Abstract
Dengue virus infection causes significant morbidity and mortality in humans world-wide. The Aedes aegypti mosquito is the major vector that spread dengue virus to humans. Interaction between dengue viruses and A. aegypti is a multi-factorial phenomena that is determined by both virus and mosquito genotypes. Although, studies have suggested significant association of mosquito vectorial capacity with population variation of dengue virus, specifications of the vector factors that may influence vector-virus compatibility have are very limited in the literature. Recently, we have shown that a large number of genes are differentially expressed between MOYO-S (susceptible) and MOYO-R (refractory) A. aegypti strains upon infection with dengue virus (JAM-1409 genotype). In the current study, we show that specific intrinsic features of A. aegypti genes are significantly associated with `responsiveness' of mosquito genes to dengue infection. Binomial logistic regression analysis further reveals differential marginal effects of these features on gene responsiveness of mosquito to the viral infection. Thus, our result shows that intrinsic features of genes significantly affect differential expression of A. aegypti genes to dengue infection. The information will benefit further investigation on evolution of genes among natural populations A. aegypti conferring differential susceptibility to dengue virus.
Keywords: Aedes aegypti, vector-virus interaction, logit, gene context, codon bias, intron, co-evolution
1. Introduction
The mosquito Aedes aegypti is the principal vector of dengue virus (DENV). DENV is a rapidly re-emerging arbovirus (Kyle and Harris 2008, Phillips 2008). According to the World Health Organization, about 2.5 billion people are at risk of contacting the virus world-wide. There is no effective vaccine available for dengue treatment at the moment and hence mosquito control remains the most used strategy for disease prevention. Understanding the basic mechanism of how the mosquito acts as sustainable carrier of DENV is important for designing control strategies against dengue transmission.
Infection of dengue virus in A. aegypti mosquitoes is regulated by activation of either permissive or defensive responses in the mosquito which is determined by the genotypes of mosquito as well as the infecting virus (Rico-Hesse 2007). After the virus enters the mosquito midgut upon feeding on viremic human, presumably a robust interaction between mosquito and virus occurs that is instrumental in determining the success or failure of establishing an infection in epithelium cells (Halstead 2008). The intrinsic ability of A. aegypti to allow the virus to complete life cycle is generally referred to as `vector competence'. Although considerable genetic variability has been observed among A. aegypti populations (Gubler et al. 1979; Rosen et al. 1985; Tardieux et al. 1990; Diallo et al. 2008), direct association of genetic variation with vector competence of the mosquito to DENV infection is not known (Woodring et al. 1996; Halstead 2008). Several quantitative trait loci associated with midgut infection barrier (MIB), midgut escape barrier (MEB) and salivary gland barrier have been described in A. aegypti (Bennett et al. 2002). However, specific genes within these QTLs have not been characterized. However, studies do suggest that interaction bewteen mosquito and dengue virus is modulated by multiple factors associated with both the infecting virus and the mosquito genotype (Black et al. 2002, Anderson and Rico-Hesse 2006, Salazar et al. 2007). Recently, we performed a genome-wide transcriptome study to identify responsive genes of susceptible (MOYO-S) and refractory (MOYO-R) mosquito strains to dengue virus (JAM-1409 genotype) infection (Behura et al. 2011). The study revealed that a large number of mosquito genes (n = 2,454) were associated with significant changes of expression (`responsive genes') in response to the viral challenge. The differentially expressed genes were coordinately expressed in distinct modules to respond to the infection.
The primary objective of the current study is to investigate if the responsive and non-responsive genes identified from the microarray experiment (Behura et al. 2011) are significantly distinct in intrinsic features related to gene structure and evolution. The features were chosen based on literature evidences that suggest influential role of specific structural and evolutionary features in differential expression of genes (Korbel et al. 2004; Chiaromonte et al. 2003; Chen et al. 2005; Sapra et al. 2004; Buckingham 1990; Coleman et al. 2008). The A. aegypti genome annotation data (www.vectorbase.org) was exploited in the present study to identify these features of genes. Binomial logistic regression analyses were then used to determine effects of gene features on the differential expression of genes. The results obtained from this study suggest that intrinsic features of A. aegypti genes have significant effects on the transcriptional response of MOYO-S/ MOYO-R mosquitoes to dengue infection.
2. Material and Methods
2.1. Gene lists
The A. aegypti genes showing statistically significant transcriptional response to DENV infection (Behura et al. 2011) are analyzed in the current study. Each of these genes was either up-regulated or down-regulated significantly in response to infection by dengue (serotype 2, JAM-1409). The differential expression of genes was measured at two early post-infection times (3hr and 18hr) in two populations (MOYO-S) and (MOYO-R) of A. aegypti. MOYO-R is refractory to dengue infection whereas MOYO-S is susceptible. The list of significant genes, categorized as up-regulated or down-regulated genes at the two time points, is provided in Supplementary Table 1. The expression data is publicly available at the Gene Expression Omnibus database with the accession number GSE16563.
2.2. Comparison of gene features
The responsive and non-responsive genes were compared for six features: 1) gene context, 2) codon usage bias and 3) introns, 4) paralogs, 5) ancestral origin (orthologs present in related mosquito species) and 6) derived origin (specific to A. aegypti with no detectable ortholog in related mosquitoes).
