Abstract
Zinc-finger nucleases (ZFNs) are versatile reagents that have redefined genome engineering. Realizing the full potential of this technology requires the development of safe and effective methods for delivering ZFNs into cells. We demonstrate the intrinsic cell-penetrating capabilities of the standard ZFN architecture and show that direct delivery of ZFNs as proteins leads to efficient endogenous gene disruption in a variety of mammalian cell types with minimal off-target effects.
Zinc-finger nucleases (ZFNs) are fusions of the non-specific cleavage domain from the FokI restriction endonuclease with custom-designed Cys2-His2 zinc-finger proteins (ZFPs)1. These chimeric nucleases induce sequence-specific DNA double-strand breaks (DSBs) that can be repaired by error-prone non-homologous end joining (NHEJ) to yield small alterations at targeted genomic loci. This strategy has enabled highly efficient gene disruption in numerous cell types2, 3 and model organisms4, 5 and has facilitated the progress of targeted gene therapy in humans6, 7. Despite these advances and more recent methodological improvements8–10, there remains a need for new methods that can improve the utility of these enzymes. The development of safe and effective ZFN delivery methods is of particular importance, as many viral and non-viral ZFN gene delivery systems may hinder the continued advancement of this technology. In particular, viral vectors11 are time-consuming to produce and can be associated with undesirable side-effects, such as insertional mutagenesis, while non-viral DNA and mRNA delivery systems are restricted to certain cell types and have been reported to show toxicity12, 13 and low efficiency14. To address this problem, we set out to develop a simple alternative to conventional ZFN delivery systems by investigating the direct delivery of purified ZFN proteins to cells.
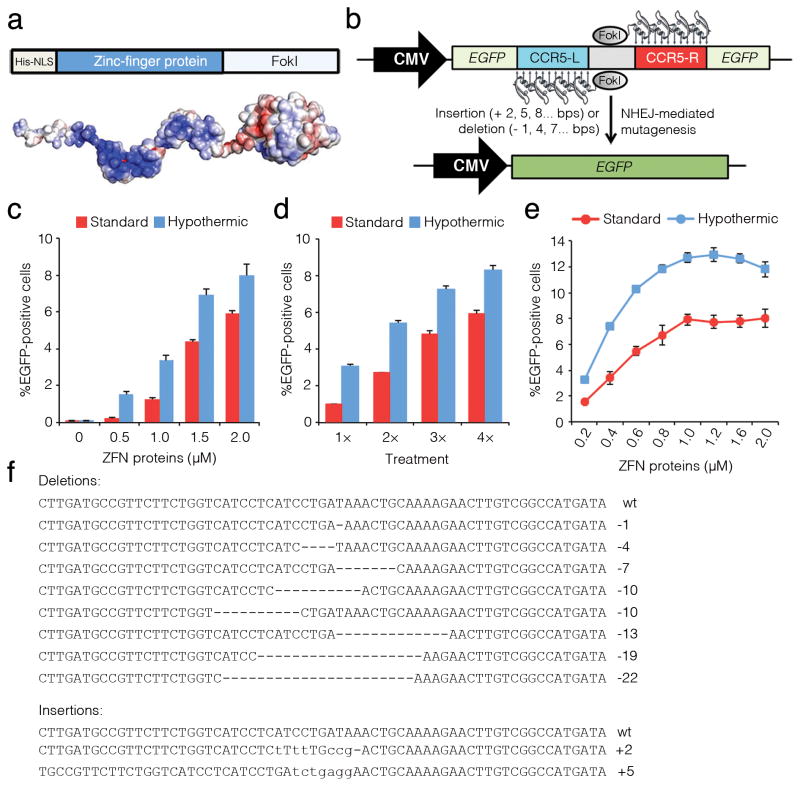
We began by introducing protein transduction domains into the established ZFN architecture. For this, the cell-penetrating peptide sequence from the HIV-1 TAT protein or a polyarginine motif was genetically fused to the N-termini of ZFNs designed to target the human CCR5 gene6. These ZFNs, however, were consistently difficult to express or purify in quantities sufficient for analysis in cell culture (data not shown). Following these results, and based on the observation that ZFP-DNA binding domains carry a net positive charge (Fig. 1a), we hypothesized that ZFNs might penetrate the cell in the absence of additional modification. ZFNs designed to target the CCR5 gene and lacking any transduction domain were expressed in Escherichia coli and purified to homogeneity from either the soluble or the insoluble fractions (Supplementary Fig. 1). In vitro analysis confirmed that functional ZFN proteins with similar DNA cleavage profiles could be obtained by either method (Supplementary Fig. 2 and Supplementary Note).
