Abstract
Pepsin was immobilized on ethyl-bridged hybrid (BEH) particles and digestion performance was evaluated in a completely online format, with the specific intent of using the particles for hydrogen deuterium exchange mass spectrometry (HDX MS) experiments. Because the BEH particles are mechanically strong, they could withstand prolonged, continuous high-pressure at 10,000 psi. Online digestion was performed under isobaric conditions with continuous solvent flow, in contrast to other approaches where the pressure or flow is cycled. As expected, digestion efficiency at 10,000 psi was increased and reproducibly produced more peptic peptides versus digestion at 1,000 psi. Prototype columns made with the BEH pepsin particles exhibited robust performance and deuterium back-exchange was similar to that of other immobilized pepsin particles. These particles can be easily incorporated in existing HDX MS workflows to provide more peptide coverage in experiments where fast, efficient, and reproducible online pepsin digestion is desired.
Keywords: Pressure, online proteolysis, enzyme, bioreactor, ultra performance liquid chromatography, UPLC, LC, ethyl-bridged hybrid, BEH, acid protease
INTRODUCTION
Enzymatic proteolysis is an important sample preparation step prior to liquid chromatography mass spectrometry (LC-MS) studies including for protein identification, posttranslational modification characterization, and protein structural characterization.1 Traditionally, protein digestion is performed offline in a solution consisting of a mixture of the enzyme and the protein(s) being digested.2 Manual preparation processes and typically lengthy digestion time(s) can be limiting factors in offline, in-solution digestion.3 There is, therefore, a desire to move protein digestion from an offline to an online format and to concomitantly improve multiple characteristics of the online platform.
Many applications have used automated online digestion with an immobilized enzyme reactor (IMER).4 An intact protein is directly introduced into an online IMER with a pump at an optimized flow rate (usually protein dependent). The eluent from the IMER contains the newly created peptides, which can then be chromatographically separated and introduced into a mass spectrometer, for example. An online IMER digests proteins with higher efficiency than solution digestion mainly as a result of the increased enzyme-substrate ratio. Online digestion can be preferred for high-throughput screening and rapid sample processes.5 A tryptic IMER for peptide mapping and a PNGase,F IMER for deglycosylation have been used in proteomics applications.3-4, 5b, 6 In order to prepare an IMER that is capable of robust protein digestion, it is very important that the covalent attachment process not interfere with or limit the activity of the enzyme. An IMER can be a particle-packed column or a microfluidic device, in which the solid media is covalently cross-linked to the protease at a high surface concentration.6-7 Various types of media, such as perfusion particles8, monoliths4b, 9, fibers10, and nanoparticles11, have been used as IMER solid supports and have utilized a variety of different covalent linkers.
Pepsin digestion is used in some important MS applications because pepsin is capable of highly active acidic proteolysis, even at low temperatures. Hydrogen deuterium exchange mass spectrometry (HDX MS) requires rapid digestion at acidic pH in order to refine the location of incorporated deuterium.12 Porcine pepsin, aspergillopepsin (protease type XIII) and other pepsin-like enzymes have been used in HDX MS, as reviewed elsewhere.13 Online digestion with a pepsin column was incorporated into the HDX workflow more than 10 years ago 12b, 14 and is now very widely utilized.
One way to improve the efficiency of enzymatic digestion is to perform the digestion at elevated pressure. High pressure often leads to inactivation of enzymes (as well as destruction of bacteria), and it is therefore utilized by some industrial food processes.15 In other cases, high pressure is known to induce stabilization and activation of some enzymes.16 Pepsin digestion of β-lactoglobulin at pressures up to 50,000 psi showed up to 270-fold increase in cleavage rate.17 There are several explanations16 for pressure-induced changes in enzyme-catalyzed reaction rates, including: (1) direct changes in enzyme structure, (2) changes in the reaction mechanisms, (3) changes in solvent physical properties (pH, density viscosity, phase).
Digestion at high pressure has been used to increase digestion efficiency in various mass spectrometry and proteomics applications. For example, Lopez-Ferrer and his colleagues introduced the fast online digestion system (FOLDS)18, a pressure cycling strategy for trypsin-based high-throughput screening19, and pressurized pepsin digestion in FOLDS for proteomics applications.20 In addition, pepsin digestion using a pressurized sample loop was utilized for HDX MS.21 These high pressure digestion approaches are (semi-)offline methods, in which pepsin was first mixed with protein samples at low pressure and then the mixture was put into a closed loop or chamber and pressurized for digestion.