Gene context refers to localization of a gene in physical proximity to another (Ciria et al. 2004, Martinez-Guerrero et al. 2008). Positional adjacency has been known to influence gene expressions in many species including fruit fly (Boutanaev et al. 2002; Spellman and Rubin 2002), C. elegans (Lercher et al. 2003), yeast (Cohen et al. 2000) as well as in humans (Elizondo et al. 2009). Thus, we wanted to know if gene context is an influential factor for regulating gene expression of mosquito to dengue infection. To know that, we identified gene contexts of responsive as well as non-responsive genes identified in MOYO-S/ MOYO-R to dengue infection (Behura et al. 2011). The intergenic distance between neighboring genes was determined from the start and end coordinates of the annotated genes from the genome sequences (Nene et al. 2007). A gene context was considered present if a gene was found within 1 kb of another gene. We chose 1 kb to identify gene context because our earlier work have shown that 1 kb is the upper limit in the distribution of distances between neighboring genes for which maximum number of genes in A. aegypti genome are closed spaced to each other (Behura SK and Severson DW, unpublished data). In fact, nearly one third of the annotated genes in A. aegypti genome are associated with intergenic distances less than 1 kb.
Furthermore, it is also well established that introns have significant role in gene expression (Hurst et al. 1996; Castillo-Davis et al. 2002; Eisenberg and Levanon 2003; Chen et al. 2005). Experimental evidences suggest that presence of intron sequences in genes enhances gene expression level significantly (Buchman and Berg 1988; Palmiter et al. 1991; Duncker et al. 1997; Le Hir et al. 2003). Thus, we wanted to know if intron presence/absence in A. aegypti genes has any association with differential gene expression in the MOYO-S/ MOYO-R strains to dengue infection. The intronless and intron-containing genes of responsive and non-responsive genes were determined from the number of exons predicted from the A. aegypti official genes. The predicted exons of A. aegypti official genes were obtained from Biomart (www.biomart.org) and were used to classify the annotated genes into two categories: 1) genes containing only one exon and 2) genes containing multiple exons. The former category represented intronless genes and the latter category represented intron-containing genes.
The role of codon usage bias in gene expression is well documented (reviewed in Hershberg and Petrov 2008). Generally, highly expressed genes are associated with high codon usage bias, a phenomenon that is also evident in insects including A. aegypti (Behura et al. 2010, Behura and Severson, 2011). Thus, we wanted to know if codon bias has a role in differential expression of genes of MOYO-S/ MOYO-R strains to dengue infection. To estimate the extent of codon bias of responsive and non-responsive genes, the CodonO software (Angellotti et al. 2007) was used to locally calculate synonymous codon usage order (SCUO) from the coding sequences of A. aegypti genes (downloaded from www.biomart.org). As SCUO values varies from 0 (no bias) to 1 (most bias), we categorized the responsive as well as the non-responsive genes as low biased genes if SCUO ≤ 0.5 and as high biased genes if SCUO > 0.5.
Links between gene expression and sequence evolution are also known (Pal et al. 2001, Subramanian and Kumar, 2004; Drummond et al. 2005). According to these studies genes that are less evolvable and hence remain highly conserved across related species are expressed at higher levels than poorly conserved genes throughout times and tissues. Also, genes that have paralogous copies in the genome tend to show more divergent spatial and temporal expressions than singleton genes (Gu et al. 2004, Gu et al. 2005). Thus, we wanted to know if different orthology and paralogy features of A. aegypti genes have association with differential expression of genes of MOYO-S/ MOYO-R strains to dengue infection. The paralogy relationships previously annotated for A. aegypti genes (downloaded from www.biomart.org) were used to classify the responsive and the non-responsive genes into two categories: 1) genes with paralog(s) and 2) genes without paralogs (singletons). The orthology relationships of A. aegypti genes with C. quinquefasciatus and A. gambiae genes were also obtained from www.biomart.org and were used to categorize the responsive and the non-responsive genes into 1) genes that have ortholog(s) in both C. quinquefasciatus and A. gambiae (ancestral genes) and 2) genes that have no ortholog in either C. quinquefasciatus or A. gambiae (derived genes).
2.3. Regression models
Binomial logistic regression models were developed to determine relationships of gene features with the observed transcriptional outcome (response/non-response) of the mosquito genes to DENV infection. The mosquito genes were assigned the value 1 if responsive and 0 if non-responsive to infection based on the microarray data. The six gene features were used as predictor variables simultaneously in the regressions to estimate the regression coefficients of each feature on the `outcome' (responsiveness or non-responsiveness). The regression analysis was performed in R using the generalized linear model (glm) function where `logit' was used as the link function for the binomial distribution.
3. Results
3.1. Comparison of gene features between responsive and non-responsive genes
The count statistics of responsive and non-responsive genes of A. aegypti to dengue infection associated with different gene features is shown in Table1. It shows the number of responsive and non-responsive genes corresponding to each gene feature. The 2×2 contingency analyses (Pearson Chi square tests) with the number of responsive genes versus non-responsive genes show significant association (p < 0.05) with the gene features.
