indoline formation by greater than 7-fold, whereas the MVC-free and Na+ forms of the enzyme show similar relaxation rates (22.9 s−1 and 10.1 s−1, respectively). In this latter comparison, it is important to notice that there is a large influence of the MVC on reaction amplitude (> 6-fold) (see the Supporting Information, Table S2).
By adjusting the forward and reverse rates for the step responsible for the formation of inactive E(A-A) according to Scheme 3B, the effects of Na+ and Cs+ binding on the formation and decay of E(Q)indoline, were simulated. Comparison of the experimental data with these simulations shows good agreement (see Figure S1 and Table S2 of the Supporting Information), confirming that the fast formation phase corresponds to the observed rate of formation of E(A-A) for the Na+, Cs+, and MVC-free forms of the enzyme (9). Therefore, these simulations support a model (Schemes 2 and 3) where there are two possible fates for E(A-A): either the electrophilic β-carbon of E(A-A) reacts with indoline to form E(Q)indoline, or E(A-A) remains in an inactive state with indoline noncovalently bound. It is our assessment that the two branches shown in Scheme 2 occur under any MVC condition, however the inactive E(A-A) branch is of little significance when the MVC site is occupied.
In contrast to the effects brought about by MVC substitutions, the absence of a bound MVC impairs the reactions of nucleophiles such as indoline and indole with the α-aminoacrylate intermediate (Figures 2–4) and significantly destabilizes quinonoid species (Figure 6). Although reaction of L-Ser initially generates a reactive E(A-A), this species decays to an inactive state within a few seconds (Figures 3, 4 and 7). Reactivity is restored by the binding of MVCs and is partially preserved by the binding of ASLs (Figures 5 and 6). We conclude that the decay of the active MVC-free E(A-A) to an inactive state results from a conformational transition that compromises the structural integrity of the indole sub-site of the β-subunit.
CONCLUSIONS
Metal ion activated proteins are present throughout nature, however, the mechanisms of activation are very diverse (25, 26). In tryptophan synthase catalysis and regulation, cation activation is achieved through an allosteric linkage connecting the PLP active site to the MVC binding site, ~ 8A away (Figures 1 and 8). The MVC site consists of backbone carbonyls from a stretch of amino acids that can twist and bend to coordinate cations of different size and incorporate ligated water molecules as needed to satisfy the electrostatic requirements of the bound MVC.
These studies establish that without a bound MVC, both the reaction of indoline with E(A-A) and the reaction of L-His with E(Ain) are strongly impaired. The reaction of L-Ser with MVC-free E(Ain) gives a reactive but relatively unstable E(A-A) species which converts to an inactive form. The binding of Na+, NH4+ or Cs+ repairs β-site reactivity in the indoline reaction (Figures 2–4; Table 1) and restores allosteric communication (8–10). In the reaction of E(Ain) with L-His, the binding of NH4+ or Cs+ restores activity to E(Ain), but the binding of Na+ does not. The binding of a high affinity ASL (Figures 4 and 6; Table 2), also restores reactivity to the MVC-free β-site and acts synergistically with MVCs to stabilize E(A-A) and E(Q) species.
Consequently, MVC binding insures the structural integrity needed by the β-active site during stage II of the β-reaction both for the nucleophilic attack of indole and indole analogues on the E(A-A), and for the conversion of E(Q) to E(Aex), and then to E(GD) and E(Ain) and finally to the new amino acid as the catalytic cycle is completed. The origins of the deleterious effects of MVC removal from the β-subunit are reasonably explained as due to the collapse or distortion of the indole ring sub-site within the closed conformation of the β-subunit. Determination of the structural details of the perturbed site must await further studies.
In the tryptophan synthase system, the efficient channeling of substrate indole between the α- and β-sites requires the switching of αβ-dimeric units of the tetrameric enzyme between open (active) and closed (inactive) conformational states (5, 33, 25, 42). Under normal in vivo conditions, the bienzyme complex likely exists in an inactive (“resting”) state with the α- and β-subunits in open conformations with the α-site unoccupied and the β-site in the form of E(Aex1). IGP binding switches the α-subunit to the closed conformation, triggering a switch in the β-subunit from the resting E(Aex1) state to a reactive closed conformation which then undergoes reaction to give E(A-A). This conformational switch activates the cleavage of IGP to indole and G3P at the α-site. The closed conformation of the αβ-dimeric unit confines indole to the indole sub-sites of the α- and β-subunits and the interconnecting tunnel, insuring reaction between indole and E(A-A) (as per Scheme 1). When the indole quinonoid is converted to E(Aex2) the dimeric unit is switched back to the open conformation, releasing G3P from the α-site as L-Trp is formed in stage II of the β-reaction (5). The results presented herein further substantiate that the roles of bound MVC include stabilization of the β-subunit indole binding sub-site and maintenance of a balance between the relative energies of the open and closed conformations of the β-subunit during catalysis (22, 25). This balance insures that synchronization of the α- and β-catalytic cycles is achieved via ligand-mediated conformational switching between open and closed states of low and high activity (5).