Figure 1. ZFN proteins are cell permeable and induce targeted mutagenesis in human cells.
(a) Diagram of ZFN protein and electrostatic potential of the ZFN protein surface colored from dark red (− 5 kT/e) to white (0 kT/e) to dark blue (+ 5 kT/e). (b) Schematic representation of the fluorescence reporter system used to evaluate cellular penetration of ZFN proteins. (c–e) Percentage of EGFP-positive cells as determined by flow cytometry in reporter cells treated with increasing amounts of ZFN proteins (c), subjected to consecutive treatments with 0.2 μM ZFN proteins (d), or subjected to three consecutive treatments with increasing amounts of ZFN proteins (e), in all cases using either standard or transient hypothermic conditions. Error bars indicate s.d. (n = 3). (f) Representative sequence analysis of the EGFP locus from isolated EGFP-positive cells. Multiple deletions (dashes) and insertions (lowercase) induced by NHEJ repair are aligned to the cleavage site (wt).
To determine the ability of ZFN proteins to penetrate cells and stimulate mutagenesis, we generated a fluorescence-based reporter system to measure ZFN-induced DSBs (Fig. 1b). This system uses an integrated enhanced green fluorescent protein (EGFP) gene whose expression has been interrupted by a frameshift mutation introduced by a strategically placed ZFN cleavage site. ZFN proteins that penetrate reporter cells can induce DSBs at this target site and drive the introduction of small insertions and deletions in the EGFP locus by NHEJ. Because NHEJ is a stochastic process, approximately one-third of these mutational events (+ 2, 5, 8 …bps or − 1, 4, 7 …bps) will restore the frame and EGFP function.
Direct application of ZFN proteins to reporter cells resulted in a dose-dependent increase in EGFP fluorescence, with maximum activity (6% EGFP-positive cells) achieved after treatment with 2 μM ZFN proteins (Fig. 1c). By comparison, transient transfection of ZFN expression plasmids under saturating conditions resulted in ~7% EGFP-positive cells (Supplementary Fig. 3). We observed no difference in activity between ZFN proteins purified from the soluble fraction or inclusion bodies (Supplementary Fig. 4). At all ZFN concentrations evaluated, the use of transient hypothermic culture conditions9 enhanced the efficiency of mutagenesis nearly twofold (Fig. 1c,d). Extended periods of incubation (>60 min) did not increase the frequency of genome editing (Supplementary Fig. 5). Consecutive protein treatments, however, did increase the percentage of EGFP-positive cells (Fig. 1d,e). Notably, repeated treatment with ZFN proteins over three days using transient hypothermic conditions yielded ~12% EGFP-positive cells (Fig. 1e). Sequence analysis of isolated EGFP-positive cells verified targeted mutagenesis, revealing the presence of the anticipated ZFN-induced insertions and deletions in the EGFP locus (Fig. 1f).
To determine the contribution of each ZFN component to cellular penetration, we incubated cells with fluorescently labeled ZFN or FokI cleavage domain proteins (Supplementary Fig. 6). Fluorescence was observed in cell lysate following treatment with ZFN – in the presence or absence of a nuclear localization sequence – but not with FokI cleavage domain, suggesting that zinc-finger domains facilitate cellular internalization.
We evaluated the efficacy of this approach for the disruption of endogenous genes by treating human embryonic kidney (HEK) 293 and human acute monocytic leukemia (THP1) cell lines, as well as primary adult human dermal fibroblast (HDF) and primary CD4+ T cells with ZFN proteins targeting the CCR5 gene. These ZFNs utilized the high-activity Sharkey cleavage domain10. Analysis of DNA isolated from each cell type with the Surveyor nuclease assay revealed efficient and dose-dependent endogenous CCR5 gene disruption (Fig. 2a). HEK293 and HDF cells subjected to three consecutive treatments with 2 μM ZFN proteins exhibited gene disruption frequencies >24%, while CD4+ cells subjected to three consecutive treatments with 0.5 μM ZFN proteins exhibited gene disruption frequencies >8%. As observed in the reporter system, the frequency of gene disruption increased with repeated protein treatments (Supplementary Fig. 7). Sequence analysis of cloned CCR5 alleles amplified from each treated cell type confirmed the presence of ZFN-induced insertions and deletions in the CCR5 gene (Supplementary Fig. 8).