Table 1.
Association of responsive and non-responsive genes with genes features. The numbers of responsive and non-responsive genes with (+) or without (−) the specific features (1st column) are shown along with associated p-value of statistical significance.
| Responsive genes | Non-response genes | ||||
|---|---|---|---|---|---|
|
|
|||||
| Feature + | Feature − | Feature + | Feature − | p-value | |
| Gene context | 967 | 1432 | 5056 | 8532 | 0.004 |
| High codon bias | 12 | 2387 | 128 | 13460 | 0.031 |
| Intronless | 2159 | 241 | 11958 | 1629 | 0.005 |
| Paralog | 1482 | 917 | 8884 | 4704 | 0.005 |
| Ancestral origin | 1410 | 989 | 7642 | 5946 | 0.021 |
| Derived origin | 1596 | 324 | 11992 | 2075 | 0.015 |
3.2. Gene features as predictors of transcriptional responsiveness to dengue infection
We performed logistic regression using the gene features as predictor variables and transcriptional responsiveness of mosquito to dengue infection as outcomes. The regression models are based on latent variable function y* = β0 + Xβ + e, y = 1[y*>0], where X represents independent variable and β represents coefficient of independent or predictor variable. The value of y equals 1 if the event occurs (i.e. gene is responsive to DENV infection) and zero otherwise (i.e. gene is non-responsive to the infection). Therefore, y = 1 if y*>0 and y = 0 if y*≤ 0. It is assumed that e is independent of X and has a standard logistic distribution with mean zero. Thus, the logit probability of our model is given by
where, `OUTCOME' represents response/non-response of genes to infection and the six independent variables of the equation represent the individual gene features. The estimated coefficient of the logistic regression represents the change in the log odds of the outcome for unit increase in the predictor variable. Because probability (p) of the outcome in logit model is estimated as the logarithm of the odds [p/(1 – p)], the estimated regression coefficient explain the variation (%) of the outcome with unit change in the predictor variable in our analysis. The estimates of coefficients of the gene features along with the significance values derived from the regression analysis are shown in Table 2. Based on the regression results, the probability of responsiveness of A. aegypti genes to DENV infection, in terms of gene features, is expressed as
where z = [(0.13 × context)+ (0.234 × intron) − (0.642 × codon bias) − (0.13 × paralog) + (0.245 × derived origin) + (0.126 × ancestral origin)] − 2.01.
Table 2.
Estimates of co-efficients and p-values of significance of logistic regressions bewteen gene features (independent variables) and responsiveness/ non-responsiveness of A. aegypti genes to dengue virus infection.
| Gene feature | Estimate | Std. error | z | Pr(>|z|) | Significance |
|---|---|---|---|---|---|
| Gene context | 0.1318 | 0.0455 | 2.896 | 0.00377 | ** |
| High codon bias | −0.64237 | 0.30472 | −2.108 | 0.03503 | * |
| Intronless | 0.23473 | 0.076 | 3.089 | 0.00201 | ** |
| Paralog | −0.1301 | 0.04773 | −2.726 | 0.00641 | ** |
| Ancestral origin | 0.1268 | 0.0509 | 2.491 | 0.01274 | * |
| Derived origin | 0.24522 | 0.07796 | 3.145 | 0.00166 | ** |
Note: The estimate values represent the coefficients of predictor variables (with std. errors shown), z value is score of the z-statistic, and Pr(>|z|) indicates the associated p-values. The significance levels P < 0.05 are shown by single asterisk and those less than 0.01 is shown by double asterisks.
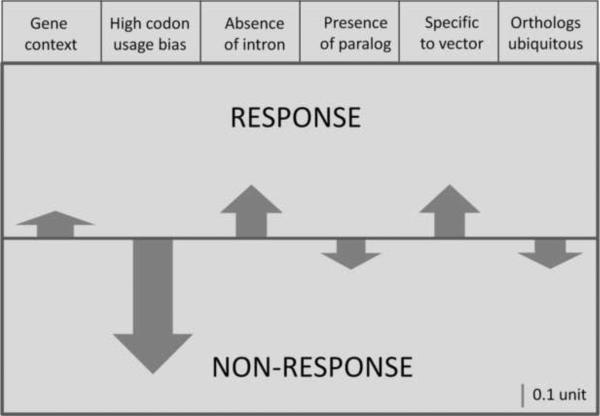
The equation shows that in A. aegypti, the context feature positively influences the responsiveness of genes to the viral infection by 0.13. Similarly, gene responsiveness to infection will also rise by 0.23 if the intronless genes in the mosquito increase by 1. If the genes are specific to the A. aegypti genome (no detectable ortholog in other two mosquitoes), that will also have a positive effect on probability of gene responsiveness of the vector to the infection. On the other hand, high codon bias has negative effect on gene responsiveness. The probability of responsiveness to infection will decrease by 0.64 with unit rise in high bias of codon usages. Similarly if genes have paralogous copies in the genome it will adversely affect responsiveness of the genes to DENV infection. The magnitudes and direction of effects of each gene feature on transcriptional responsiveness or non-responsiveness to dengue infection is illustrated in figure 1. It is apparent from these results that codon usage bias has the maximum influence than other features on the transcriptional response of A. aegypti genes to dengue infection.