Supplementary Material
Footnotes
Supported by NIH grant GM5574 (M.F.D.) and Deutsche Forschungsgemeinschaft (I.S.).
Abbreviations: α2β2, native form of tryptophan synthase from S. typhimurium; α, the alpha subunit; β, the beta subunit; E(Ain), the internal aldimine (Schiff base) species; E(Aex1), the external aldimine species formed between the PLP cofactor and L-Ser; E(GD), the gem diamine species; E(A-A), the α-aminoacrylate Schiff base species; E(Q1), the L-Ser quinonoid species; E(Q3), and E(Q)indoline the quinonoid species that accumulate, respectively, in the reaction between E(A-A) and indole; and E(A-A) and indoline; E(Aex2), the L-Trp external aldimine species; PLP, pyridoxal phosphate; L-Ser, L-serine; L-Trp, L-tryptophan; IGP, 3-indole-D-glycerol 3’-phosphate; GP, α-D,L-glycerol-phosphate; G3P, D-glyceraldehyde 3-phosphate; DIT, dihydroiso-L-tryptophan; ASL, α-site ligand; F9, N-(4'-trifluoromethoxybenzenesulfonyl)-2-aminoethyl phosphate; ANS, 8-anilino-1-naphthalene sulfonate; β-me, β-mercaptoethanol; DTNB, Dithio-bis(2-nitrobenzoic acid); TNB, 2-nitro-5-thiobenzoic acid; TEA, triethanolamine; MVC, monovalent cation; KDapp, apparent dissociation constant. SWSF, single-wavelength stopped-flow; RSSF, rapid-scanning stopped-flow. Structural elements of tryptophan synthase are designated as follows: loop αL2, α53-a60; loop αL6, α179–α193; helix αH8, α249–α265; COMM domain, β102–β189; helix βH5, β145–β150; helix βH6, β165–β181.
Although not stated in Rhee et al. (6), the Na+ structure used in the comparisons of conformation appears to be PDB code 1BKS (a refined version of the original E(Ain) structure, PDB code 1WSY).
The structure file PDB code 1KFK used in place of PDB code 1BKS (Figure 7) differs only in the position of the βAsp305 side chain. The likely origins of this difference stem from the different temperatures used in the x-ray diffraction experiments (300°K for PDB code 1BKS vs. 100° K for PDB code 1KFK). In PDB code 1BKS, the side chain takes up two positions, a swing in position toward the active site, and a swing out position pointed away. The more favored, swing-in, position makes a salt bridge to βLys167. In the PDB code 1KFK structure, the βAsp305 side chain takes up a somewhat different swing out position with Oδ1 contacting the carbonyl O of βAla302.
SUPPORTING INFORMATION AVAILABLE
The Results of the detailed kinetic simulation study consisting of Figure S1, Table S1 and Table S2 are presented. This material is available free of charge via the Internet at http://pubs.acs.org.