Figure 2. Modification of endogenous human genes by direct delivery of ZFN proteins.
(a) Frequency of endogenous CCR5 gene disruption in HEK293, THP1, HDF and CD4+ T cells subjected to three consecutive treatments with ZFN proteins using transient hypothermic conditions, as determined by the Surveyor nuclease assay. Black arrow indicates specific cleavage product. (b) DHFR production measured by fluorescein-methotrexate binding in CHO cells subjected to three consecutive treatments with ZFN proteins using standard or transient hypothermic conditions. Mock treated CHO cells exhibited approximately 1% reduction in functional DHFR (not shown). (c) Proliferation of HEK293, HDF and THP1 cells treated with ZFN proteins targeting the CCR5 gene and containing Sharkey mutations. (d) Proliferation of CHO cells treated with ZFN proteins targeting the DHFR gene and utilizing a variety of specialized cleavage domains. Values normalized to mock treated cells. Error bars indicate s.d. (n = 3).
To investigate the cleavage specificity of ZFNs using this approach, we evaluated the activity of the CCR5 ZFN proteins against nine previously described6, 15 off-target cleavage sites in HEK293 cells (Supplementary Fig. 9). In direct comparison to Lipofectamine-mediated transient transfection of ZFN expression plasmids, we found that cells subjected to consecutive protein treatments demonstrated a marked decrease in ZFN activity at every off-target site, including the CCR2 locus. Notably, no detectable ZFN activity was observed at three of these loci. Western blot analysis revealed complete degradation of delivered ZFN proteins less than 4 h after application, while cells transfected with ZFN expression plasmids produced high-levels of protein continuously from 16 to 72 h post-transfection (Supplementary Fig. 10), indicating that the differences in cleavage specificity could be attributed to the short half-lives of transduced ZFN proteins and that limiting the duration of ZFN exposure inside cells is a viable method for minimizing toxicity16. Consistent with these degradation kinetics, cells treated with ZFN proteins exhibited maximum activity at 8 h, whereas cells expressing ZFNs from plasmid DNA showed maximum activity at 48 h (Supplementary Fig. 10).
In order to examine the breadth of this technique, we treated Chinese hamster ovary (CHO) cells with ZFN proteins designed to target the DHFR gene2. These ZFNs utilized various specialized cleavage domains, including Sharkey and the evolutionarily optimized DS/RR obligate heterodimeric architecture10. Reduced levels of functional DHFR protein, as determined by fluorescein-labeled methotrexate-based flow cytometry analysis, were observed in CHO cells following three consecutive treatments with DHFR ZFN proteins (Fig. 2b). Notably, CHO cells incubated with ZFNs containing Sharkey mutations exhibited a >12% reduction in functional DHFR. Sequence analysis of cloned DHFR alleles amplified from cells treated with ZFN proteins validated these percentages and confirmed the presence of ZFN-induced insertions and deletions in the DHFR gene (Supplementary Fig. 11). Examination of DHFR protein levels in expanded clonal populations indicated biallelic DHFR gene disruption frequencies >7% (Supplementary Fig. 12), demonstrating that constitutive ZFN expression from plasmid DNA is not required for high-frequency biallelic modifications and instead can be achieved using directly applied ZFN proteins.
We observed no appreciable toxicity in HEK293 or HDF cells treated with ZFN proteins (Fig. 2c). Toxicity was also not detected in CHO cells incubated with ZFN proteins containing either the wild-type cleavage domain or the DS/RR architecture (Fig. 2d). However, we measured decreased proliferation in CHO and THP1 suspension cells incubated with >1 μM ZFN proteins containing Sharkey mutations (Fig. 2c,d). Toxicity was also observed, qualitatively, in CD4+ cells subjected to consecutive treatments with >1 μM ZFN proteins, suggesting that sensitive cell types may require protein to be administered in consecutive low doses to minimize potential toxic effects.