Figure 1.
Schematic representation of positive and negative effect of different gene features on transcriptional responsiveness of A. aegypti genes to dengue virus infection. The upwards and downward arrows show positive and negative effects on gene responsiveness respectively. The lengths of arrow represent magnitude of effect, the scale of which is shown in the bottom right of the figure.
4. Discussion
The results from this investigation suggest that dengue-2 susceptibility have significant association with structural/ evolutionary features of the responsive genes in MOYO-S/ MOYO-R strains of A. aegypti. Although several gene expression studies have been performed in A. aegypti to identify genes up-regulated/ down-regulated upon dengue infection (Souza-Neto et al. 2009, Sim and Dimopoulos 2010, Ramirez and Dimopoulos 2010, Colpitts et al. 2011), no attempt have been made if these genes are differentially expressed in refractory strains of the mosquito. The Behura et al. 2011 study compared expression changes between susceptible and refractory mosquitoes upon dengue infection. Thus, results of the present study deals with genes relevant to the intrinsic ability of the mosquito to either host or defend dengue infection. We also think that, expression patterns observed in the two specific strains (MOYO-S and MOYO-R) may differ in other susceptible and refractory populations of A. aegypti. Therefore, the information of this study may be most relevant to the populations investigated (MOYO-S and MOYO-R). Thus, it is important to explore natural populations of A. aegypti mosquito those have differential intrinsic ability to either host or defend the viral infection to further investigate the role of these gene features in vector-virus interaction.
In A. aegypti, a large number of genes (about 6,000) are present in close proximity to each other wherein the intergenic distance between the neighboring genes is less than 1 kb (data not shown). Although gene context is widespread in prokaryotic genomes, eukaryotic genomes also show such pattern albeit with lower frequency. Evidences show that physical proximity of gene localization in the genome is strongly associated with expression of genes (Chiaromonte et al. 2003). Genes localized close to each other generally show co-regulated expression (Fukuoka et al. 2004; Tsai et al. 2007). In the MOYO-S/ MOYO-R strains of A. aegypti, genes those respond to dengue infection are expressed in highly modular manner and many of these expression modules are enriched with specific biochemical and signaling pathways (Behura et al. 2011). Thus, it is likely that in A. aegypti mosquito the gene context feature has a significant role for genes to respond coordinately to the infection. Moreover, literature evidences also suggest that gene contexts have significant association with coordinated function of genes in the form of biochemical pathways (Lee and Sonnhammer, 2003 and Hurst et al. 2004).
The results of this study further show that intronless genes of A. aegypti have higher propensity than intron-containing genes to positively influence transcriptional responsiveness of genes to DENV infection. In a compatible reaction (susceptible), the virus is likely to recruit less evolvable genes of the mosquito to sustain infection. On the other hand, the mosquito must evolve counter strategies to recruit highly evolvable genes to defend the virus adaptation (refractory response). This mechanism is fairly universal for evolution of most host-virus interactions (Marques and Carthew 2007). In this context, it is highly likely that our findings on role of intron on vector gene expressions corroborate to a co-evolutionary strategy between A. aegypti and dengue virus adaptability. Intronless genes generally evolve at a faster rate than intron-containing genes (Shabalina et al. 2010). Also, expression of intron-poor genes is more variable with time compared to that of intron-rich genes (Jeffares et al. 2008). Thus, we hypothesized that expression of intronless genes are more variable in the post-infection time periods than the intron-containing genes. To test that possibility, we reconstructed the expression networks reported in Behura et al. 2011 separately by using either the intronless or intron-containing genes. It was found that network pattern of the intron-containing genes is more variable than that of the intronless genes (data not shown). Furthermore, the intron-containing genes were differentially expressed at both 3hr and 18hr post infection time points whereas the intronless genes were differentially expressed at either 3hr or 18hr time point (Behura et al. 2011). This further suggests that intronless genes are more variable in expression between post-infection time points than intron-containing genes. From the results of regression analysis, it is also observed that increase of intronless genes shall increase responsiveness of A. aegypti genes to dengue infection. Moreover, as intronless genes are more evolvable than intron-containing genes, recruitment of such genes may also counteract virus adaptation by co-evolution of vector genes. Such an observation is also reported by independent study (Ruvolo et al. 1998).
We also observed a significant association between codon usage bias and expression of A. aegypti genes to dengue infection. Codon usage bias is known to have an association with translational efficiency and accuracy of genes in many species including Aedes aegypti mosquito (Behura and Severson, 2011). Furthermore, codon bias is likely to influence the efficiency of gene translation so that the gene can quickly respond to the stress (Lobo et al. 2009). The study by (Lobo et al. 2009) reveals that Flaviviridae viruses and their mosquito and vertebrate hosts undergo co-evolutionary changes in the codon usage patterns. In case of dengue infection, the A. aegypti translational machineries are exploited by dengue virus to synthesize viral proteins to complete its life cycle within the mosquito. Thus, it is likely that codon optimization of mosquito genes may have a role in facilitating virus replication. Such a hypothesis has been tested in poliovirus where it was found that replacement of optimized codons of the virus with rarely used synonymous codons attenuated the virus within the host (Coleman et al. 2008).