In the study reported herein, we introduce online high-pressure digestion with continuous flow. A column was packed with newly prepared, high-strength hybrid silica particles onto which pepsin had been conjugated. These particles can withstand prolonged high-pressure, enabling continuous flow pepsin digestion at 10,000 psi. The new hybrid particle immobilized pepsin column was compared with a conventional POROS pepsin column typically used for online pepsin digestion at ~1,000 psi. We also determined the digestion efficiency of the new particles with online pepsin digestions of various proteins.
EXPERIMENTAL SECTION
Chemicals and Materials
Ethyl-bridged hybrid (BEH) 5 μm particles (pore size: 300 Å, surface area: 90 m2/g) were obtained from Waters Corporation (Milford, MA, USA) and POROS AL 20 μm particles and immobilized pepsin POROS resins were purchased from Life Technologies/Applied Biosystems (Carlsbad, CA, USA). Pepsin (procine gastric mucosa, part #P7012), phosphorylase b (rabbit muscle, part #P7012), and cytochrome c (equine heart, part #C7752) were purchased from Sigma (St. Louis, MO). Triethoxysilyl butyraldehyde Tech-90 (part # SIT8185.3) was from Gelest, Morrisville, PA, USA). Deuterium oxide (99.9%) and deuterium chloride (20%) were from Cambridge Isotope Laboratories, Inc. (Andover, MA, USA). All other common lab chemicals were purchased from Sigma.
Strength Testing
All particles were packed into an empty 2.1 × 30 mm column housing. The packed columns were pressurized up to 24,000 psi by pumping 0.1% aqueous formic acid solution into the column while a 40 μm × 10 cm fused silica capillary piece was inline at the outlet of the column as a flow restrictor. The flow rate of the solvent was determined at 2,000 psi increments, up to 24,000 psi. Particles exposed at several pressures were carefully collected for examination with a scanning electron microscope (JEOL, Japan) after platinum sputter coating. The SEM examination was performed on particles exposed to no pressure (control), pressure at 2,000 psi for 21 seconds, and then a series of higher pressures wherein the pressure was ramped from 2,000 psi such that a total of 1 min, 2 min, and 2.4 min were taken to reach 3,600, 17,000, and 24,000 psi, respectively.
Immobilization
A prototype BEH pepsin digestion column was made by immobilizing pepsin onto BEH particles via coupling chemistry available from Sterogene Bioseparations (Carlsbad, CA) wherein a aldehyde containing linker (triethoxysilyl butyraldehyde, see Figure 1) reacts with a primary amine in the protein, forms a temporary Schiffs base which is then rapidly reduced with sodium cyanoborohydride. Specifically, a solution containing 80 mg of pepsin was prepared in 2 mL of 50 mM trisodium citrate at pH 5.0. One mL of ALD coupling solution (Sterogene Bioseparations, Carlsbad, CA, USA) containing 1 M sodium cyanoborohydride and 106 mg triethoxysilyl butyraldehyde (Tech-90) was added to the pepsin solution. The mixture was rotated for 2 hours at room temperature. 0.6 g of BEH particles were then added to the solution and mixed for 30 min. The mixture was salted out by adding 2.2 mL of 2 M sodium sulfate in 50 mM trisodium citrate at pH 5.0, followed by rotation overnight at room temperature. The immobilization reaction was quenched by adding 1 mL of 1 M ethanolamine and rotated for 2 hours at room temperature. Five aliquots of 10 mL each of 50 mM trisodium citrate were used to wash the pepsin immobilized BEH particles; the particles were then washed with 30 mL of 1 M sodium chloride. Finally, the particles were washed with 50 mL of 50 mM trisodium citrate followed by 20 mL of 0.08% trifluoroacetic acid in water. The batch of pepsin immobilized BEH particles was stored in 5 mL of 0.08% TFA at 5 °C until they were packed into an empty 2.1 × 30 mm column housing.
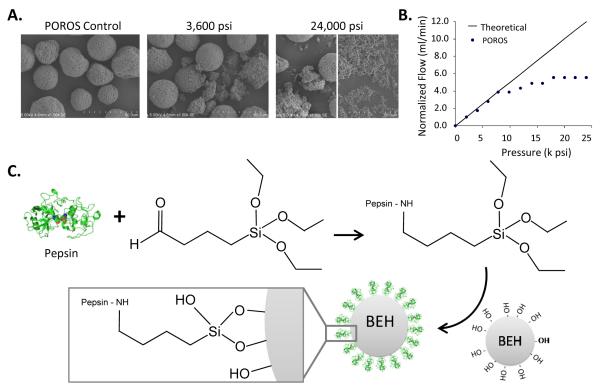
Figure 1.