REFERENCES
- 1.Yanofsky C, Crawford IP. Tryptophan synthase. In: Boyer PD, editor. The Enzymes. 3rd ed. New York: Academic Press; 1972. pp. 1–31. [Google Scholar]
- 2.Miles EW. Tryptophan synthase: structure, function, and subunit interaction. Adv. Enzymol. Relat Areas Mol. Biol. 1979;49:127–186. doi: 10.1002/9780470122945.ch4. [DOI] [PubMed] [Google Scholar]
- 3.Miles EW. Structural basis for catalysis by tryptophan synthase. Advances in Enzymology and Related Areas of Molecular Biology. 1991;64:93–172. doi: 10.1002/9780470123102.ch3. [DOI] [PubMed] [Google Scholar]
- 4.Miles EW, Rhee S, Davies DR. The molecular basis of substrate channeling. Journal of Biological Chemistry. 1999;274:12193–12196. doi: 10.1074/jbc.274.18.12193. [DOI] [PubMed] [Google Scholar]
- 5.Dunn MF, Niks D, Ngo H, Barends TRM, Schlichting I. Tryptophan synthase: the workings of a channeling nanomachine. Trends in Biochemical Sciences. 2008;33:254–264. doi: 10.1016/j.tibs.2008.04.008. [DOI] [PubMed] [Google Scholar]
- 6.Rhee S, Parris KD, Ahmed SA, Miles EW, Davies DR. Exchange of K+ or Cs+ for Na+ induces local and long-range changes in the three-dimensional structure of the tryptophan synthase α2β2 complex. Biochemistry. 1996;35:4211–4221. doi: 10.1021/bi952506d. [DOI] [PubMed] [Google Scholar]
- 7.Peracchi A, Mozzarelli A, Rossi GL. Monovalent cations affect dynamic and functional properties of the tryptophan synthase α2β2 complex. Biochemistry. 1995;34:9459–9465. doi: 10.1021/bi00029a022. [DOI] [PubMed] [Google Scholar]
- 8.Woehl EU, Dunn MF. Monovalent metal ions play an essential role in catalysis and intersubunit communication in the tryptophan synthase bienzyme complex. Biochemistry. 1995;34:9466–9476. doi: 10.1021/bi00029a023. [DOI] [PubMed] [Google Scholar]
- 9.Woehl E, Dunn MF. Mechanisms of monovalent cation action in enzyme catalysis: the first stage of the tryptophan synthase β-reaction. Biochemistry. 1999;38:7118–7130. doi: 10.1021/bi982918x. [DOI] [PubMed] [Google Scholar]
- 10.Woehl E, Dunn MF. Mechanisms of monovalent cation action in enzyme catalysis: the tryptophan synthase α-, β-, and αβ-reactions. Biochemistry. 1999;38:7131–7141. doi: 10.1021/bi982919p. [DOI] [PubMed] [Google Scholar]
- 11.Fan YX, McPhie P, Miles EW. Guanidine hydrochloride exerts dual effects on the tryptophan synthase α2β2 complex as a cation activator and as a modulator of the active site conformation. Biochemistry. 1999;38:7881–7890. doi: 10.1021/bi990307e. [DOI] [PubMed] [Google Scholar]
- 12.Drewe WF, Jr, Dunn MF. Detection and identification of intermediates in the reaction of L-serine with Escherichia coli tryptophan synthase via rapid-scanning ultraviolet- visible spectroscopy. Biochemistry. 1985;24:3977–3987. doi: 10.1021/bi00336a027. [DOI] [PubMed] [Google Scholar]
- 13.Drewe WF, Jr, Dunn MF. Characterization of the reaction of L-serine and indole with Escherichia coli tryptophan synthase via rapid-scanning ultraviolet-visible spectroscopy. Biochemistry. 1986;25:2494–2501. doi: 10.1021/bi00357a032. [DOI] [PubMed] [Google Scholar]
- 14.Hur O, Niks D, Casino P, Dunn MF. Proton transfers in the β-reaction catalyzed by tryptophan synthase. Biochemistry. 2002;41:9991–10001. doi: 10.1021/bi025568u. [DOI] [PubMed] [Google Scholar]
- 15.Phillips RS, Miles EW, Holtermann G, Goody RS. Hydrostatic pressure affects the conformational equilibrium of Salmonella typhimurium tryptophan synthase. Biochemistry. 2005;44:7921–7928. doi: 10.1021/bi050056b. [DOI] [PubMed] [Google Scholar]
- 16.Osborne A, Teng Q, Miles EW, Phillips RS. Detection of open and closed conformations of tryptophan synthase by 15N-heteronuclear single-quantum coherence nuclear magnetic resonance of bound 1-15N-L-tryptophan. J. Biol. Chem. 2003;278:44083–44090. doi: 10.1074/jbc.M308276200. [DOI] [PubMed] [Google Scholar]