Furthermore, our study shows that ancestral genes of A. aegypti have a negative relationship with transcriptional responsiveness to dengue infection. In other words, genes that tend to be evolutionarily conserved in closely related species are not the preferred genes deployed by A. aegypti in a susceptible interaction with dengue virus. On the other hand, derived genes are more likely to respond to the infection. We argue that this is another form of counter-selection of the vector mosquito against survival strategies the virus. The evolutionary conserved genes (orthologs those are ubiquitously present across related species) are primarily house-keeping genes (She et al. 2009). If the virus is successful in exploiting such genes in the mosquito, establishing infection in the mosquito could be highly sustainable because these genes are also essential for the survival of the vector. Literature evidences suggest that house-keeping genes generally evolve slow (Zhang and Li, 2004) and are largely underrepresented in the genes responsible for disease susceptibility (Winter et al. 2004). On the other hand, genes encoding immunity related proteins generally evolve fast (Waterhouse et al. 2007). Thus, it is likely that the mosquito tends to activate genes that are highly evolving in nature.
Furthermore, our data also shows that A. aegypti genes with paralogous copies in the genome negatively influence gene responsiveness to dengue infection. The paralogs are products of gene duplication within the genome that are generally associated with functional divergence of the parental gene (Conant and Wolfe 2008). These genes tend to have higher variability in the expression than singleton genes and are believed to have important roles for the organism to respond and adapt to fluctuating environment (Dong et al. 2011). In mosquitoes, immune-related gene families display significant increases in numbers, most commonly as a result of gene duplication events that generate paralogous copies (Waterhouse et al. 2007). Thus, in A. aegypti, these genes are likely to be employed to counteract susceptible reaction with dengue virus.
Supplementary Material
Highlights
-
➢
Probability of Aedes aegypti gene expression to dengue infection is studied.
-
➢
Gene context positively influences expression of genes to dengue infection.
-
➢
Probability of gene expression increases with increase of intronless feature.
-
➢
High codon bias has negative effect on gene responsiveness to dengue infection.
-
➢
Derived genes in A. aegypti are more likely to respond to dengue infection.
Acknowledgement
The research was supported in part from a NIH-NIAID grant (RO1-AI059342). The authors are thankful to Dr. Roberta Angel for reading the manuscript and also to two anonymous reviewers for helpful suggestions to improve the paper.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Anderson JR, Rico-Hesse R. Aedes aegypti vectorial capacity is determined by the infecting genotype of dengue virus. Am J Trop Med Hyg. 2006;75:886–892. [PMC free article] [PubMed] [Google Scholar]
- Angellotti MC, Bhuiyan SB, Chen G, Wan XF. CodonO: codon usage bias analysis within and across genomes. Nucleic Acids Res. 2007;35(Web Server issue):W132–136. doi: 10.1093/nar/gkm392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennett KE, Olson KE, Muñoz Mde L, Fernandez-Salas I, Farfan-Ale JA, Higgs S, Black WC, 4th, Beaty BJ. Variation in vector competence for dengue 2 virus among 24 collections of Aedes aegypti from Mexico and the United States. Am J Trop Med Hyg. 2002;67:85–92. doi: 10.4269/ajtmh.2002.67.85. [DOI] [PubMed] [Google Scholar]
- Behura SK, Gomez-Machorro C, Harker BW, Debruyn B, Lovin DD, Hemme RR, Mori A, Romero-Severson J, Severson DW. Global Cross-Talk of Genes of the Mosquito Aedes aegypti in Response to Dengue Virus Infection. PLoS Negl Trop Dis. 2011;5:e1385. doi: 10.1371/journal.pntd.0001385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Behura SK, Severson DW. Coadaptation of isoacceptor tRNA genes and codon usage bias for translation efficiency in Aedes aegypti and Anopheles gambiae. Insect Mol Biol. 2011;20:177–187. doi: 10.1111/j.1365-2583.2010.01055.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Behura SK, Stanke M, Desjardins CA, Werren JH, Severson DW. Comparative analysis of nuclear tRNA genes of Nasonia vitripennis and other arthropods, and relationships to codon usage bias. Insect Mol Biol. 2010;19:49–58. doi: 10.1111/j.1365-2583.2009.00933.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Black WC, 4th, Bennett KE, Gorrochótegui-Escalante N, Barillas-Mury CV, Fernández-Salas I, de Lourdes Muñoz M, Farfán-Alé JA, Olson KE, Beaty BJ. Flavivirus susceptibility in Aedes aegypti. Arch Med Res. 2002;33:379–388. doi: 10.1016/s0188-4409(02)00373-9. [DOI] [PubMed] [Google Scholar]