Scanning electron microscopy images of POROS AL particles before (A) and after pressurization (B). The control did not experience any pressure prior to the SEM image while 3,600, and 24,000 psi were applied in the middle and right panels. For the 24,000 psi experiment, two different fields of particles are shown. (B) POROS particle strength in normalized flow vs. pressure. The results for POROS particles begin to deviate from the theoretical value assuming no particle destruction around 8,000 psi. (C) Scheme for pepsin immobilization onto BEH particles. Note that in the attachment of the triethoxysilyl butyraldehyde to pepsin, only the final product is shown. The chemistry involving the aldehyde reaction with the amines in pepsin, Schiff base formation and subsequent reduction are not show in the diagram but are explained in the Experimental Section.
HDX Experimental Method
Phosphorylase b and cytochrome c were used as standard proteins for online digestion with and without pressurization. Online pepsin digestion was performed with a nanoACQUITY UPLC™ system with HDX technology (Waters).22 The HDX manager of this system provides independent temperature control for the digestion and holds the temperature of the peptide trapping and ultra performance liquid chromatography (UPLC) separation at 0 °C. The temperature of the pepsin column compartment inside the HDX manager was set at 0, 10, 25, or 37 °C for cytochrome c. For both non-pressurized and pressurized digestion, the protein sample was passed through the BEH pepsin column (kept at 10 °C for phosphorylase b) at 100 μL/min for 3 min in 0.2% formic acid in water, pH 2.5. A pressure restrictor (2.1 × 150 mm column packed with naked 1.7 μm UPLC particles) was placed before the waste line in the trap valve (Figure 2) so that the whole BEH pepsin column was pressurized. The digests were trapped and desalted online using an ACQUITY UPLC® BEH C18 1.7 μm VanGuard™ Pre-Column (Waters) at 0 °C. The flow was diverted by switching valves, and trapped peptides were eluted into an ACQUITY UPLC BEH C18 1.7μm, 1 mm × 100 mm column (Waters) held at 0 °C. Peptides were separated with a 6 min linear acetonitrile gradient (7-40%) containing 0.1% formic acid at 40 μL/min. The eluent was directed into a SYNAPT® MS mass spectrometer (Waters) with electrospray ionization and lock-mass correction using Glu-Fibrionogen peptide. Mass spectra were acquired in MSE mode23 over the m/z range of 50-2000. Peptides were identified prior to deuteration using ProteinLynx Global Server software (Waters).24 The sequence coverage map of phosphorylase b and cytochrome c were plotted using MSTools.25
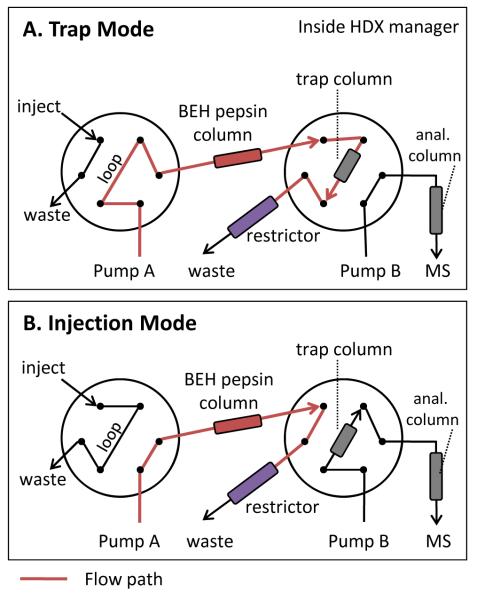
Figure 2.
Valve scheme for the flow path in Trap (A) and Injection (B) modes. The red line indicates the flow path. The BEH pepsin column, the trap/analytical columns, and the pressure restrictor, are colored in red, grey, and purple, respectively. Pump A delivers mobile phase [0.2% formic acid in water, pH 2.5] for pepsin digestion and Pump B drives the reversed phase gradient [0.1% formic acid in water and 0.1% formic acid in acetonitrile] for peptide trapping and separation.