We have demonstrated the intrinsic cell-penetrating capabilities of the standard ZFN architecture and have shown that direct delivery of ZFNs as proteins can be used to disrupt the expression of endogenous genes in a variety of mammalian cell types, including primary CD4+ T cells and primary adult human dermal fibroblasts, which are frequently used to generate induced pluripotent stem cells. In contrast to methods that require ZFN expression from DNA, ZFN protein delivery leads to comparatively fewer off-target cleavage events and does not carry the risk of insertional mutagenesis, making this method suitable for genome editing applications in which minimizing cellular toxicity or maintaining genetic integrity is of particular importance, such as the in vitro modeling of human diseases and the ex vivo modification of non-transformed human cell types. We show that this method can also be used to modify difficult-to-transfect cell types, including patient-derived leukemia cell lines and primary human lymphocytes, supporting the use of this technique in place of viral-mediated gene delivery for inducing gene knockouts in cultured cells for reverse genetics and drug discovery. As methods for engineering cell-permeability into proteins improve, we anticipate that protein delivery and the benefits afforded therein will be extended to other designer nucleases, including TALENs.
ONLINE METHODS
Plasmid construction
The left (L) and right (R) zinc-finger nuclease (ZFN) proteins designed to target the human C-C chemokine receptor type 5 (CCR5)6 and Chinese hamster dihydrofolate reductase (DHFR)2 genes were PCR amplified from their respective mammalian expression vectors (previously constructed in our laboratory) pVAX1-NH.CCR5.L/R.FN and pVAX1-NH.DHFR.L/R.FN with the primers 5′ NdeI-NLS and 3′ FN-SacI-SalI. All primer sequences are provided in Supplementary Table 1. PCR products were digested with NdeI and SalI and ligated into the NdeI and XhoI restriction sites of the pET-28 (+) expression vector (Novagen) to generate the plasmids pET.CCR5.L/R.FN and pET.DHFR.L/R.FN. Specialized FokI cleavage domain variants, including the high-activity Sharkey cleavage domain and the evolutionarily optimized obligate heterodimer architecture DS and RR, were isolated from previously described pPDAZ templates10 and cloned into pET.CCR5.L/R.FN and pET.DHFR.L/R.FN with the restriction sites BamH1 and SacI to generate pET.CCR5.L.R/Sh, pET.DHFR.L/R.Sh, pET.DHFR.L/R.DS/RR and pET.DHFR.L/R.Sh.DS/RR (L: Left ZFN; R: Right ZFN; FN: FokI cleavage domain; Sh: Sharkey cleavage domain; DS/RR: DS or RR architecture; Sh.DS/RR: Sharkey cleavage domain with DS or RR architecture). Correct construction of each ZFN expression cassette was verified by sequence analysis (Supplementary Table 2).
ZFN protein expression and purification
ZFN expression plasmids were transformed into chemically competent E. coli BL21 (DE3) pGro7 cells (Stratagene). A single colony was added to 10 ml LB medium in the presence of 50 μg ml−1 kanamycin, 100 μg ml−1 chloramphenicol and 1% glucose. Bacteria were grown overnight at 37°C with shaking. The following day, 500 ml LB medium supplemented with 100 μM ZnCl2, 50 μg ml−1 kanamycin, 100 μg ml−1 chloramphenicol and 0.2% glucose was inoculated with 5 ml of the overnight culture and incubated at 30°C with shaking. The culture was grown to an OD600 of 0.4 and then incubated at room temperature with shaking until the OD600 reached 0.8. Protein synthesis was then induced with 0.1 mM isopropyl β-D-1-thiogalactopyranoside (IPTG). After 4 h, cells were harvested by centrifugation at 2,000 rcf for 20 min at 4°C and the pellet was resuspended in native lysis buffer (50 mM Tris-HCl, 500 mM NaCl, 100 μM ZnCl2, 1 mM PMSF, 1 mM MgCl2, 1 mM β-mercaptoethanol, 10 mM imidazole, 0.2% Triton X-100 and 10% glycerol, pH 8.0). Cells were lysed using standard sonication conditions and the soluble fraction was isolated by centrifugation at 18,000 rcf for 30 min at 4°C. ZFN proteins were purified using Ni-NTA agarose resin (QIAGEN) and eluted with 500 mM imidazole in the presence of 50 mM Tris-HCl, 500 mM NaCl, 100 μM ZnCl2 and 10% glycerol at pH 8.0. Each protein was concentrated and dialyzed against storage buffer (20 mM HEPES, 90 μM ZnCl2, 150 mM NaCl, 5 mM KCl, 1 mM MgCl2, pH 7.4) and assessed for purity by SDS-PAGE. The protein yield after purification was determined to be between 0.5–1.0 mg l−1.