- 17.Roy M, Keblawi S, Dunn MF. Stereoelectronic control of bond formation in Escherichia coli tryptophan synthase: substrate specificity and enzymatic synthesis of the novel amino acid dihydroisotryptophan. Biochemistry. 1988;27:6698–6704. doi: 10.1021/bi00418a009. [DOI] [PubMed] [Google Scholar]
- 18.Harris RM, Dunn MF. Intermediate trapping via a conformational switch in the Na+-activated tryptophan synthase bienzyme complex. Biochemistry. 2002;41:9982–9990. doi: 10.1021/bi0255672. [DOI] [PubMed] [Google Scholar]
- 19.Harris RM, Ngo H, Dunn MF. Synergistic effects on escape of a ligand from the closed tryptophan synthase bienzyme complex. Biochemistry. 2005;44:16886–16895. doi: 10.1021/bi0516881. [DOI] [PubMed] [Google Scholar]
- 20.Barends TRM, Domratcheva T, Kulik V, Blumenstein L, Niks D, Dunn MF, Schlichting I. Structure and mechanistic implications of a tryptophan synthase quinonoid intermediate. ChemBiochem. 2008;9:261–266. doi: 10.1002/cbic.200700703. [DOI] [PubMed] [Google Scholar]
- 21.Ngo H, Harris R, Kimmich N, Casino P, Niks D, Blumenstein L, Barends TR, Kulik V, Weyand M, Schlichting I, Dunn MF. Synthesis and characterization of allosteric probes of substrate channeling in the tryptophan synthase bienzyme complex. Biochemistry. 2007;46:7713–7727. doi: 10.1021/bi700385f. [DOI] [PubMed] [Google Scholar]
- 22.Ngo H, Kimmich N, Harris R, Niks D, Blumenstein L, Kulik V, Barends TR, Schlichting I, Dunn MF. Allosteric regulation of substrate channeling in tryptophan synthase: modulation of the L-serine reaction in stage I of the β-reaction by α-site ligands. Biochemistry. 2007;46:7740–7753. doi: 10.1021/bi7003872. (2007). [DOI] [PubMed] [Google Scholar]
- 23.Suelter CH. Enzymes activated by monovalent cations. Science. 1970;168:789–795. doi: 10.1126/science.168.3933.789. [DOI] [PubMed] [Google Scholar]
- 24.Evans HJ, Sorger GJ. Role of mineral elements with emphasis on the univalent cations. Annu. Rev. Plant Physiol. 1966;17:47–76. [Google Scholar]
- 25.Woehl EU, Dunn MF. The roles of Na+ and K+ in pyridoxal phosphate enzyme catalysis. Coord. Chem. Rev. 1995;144:147–197. [Google Scholar]
- 26.Paige MJ, Di Cera E. Role of Na+ and K+ in Enzyme Function. Physiol. Rev. 2006;86:1049–1092. doi: 10.1152/physrev.00008.2006. 2006. [DOI] [PubMed] [Google Scholar]
- 27.Weber-Ban E, Hur O, Bagwell C, Banik U, Yang L-H, Miles EW, Dunn MF. Investigation of allosteric linkages in the regulation of tryptophan synthase: the roles of salt bridges and monovalent cations probed by site-directed mutation, optical spectroscopy, and kinetics. Biochemistry. 2001;40:3497–3511. doi: 10.1021/bi002690p. [DOI] [PubMed] [Google Scholar]
- 28.Yang L, Ahmed SA, Miles EW. PCR mutagenesis and overexpression of tryptophan synthase from Salmonella typhimurium: on the roles of β2 subunit Lys-382. Protein Expr. Purif. 1996;8:126–136. doi: 10.1006/prep.1996.0082. [DOI] [PubMed] [Google Scholar]
- 29.Riddles PW, Blakeley RL, Zerner B. Ellman's reagent: 5,5'-dithiobis(2-nitrobenzoic acid)--a reexamination. Anal. Biochem. 1979;94:75–81. doi: 10.1016/0003-2697(79)90792-9. [DOI] [PubMed] [Google Scholar]
- 30.Milescu L. Informatics Laboratory, National Insititute of Biotechnology, P.O. Box 60-45. Romania: Bucharest; copyright 1994, 1998. [Google Scholar]
- 31.Drewe WF, Jr, Dunn MF. Characterization of the reaction of L-serine and indole with Escherichia coli tryptophan synthase via rapid-scanning ultraviolet-visible spectroscopy. Biochemistry. 1986;25:2494–2501. doi: 10.1021/bi00357a032. [DOI] [PubMed] [Google Scholar]
- 32.Brzovic PS, Sawa Y, Hyde CC, Miles EW, Dunn MF. Evidence that mutations in a loop region of the α-subunit inhibit the transition from an open to a closed conformation in the tryptophan synthase bienzyme complex. J. Biol. Chem. 1992;267:13028–13038. [PubMed] [Google Scholar]