- Boutanaev AM, Kalmykova AI, Shevelyov YY, Nurminsky DI. Large clusters of co-expressed genes in the Drosophila genome. Nature. 2002;420:666–669. doi: 10.1038/nature01216. [DOI] [PubMed] [Google Scholar]
- Buchman AR, Berg P. Comparison of intron-dependent and intron-independent gene expression. Mol Cell Biol. 1998;8:4395–4405. doi: 10.1128/mcb.8.10.4395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castillo-Davis CI, Mekhedov SL, Hartl DL, Koonin EV, Kondrashov FA. Selection for short introns in highly expressed genes. Nat Genet. 2002;31:415–418. doi: 10.1038/ng940. [DOI] [PubMed] [Google Scholar]
- Chen J, Sun M, Rowley JD, Hurst LD. The small introns of antisense genes arebetter explained by selection for rapid transcription than by “genomic design”. Genetics. 2005;171:2151–2155. doi: 10.1534/genetics.105.048066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiaromonte F, Miller W, Bouhassira EE. Gene length and proximity to neighbors affect genome-wide expression levels. Genome Res. 2003;13:2602–2608. doi: 10.1101/gr.1169203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciria R, Abreu-Goodger C, Morett E, Merino E. GeConT: gene context analysis. Bioinformatics. 2004;20:2307–2308. doi: 10.1093/bioinformatics/bth216. [DOI] [PubMed] [Google Scholar]
- Cohen BA, Mitra RD, Hughes JD, Church GM. A computational analysis of whole-genome expression data reveals chromosomal domains of gene expression. Nat Genet. 2000;26:183–186. doi: 10.1038/79896. [DOI] [PubMed] [Google Scholar]
- Coleman JR, Papamichail D, Skiena S, Futcher B, Wimmer E, Mueller S. Virus attenuation by genome-scale changes in codon pair bias. Science. 2008;320:1784–1787. doi: 10.1126/science.1155761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colpitts TM, Cox J, Vanlandingham DL, Feitosa FM, Cheng G, Kurscheid S, Wang P, Krishnan MN, Higgs S, Fikrig E. Alterations in the Aedes aegypti transcriptome during infection with West Nile, dengue and yellow fever viruses. PLoS Pathog. 2011;7:e1002189. doi: 10.1371/journal.ppat.1002189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conant GC, Wolfe KH. Turning a hobby into a job: how duplicated genes find new functions. Nat Rev Genet. 2008;9:938–950. doi: 10.1038/nrg2482. [DOI] [PubMed] [Google Scholar]
- Diallo M, Ba Y, Faye O, Soumare ML, Dia I, Sall AA. Vector competence of Aedes aegypti populations from Senegal for sylvatic and epidemic dengue 2 virus isolated in West Africa. Trans R Soc Trop Med Hyg. 2008;102:493–498. doi: 10.1016/j.trstmh.2008.02.010. [DOI] [PubMed] [Google Scholar]
- Dong D, Yuan Z, Zhang Z. Evidences for increased expression variation of duplicate genes in budding yeast: from cis- to trans-regulation effects. Nucleic Acids Res. 2011;39:837–847. doi: 10.1093/nar/gkq874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drummond DA, Bloom JD, Adami C, Wilke CO, Arnold FH. Why highly expressed proteins evolve slowly. Proc Natl Acad Sci U S A. 2005;10240:14338–14343. doi: 10.1073/pnas.0504070102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duncker BP, Davies PL, Walker VK. Introns boost transgene expression in Drosophila melanogaster. Mol Gen Genet. 1997;254:291–296. doi: 10.1007/s004380050418. [DOI] [PubMed] [Google Scholar]
- Eisenberg E, Levanon EY. Human housekeeping genes are compact. Trends Genet. 2003;19:362–365. doi: 10.1016/S0168-9525(03)00140-9. [DOI] [PubMed] [Google Scholar]
- Elizondo LI, Jafar-Nejad P, Clewing JM, Boerkoel CF. Gene clusters, molecular evolution and disease: a speculation. Curr Genomics. 2009;10:64–75. doi: 10.2174/138920209787581271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukuoka Y, Inaoka H, Kohane IS. Inter-species differences of co-expression of neighboring genes in eukaryotic genomes. BMC Genomics. 2004;5:4. doi: 10.1186/1471-2164-5-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gubler DJ, Nalim S, Tan R, Saipan H, Saroso JS. Variation in susceptibility to oral infection with dengue viruses among geographic strains of Aedes aegypti. Am J Trop Med Hyg. 1979;28:1045–1052. doi: 10.4269/ajtmh.1979.28.1045. [DOI] [PubMed] [Google Scholar]
- Gu X, Zhang Z, Huang W. Rapid evolution of expression and regulatory divergences after yeast gene duplication. Proc Natl Acad Sci U S A. 2005;102:707–712. doi: 10.1073/pnas.0409186102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu Z, Rifkin SA, White KP, Li WH. Duplicate genes increase gene expression diversity within and between species. Nat Genet. 2004;36:577–579. doi: 10.1038/ng1355. [DOI] [PubMed] [Google Scholar]