Inclusion body purification
ZFN proteins were also purified from inclusion bodies using an alternative method. Following the inoculation of 500 ml LB medium with 5 ml of the overnight culture, cells were incubated at 37°C with shaking until an OD600 of 0.4 was reached. Cells were then incubated at room temperature with shaking until the OD600 reached 0.9. Protein synthesis was induced by addition of 0.4 mM IPTG and 4 mM arabinose. After 16 h, cells were harvested by centrifugation at 2,000 rcf for 20 min at 4°C, and the pellet was resuspended in lysis buffer (50 mM Tris-HCl, 150 mM NaCl, 0.1% Triton X-100, pH 8.0). Cells were lysed using standard sonication conditions and centrifuged at 10,000 rcf for 30 min at 4°C. Inclusion bodies were then washed with lysis buffer and centrifuged at 10,000 rcf for 30 min at 4°C. This process was repeated twice, followed by a procedure designed to enhance protein yield and purity: inclusion bodies were: (i) washed with cold water, (ii) centrifuged at 10,000 rcf for 30 min at 4°C, (iii) resuspended in PBS, (iv) centrifuged as in step 2, (v) incubated with solubilization buffer (PBS, 500 mM L-Arg, 90 μM ZnCl2, 1 mM MgCl2, pH 7.4) for 3 hr at 4°C and (vi) centrifuged as in step 2. L-Arg was included to facilitate refolding of recombinant proteins from inclusion bodies17. Following this procedure, the inclusion body supernatant was isolated and protein purity was assessed by SDS-PAGE. The protein yield after purification was determined to be between 3 – 5 mg l−1.
In vitro cleavage analysis
Cleavage analysis was performed as previously described10.
Reporter system construction and analysis
The EGFP reporter systems used in this study were generated using the Flp-In System (Invitrogen) and constructed as previously described10 with the following exception: The CCR5 ZFN cleavage site was amplified with the overlapping oligonucleotides 5′ midEGFP-CCR5 and 3′ midEGFP-CCR5. Reporter cells were analyzed by flow cytometry as previously described10.
Cell culture
Human embryonic kidney (HEK) 293, human acute monocytic leukemia (THP1), primary adult human dermal fibroblast (HDF; Invitrogen) and Chinese hamster ovary (CHO) cells were seeded onto 24-well plates at a density of 1 × 105 cells per well and established in a humidified 5% CO2 atmosphere at 37°C. Peripheral blood mononuclear cells (PBMCs) were obtained from anonymous healthy blood donors through the Scripps Research Institute Normal Blood Donor Program. Primary CD4+ T cells were purified from PBMCs using the RosetteSep Human CD4+ T Cell Enrichment Cocktail (Stem Cell Technologies) according to the manufacturer’s instructions. CD4+ cells were seeded onto 96-well plates at a density of 1 × 105 cells per well and established in a humidified 5% CO2 atmosphere at 37°C. HEK293 and HDF cells were maintained in DMEM containing 10% (v/v) FBS and 1% Antibiotic-Antimycotic (Anti-Anti; Gibco). THP1, CHO and CD4+ cells were maintained in RPMI containing 10% (v/v) FBS and 1% (v/v) Anti-Anti. CHO cells were maintained with HT supplement (100 μM sodium hypoxanthine and 160 μM thymidine; Invitrogen) after treatment with ZFN proteins.
ZFN protein treatments
ZFN proteins were prepared for treatment as follows: ZFNs purified from the soluble fraction were diluted into serum-free medium containing 90 μM ZnCl2 at pH 7.4. ZFNs purified from inclusion bodies were diluted into serum-free medium containing 100 mM L-Arg and 90 μM ZnCl2 at pH 7.4. Twenty-four h after seeding, cells were washed with serum-free medium and treated with ZFN proteins for 1 h at 37°C. After treatment, cells were washed and maintained at 37°C with serum-containing medium for 24 h before the next treatment or up to three days before cells were harvested for analysis. This process was repeated up to four times over four consecutive days.