- 33.Dunn MF, Aguilar V, Brzovic P, Drewe WF, Jr, Houben KF, Leja CA, Roy M. The tryptophan synthase bienzyme complex transfers indole between the α- and β-sites via a 25–30 Å long tunnel. Biochemistry. 1990;29:8598–8607. doi: 10.1021/bi00489a015. [DOI] [PubMed] [Google Scholar]
- 34.Ferrari D, Yang LH, Miles EW, Dunn MF. beta D305A mutant of tryptophan synthase shows strongly perturbed allosteric regulation and substrate specificity. Biochemistry. 2001;40:7421–7432. doi: 10.1021/bi002892l. [DOI] [PubMed] [Google Scholar]
- 35.Pan P, Dunn MF. β-site covalent reactions trigger transitions between open and closed conformations of the tryptophan synthase bienzyme complex. Biochemistry. 1996;35:5002–5013. doi: 10.1021/bi960033k. [DOI] [PubMed] [Google Scholar]
- 36.Houben KF, Kadima W, Roy M, Dunn MF. L-serine analogues form Schiff base and quinonoidal intermediates with Escherichia coli tryptophan synthase. Biochemistry. 1989;28:4140–4147. doi: 10.1021/bi00436a003. [DOI] [PubMed] [Google Scholar]
- 37.Houben KF, Dunn MF. Allosteric effects acting over a distance of 20–25 A in the Escherichia coli tryptophan synthase bienzyme complex increase ligand affinity and cause redistribution of covalent intermediates. Biochemistry. 1990;29:2421–2429. doi: 10.1021/bi00461a028. [DOI] [PubMed] [Google Scholar]
- 38.Rhee S, Parris KD, Hyde CC, Ahmed SA, Miles EW, Davies DR. Crystal structures of a mutant (βK87T) tryptophan synthase α2β2 complex with ligands bound to the active sites of the α- and β-subunits reveal ligand-induced conformational changes. Biochemistry. 1997;36:7664–7680. doi: 10.1021/bi9700429. [DOI] [PubMed] [Google Scholar]
- 39.Blumenstein L, Domratcheva T, Niks D, Ngo H, Seidel R, Dunn MF, Schlichting I. βQ114N and βT110V Mutations reveal a critically important role of the substrate carboxylate site in the reaction specificity of tryptophan synthase. Biochemistry. 2007;46:14100–14116. doi: 10.1021/bi7008568. [DOI] [PubMed] [Google Scholar]
- 40.Kulik V, Weyand M, Seidel R, Niks D, Arac D, Dunn MF, Schlichting I. On the role of αThr183 in the allosteric regulation and catalytic mechanism of tryptophan synthase. J. Mol. Biol. 2002;324:677–690. doi: 10.1016/s0022-2836(02)01109-9. [DOI] [PubMed] [Google Scholar]
- 41.Ferrari D, Niks D, Yang L-H, Miles EW, Dunn MF. Allosteric communication in the tryptophan synthase bienzyme complex: Roles of the β-subunit aspartate 305-arginine 141 salt bridge. Biochemistry. 2003;42:7807–7818. doi: 10.1021/bi034291a. [DOI] [PubMed] [Google Scholar]
- 42.Pan P, Woehl E, Dunn MF. Protein architecture, dynamics and allostery in tryptophan synthase channeling. Trends Biochem. Sci. 1997;22:22–27. doi: 10.1016/s0968-0004(96)10066-9. [DOI] [PubMed] [Google Scholar]
- 43.Faeder EJ, Hammes GG. Kinetic studies of tryptophan synthase. Interaction of L-serine, indole, and tryptophan with the native enzyme. Biochemistry. 1971;10:1041–1045. doi: 10.1021/bi00782a016. [DOI] [PubMed] [Google Scholar]
- 44.York S. Kinetic spectroscopic studies of substrate and subunit interactions of tryptophan synthase. Biochemistry. 1972;11:2733–2740. doi: 10.1021/bi00764a029. [DOI] [PubMed] [Google Scholar]
- 45.Lane AN, Kirschner K. The catalytic mechanism of tryptophan synthase from Escherichia coli. Kinetics of the reaction of indole with the enzyme--L-serine complexes. European Journal of Biochemistry. 1983;129:571–582. doi: 10.1111/j.1432-1033.1983.tb07087.x. [DOI] [PubMed] [Google Scholar]
- 46.Lane AN, Kirschner K. The Mechanism of binding of L-serine to tryptophan synthase from Escherichia coli. European Journal of Biochemistry. 1983;129:561–570. doi: 10.1111/j.1432-1033.1983.tb07086.x. [DOI] [PubMed] [Google Scholar]
- 47.Leja CA, Woehl EU, Dunn MF. Allosteric linkages between β-site covalent transformations and α-site activation and deactivation in the tryptophan synthase bienzyme complex. Biochemistry. 1995;34:6552–6561. doi: 10.1021/bi00019a037. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.