- Halstead SB. Dengue virus-mosquito interactions. Annu Rev Entomol. 2008;53:273–291. doi: 10.1146/annurev.ento.53.103106.093326. [DOI] [PubMed] [Google Scholar]
- Hershberg R, Petrov DA. Selection on codon bias. Annu Rev Genet. 2008;42:287–299. doi: 10.1146/annurev.genet.42.110807.091442. [DOI] [PubMed] [Google Scholar]
- Hurst LD, Pal C, Lercher MJ. The evolutionary dynamics of eukaryotic gene order. Nat Rev Genet. 2004;5:299–310. doi: 10.1038/nrg1319. [DOI] [PubMed] [Google Scholar]
- Jeffares DC, Penkett CJ, Bähler J. Rapidly regulated genes are intron poor. Trends Genet. 2008;24:375–378. doi: 10.1016/j.tig.2008.05.006. [DOI] [PubMed] [Google Scholar]
- Korbel JO, Jensen LJ, von Mering C, Bork P. Analysis of genomic context: prediction of functional associations from conserved bidirectionally transcribed gene pairs. Nat Biotechnol. 2004;22:911–917. doi: 10.1038/nbt988. [DOI] [PubMed] [Google Scholar]
- Kyle JL, Harris E. Global spread and persistence of dengue. Annu Rev Microbiol. 2008;62:71–92. doi: 10.1146/annurev.micro.62.081307.163005. [DOI] [PubMed] [Google Scholar]
- Le Hir H, Nott A, Moore MJ. How introns influence and enhance eukaryotic gene expression. Trends Biochem Sci. 2003;28:215–220. doi: 10.1016/S0968-0004(03)00052-5. [DOI] [PubMed] [Google Scholar]
- Lee JM, Sonnhammer EL. Genomic gene clustering analysis of pathways in eukaryotes. Genome Res. 2003;13:875–82. doi: 10.1101/gr.737703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lercher MJ, Blumenthal T, Hurst LD. Coexpression of neighboring genes in Caenorhabditis elegans is mostly due to operons and duplicate genes. Genome Res. 2003;13:238–243. doi: 10.1101/gr.553803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lobo FP, Mota BE, Pena SD, Azevedo V, Macedo AM, Tauch A, Machado CR, Franco GR. Virus-host coevolution: common patterns of nucleotide motif usage in Flaviviridae and their hosts. PLoS One. 2009;4:e6282. doi: 10.1371/journal.pone.0006282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez-Guerrero CE, Ciria R, Abreu-Goodger C, Moreno-Hagelsieb G, Merino E. GeConT 2: gene context analysis for orthologous proteins, conserved domains and metabolic pathways. Nucleic Acids Res. 2008;36(Web Server issue):W176–180. doi: 10.1093/nar/gkn330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marques JT, Carthew RW. A call to arms: coevolu tion of animal viruses and host innate immune responses. Trends Genet. 2007;23:359–364. doi: 10.1016/j.tig.2007.04.004. [DOI] [PubMed] [Google Scholar]
- Nene V, Wortman JR, Lawson D, Haas B, Kodira C, Tu ZJ, Loftus B, Xi Z, Megy K, Grabherr M, Ren Q, Zdobnov EM, Lobo NF, Campbell KS, Brown SE, Bonaldo MF, Zhu J, Sinkins SP, Hogenkamp DG, Amedeo P, Arensburger P, Atkinson PW, Bidwell S, Biedler J, Birney E, Bruggner RV, Costas J, Coy MR, Crabtree J, Crawford M, Debruyn B, Decaprio D, Eiglmeier K, Eisenstadt E, El-Dorry H, Gelbart WM, Gomes SL, Hammond M, Hannick LI, Hogan JR, Holmes MH, Jaffe D, Johnston JS, Kennedy RC, Koo H, Kravitz S, Kriventseva EV, Kulp D, Labutti K, Lee E, Li S, Lovin DD, Mao C, Mauceli E, Menck CF, Miller JR, Montgomery P, Mori A, Nascimento AL, Naveira HF, Nusbaum C, O'leary S, Orvis J, Pertea M, Quesneville H, Reidenbach KR, Rogers YH, Roth CW, Schneider JR, Schatz M, Shumway M, Stanke M, Stinson EO, Tubio JM, Vanzee JP, Verjovski-Almeida S, Werner D, White O, Wyder S, Zeng Q, Zhao Q, Zhao Y, Hill CA, Raikhel AS, Soares MB, Knudson DL, Lee NH, Galagan J, Salzberg SL, Paulsen IT, Dimopoulos G, Collins FH, Birren B, Fraser-Liggett CM, Severson DW. Genome sequence of Aedes aegypti, a major arbovirus vector. Science. 2007;316:1718–1723. doi: 10.1126/science.1138878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pál C, Papp B, Hurst LD. Highly expressed genes in yeast evolve slowly. Genetics. 2001;158:927–231. doi: 10.1093/genetics/158.2.927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palmiter RD, Sandgren EP, Avarbock MR, Allen DD, Brinster RL. Heterologous introns can enhance expression of transgenes in mice. Proc Natl Acad Sci USA. 1991;88:478–82. doi: 10.1073/pnas.88.2.478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phillips ML. Dengue reborn: widespread resurgence of a resilient vector. Environ Health Perspectives. 2008;116:382–389. doi: 10.1289/ehp.116-a382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramirez JL, Dimopoulos G. The Toll immune signali ng pathway control conserved anti-dengue defenses across diverse Ae. aegypti strains and against multiple dengue virus serotypes. Dev Comp Immunol. 2010;34:625–629. doi: 10.1016/j.dci.2010.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rico-Hesse R. Dengue virus evolution and virulence models. Clin Infect Dis. 2007;44:1462–1466. doi: 10.1086/517587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosen L, Roseboom LE, Gubler DJ, Lein JC, Chaniotis BN. Comparative susceptibility of mosquito species and strains to oral and parenteral infection with dengue and Japanese encephalitis viruses. Am J Trop Med Hyg. 1985;34:603–615. doi: 10.4269/ajtmh.1985.34.603. [DOI] [PubMed] [Google Scholar]