Transient hypothermic conditions were achieved as follows: Twenty-four h after seeding, cells were washed with serum-free medium and treated with ZFN proteins for 1 h at 37°C. After treatment, cells were washed and incubated at 30°C with serum-containing medium for 6–24 h. Cells were then returned to and maintained at 37°C for the remaining 24 h before the next treatment or up to three days before cells were harvested for analysis. This process was repeated up to four times over four consecutive days.
Internalization of fluorescently labeled proteins
Purified ZFN and FokI cleavage domain proteins were labeled with fluorescein-5-maleimide (Invitrogen) under conditions specified by the manufacturer. Following conjugation, 2 μM ZFN or FokI cleavage domain proteins were dialyzed against 20 mM HEPES and 90 μM ZnCl2 at pH 7.4 and then incubated with HEK293 cells for 1 h. Cells were then washed three times with 20 mM HEPES, 90 μM ZnCl2 and 1 mg ml−1 heparin at pH 7.4 to remove surface-bound ZFN proteins. Cells were lysed in SDS-PAGE loading buffer (0.2 M Tris-HCl, 10% (v/v) sodium dodecyl sulfate, 10 mM β-mercaptoethanol, 0.05% (w/v) bromophenolblue, 20% (v/v) glycerol, pH 6.8). Relative quantities of internalized protein were determined by fluorescent illumination of SDS-PAGE gels using the GelDoc XR Imaging System (Bio-Rad).
Surveyor nuclease assay
Genomic DNA was extracted with Quick Extract DNA Extraction Solution (Epicentre). The CCR5 gene was amplified by nested PCR with the following primer pairs: 5′ hCCR5 External with 3′ hCCR5 External, and 5′ hCCR5 Internal with 3′ hCCR5 Internal. Following PCR amplification of the CCR5 locus using the Expand High Fidelity Taq System (Roche), the Surveyor mutation detection kit was used according to the manufacturer’s instructions (Transgenomics). Cleavage products were visualized by PAGE and the frequency of gene disruption was determined by measuring the ratio of cleaved to uncleaved substrate, as described18.
Western blot analysis
Western blot analysis was performed as previously described19 with the following exception: The internal loading control was β-actin detected with peroxidase-conjugated anti-β-actin antibody.
Cellular proliferation assay
HEK293, HDF, THP and CHO cells were seeded onto 96-well plates at a density of 1 × 105 cells per well. Twenty-four h after seeding, cells were treated with ZFN proteins as described above. Decreased cellular proliferation in response to ZFN protein was measured using the Cell Proliferation Kit II (XTT; Roche Applied Science) according to the manufacturer’s instructions.
Fluorescein methotrexate assay
The fluorescein methotrexate assay was performed as described2, 20.
Clonal analysis of CHO cells
Sixteen days after ZFN treatment, CHO cells were seeded onto 96-well plates by limiting dilution with 10% FBS-RPMI, 1% Anti-Anti and HT supplement. Each well was visually inspected to ensure single colony formation. The fluorescein methotrexate flow cytometry assay was used to determine the level of functional DHFR protein. Genomic DNA from clones determined to be DHFR−/− was isolated using the QIAmp DNA MiniKit (QIAGEN). The DHFR alleles were PCR amplified with the primers 5′ DHFR BssHII and 3′ DHFR EcoRI. PCR products were ligated into the MluI and EcoRI restriction sites of the plasmid pcDNA 3.1 (Invitrogen). Sequence analysis of transformants was used to confirm biallelic gene modification.
Sequence analysis
Genomic DNA from EGFP-positive reporter cells (isolated by FACS; FACScan Dual Laser Flow Cytometer, BD Biosciences) was purified with the QIAmp DNA MiniKit (QIAGEN) and the EGFP locus was PCR amplified with the primers 5′ EGFP 443–466 BamH1 and 3′ EGFP 497–520 EcoRI. Genomic DNA from ZFN-treated HEK293, HDF, THP1 and CD4+ cells was isolated with Quick Extract DNA Extraction Solution (Epicentre). The CCR5 locus was PCR amplified with the primers 5′ hCCR5 BamH1 and 3′ hCCR5 EcoRI. EGFP and CCR5 PCR products were cloned into the plasmid pUC19 with the restriction sites EcoRI and BamH1. Sequence analysis was performed on individual cloned transformants.