- Ruvolo V, Wang E, Boyle S, Swaminathan S. The Epstein-Barr virus nuclear protein SM is both a post-transcriptional inhibitor and activator of gene expression. Proc Natl Acad Sci U S A. 1998;95:8852–8857. doi: 10.1073/pnas.95.15.8852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salazar MI, Richardson JH, Sánchez-Vargas I, Olson KE, Beaty BJ. Dengue virus type 2: replication and tropisms in orally infected Aedes aegypti mosquitoes. BMC Microbiol. 2007;7:9. doi: 10.1186/1471-2180-7-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shabalina SA, Ogurtsov AY, Spiridonov AN, Novichkov PS, Spiridonov NA, Koonin EV. Distinct patterns of expression and evolution of intronless and intron-containing mammalian genes. Mol Biol Evol. 2010;27:1745–1749. doi: 10.1093/molbev/msq086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sapra AK, Arava Y, Khandelia P, Vijayraghavan U. Genome-wide analysis of pre-mRNA splicing: intron features govern the requirement for the second-step factor, Prp17 in Saccharomyces cerevisiae and Schizosaccharomyces pombe. J Biol Chem. 2004;279:52437–52446. doi: 10.1074/jbc.M408815200. [DOI] [PubMed] [Google Scholar]
- She X, Rohl CA, Castle JC, Kulkarni AV, Johnson JM, Chen R. Definition, conservation and epigenetics of housekeeping and tissue-enriched genes. BMC Genomics. 2009;10:269. doi: 10.1186/1471-2164-10-269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sim S, Dimopoulos G. Dengue virus inhibits immune responses in Aedes aegypti cells. PLoS One. 2010;5:e10678. doi: 10.1371/journal.pone.0010678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Souza-Neto JA, Sim S, Dimopoulos G. An evolutionary conserved function of the JAK-STAT pathway in anti-dengue defense. Proc Natl Acad Sci U S A. 2009;106:17841–17846. doi: 10.1073/pnas.0905006106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spellman PT, Rubin GM. Evidence for large domains of similarly expressed genes in the Drosophila genome. J. Biol. 2002;1:5. doi: 10.1186/1475-4924-1-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subramanian S, Kumar S. Gene expression intensity shapes evolutionary rates of the proteins encoded by the vertebrate genome. Genetics. 2004;168:373–381. doi: 10.1534/genetics.104.028944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tardieux I, Poupel O, Lapchin L, Rodhain F. Variation among strains of Aedes aegypti in susceptibility to oral infection with dengue virus type 2. Am J Trop Med Hyg. 1990;43:308–313. doi: 10.4269/ajtmh.1990.43.308. [DOI] [PubMed] [Google Scholar]
- Tsai HK, Su CP, Lu MY, Shih CH, Wang D. Co-expression of adjacent genes in yeast cannot be simply attributed to shared regulatory system. BMC Genomics. 2007;8:352. doi: 10.1186/1471-2164-8-352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waterhouse RM, Kriventseva EV, Meister S, Xi Z, Alvarez KS, Bartholomay LC, Barillas-Mury C, Bian G, Blandin S, Christensen BM, Dong Y, Jiang H, Kanost MR, Koutsos AC, Levashina EA, Li J, Ligoxygakis P, Maccallum RM, Mayhew GF, Mendes A, Michel K, Osta MA, Paskewitz S, Shin SW, Vlachou D, Wang L, Wei W, Zheng L, Zou Z, Severson DW, Raikhel AS, Kafatos FC, Dimopoulos G, Zdobnov EM, Christophides GK. Evolutionary dynamics of immune-related genes and pathways in disease-vector mosquitoes. Science. 2007;316:1738–1743. doi: 10.1126/science.1139862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winter EE, Goodstadt L, Ponting CP. Elevated rates of protein secretion, evolution, and disease among tissue-specific genes. Genome Res. 2004;14:54–61. doi: 10.1101/gr.1924004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woodring JL, Higgs S, Beaty BJ. Natural cycles of vector-borne pathogens. In: Marquardt WC, Beaty BJ, editors. The biology of disease vectors. University Press of Colorado; Boulder: 1996. pp. 51–72. [Google Scholar]
- Zhang L, Li WH. Mammalian housekeeping genes evolve more slowly than tissue-specific genes. Mol Biol Evol. 2004;21:236–239. doi: 10.1093/molbev/msh010. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.