Supplementary Material
Acknowledgments
We thank C. Gersbach for contributing to preliminary studies, P. Ikrenyi for technical assistance and A. Mercer for discussion of the manuscript. This research was supported by the National Institute of Health (NIH) grants GM065059, DP1OD006990, and The Skaggs Institute for Chemical Biology. T.G. was supported by a National Institute of General Medicine Sciences fellowship (T32GM080209). Y.K. was supported by the Japan Society for the Promotion of Science Research Fellowships for Young Scientists.
Footnotes
AUTHOR CONTRIBUTIONS
T.G., J.G., Y.K., S.J.S and C.F.B. designed research; J.G. and Y.K. purified ZFN proteins; T.G., J.G., Y.K., and S.J.S. performed experiments; T.G., J.G., Y.K., S.J.S and C.F.B. analyzed data; T.G., S.J.S and C.F.B. wrote the manuscript.
COMPETING FINALCIAL INTERESTS
The authors declare competing financial interests: details are available in the online version of the paper.
References
- 1.Kim YG, Chandrasegaran S. Proc Natl Acad Sci U S A. 1994;91:883–887. doi: 10.1073/pnas.91.3.883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Santiago Y, et al. Proc Natl Acad Sci U S A. 2008;105:5809–5814. doi: 10.1073/pnas.0800940105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hockemeyer D, et al. Nat Biotechnol. 2009;27:851–857. doi: 10.1038/nbt.1562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Doyon Y, et al. Nat Biotechnol. 2008;26:702–708. doi: 10.1038/nbt1409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Geurts AM, et al. Science. 2009;325:433. [Google Scholar]
- 6.Perez EE, et al. Nat Biotechnol. 2008;26:808–816. doi: 10.1038/nbt1410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Holt N, et al. Nat Biotechnol. 2010;28:839–847. doi: 10.1038/nbt.1663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sander JD, et al. Nat Methods. 2011;8:67–69. doi: 10.1038/nmeth.1542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Doyon Y, et al. Nat Methods. 2010;7:459–460. doi: 10.1038/nmeth.1456. [DOI] [PubMed] [Google Scholar]
- 10.Guo J, Gaj T, Barbas CF., 3rd J Mol Biol. 2010;400:96–107. doi: 10.1016/j.jmb.2010.04.060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Thomas CE, Ehrhardt A, Kay MA. Nat Rev Genet. 2003;4:346–358. doi: 10.1038/nrg1066. [DOI] [PubMed] [Google Scholar]
- 12.Warren L, et al. Cell Stem Cell. 2010;7:618–630. doi: 10.1016/j.stem.2010.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Diebold SS, et al. Science. 2004;303:1529–1531. doi: 10.1126/science.1093616. [DOI] [PubMed] [Google Scholar]
- 14.Van Tendeloo VF, et al. Gene Ther. 2000;7:1431–1437. doi: 10.1038/sj.gt.3301252. [DOI] [PubMed] [Google Scholar]
- 15.Gabriel R, et al. Nat Biotechnol. 2011;29:816–823. doi: 10.1038/nbt.1948. [DOI] [PubMed] [Google Scholar]
- 16.Pruett-Miller SM, Reading DW, Porter SN, Porteus MH. PLoS Genet. 2009;5:e1000376. doi: 10.1371/journal.pgen.1000376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Golovanov AP, Hautbergue GM, Wilson SA, Lian LY. J Am Chem Soc. 2004;126:8933–8939. doi: 10.1021/ja049297h. [DOI] [PubMed] [Google Scholar]
- 18.Guschin DY, et al. Methods Mol Biol. 2010;649:247–256. doi: 10.1007/978-1-60761-753-2_15. [DOI] [PubMed] [Google Scholar]
- 19.Gordley RM, Gersbach CA, Barbas CF., 3rd Proc Natl Acad Sci U S A. 2009;106:5053–5058. doi: 10.1073/pnas.0812502106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gaudray P, Trotter J, Wahl GM. J Biol Chem. 1986;261:6285–6292. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.