For the HDX undeuterated controls, solutions of 32 pmol/μL Phosphorylase b and 40 pmol/μL cytochrome c in 10 mM phosphate in H2O, pH 7.00 were prepared. These solutions were quenched by reducing the pH to 2.66 with a 1:1 dilution with 100% H2O quench buffer [100 mM phosphate, pH 2.66]. The totally deuterated angiotensin II and bradykinin peptides were prepared by fully labeling in 99.9% deuterium oxide containing 0.5% deuterium chloride, at 37 °C overnight. This fully deuterated mixture was used to determine the loss of deuterium in the system as described previously.26
RESULTS AND DISCUSSION
We sought to determine if continuous flow online pepsin digest at 10,000 psi could be achieved for HDX applications. The advantage of a continuous flow strategy is that the pressure in the pepsin column is isobaric and there is no need for pressure cycling, although the particles must be able to withstand prolonged high pressure. Previous approaches 18-21 did not use particles that could withstand high pressure and usually involved some kind of pressure cycling. Common commercial sources of immobilized pepsin include pepsin on agarose beads (Thermo Scientific/Pierce) and pepsin immobilized onto POROS particles (Life Technologies/Applied Biosystems). Agarose beads certainly do not have the ability to withstand pressures of 10,000 psi so such material was removed from further consideration. Although POROS pepsin columns have many advantages and are used by many HDX research groups, including us, perfusion chromatography media also has relatively low pressure stability. POROS is made from poly(styrene divinylbenzene) particles with transecting gigapores.27 The flow-through gigapores provide a large surface area, thus enabling a high concentration of biomolecules to be immobilized. Gigapores also result in low back-pressure of approximately 600–1000 psi at flow rates of 75–100 μL/min. The manufacturer of POROS particles (Life Technologies/Applied Biosystems) has suggested that 2,500 psi is the maximum pressure limit for POROS particles.
The mechanical strength of POROS particles was tested by applying pressure to the particles and then inspecting them with electron microscopy. At a pressure of 2,000 psi, the particles remained essentially unchanged (data not shown). Pressurization up to 3,600 psi for 1 min partially damaged the particles, visualized by the SEM images in Figure 1A. The SEM images of particles pressurized to 8,000 and 17,000 psi showed that the pressure crushed most particles significantly (data not shown). Some particles at pressure of 24,000 psi and above were completely broken into nanosized pieces. Figure 1B illustrates the correlation between the flow rate and the pressure applied to the POROS particles. Theoretically, if particles in a packed bed retain their shapes upon pressurization, there should be a linear correlation as predicted by the Kozeny-Carman equation.28 However, the dotted line in Figure 1B started to fall off from the theoretical line at pressures >8,000 psi. This result is an indication of particle destruction and compaction, in which small fragments clog the filter of the column outlet and inhibit the fluid flow. Note that particle destruction depends on the flow rate, the time span, and the shear force from the solvent flowing through particles, in addition to the back pressure.28 In this test, packed particles were held at each pressure level for less than 2.4 min and this pressurization time is much shorter than what is typically performed for routine HDX digestion conditions. As a whole, these results demonstrate definitively that high-pressure digestion with POROS particles is not practical.
Ethyl-bridged hybrid (BEH) particles are an ultra-high-pressure packing sorbent that can resist high pressures.29 Wyndham and colleagues demonstrated 30 with a pressure-flow diagram similar to Figure 1B that BEH particles are stable at pressures up to 9,000 psi. The mechanical strength of BEH particles was further tested 31 at pressures up to 65,000 psi. In these tests, there was no change to flow resistance in the packed bed, indicating that there was no crushing of the particles, and microscopy showed no visible gaps in the bed after exposure to 65,000 psi, indicating that the bed did not collapse even at this high pressure. Based on these prior observations indicating high-pressure stability of BEH particles, as well as a large body of work since 2004 in which BEH particles have been used in UPLC applications at pressures up to 10,000 psi, we chose to immobilize pepsin on BEH particles for continuous-flow high-pressure online digestion.
The size of particles to use for a pepsin column was determined by considering both back-pressure and pressure drop. Pressure drop is the pressure differential between the inlet and outlet of the column, and a large pressure drop creates uneven pressurization across the column bed. When small particles are used, pressure drop can become an issue. For example, 1.25 μm particles packed in a 2.1 × 30 mm column are expected to have an approximately 1,000 psi pressure drop. It was thus important to select a particle size that would maintain a small pressure drop across the column during pressurized digestion. We chose 5 Gm BEH particles, in which the estimated pressure drop was only 64 psi in a 2.1 × 30 mm column. It was not possible to accurately measure how much pepsin was immobilized. Determining the potential surface area for enzyme immobilization depends on both the available surface area of particles and the size of the active enzyme to enter into the pores. Using a combination of the per gram particle size, surface area, and pore diameter of BEH particles, a value of ~5 μmol/m2 was derived as an estimate of the theoretical maximum surface coverage for pepsin. Due to the various ranges of pore size and pore volume on the particle surface, the actual coverage of pepsin was expected to be much less than the theoretical value. An excess molar ratio of a cross-linker over pepsin (~200:1) was used to push forward the linking reaction in order to achieve maximal pepsin coverage.
The pepsin immobilization protocol for BEH particles was modified from a previous publication 14 describing immobilization onto POROS. Figure 1C shows the overall scheme wherein triethoxysilyl butyraldehyde [a cross-linker] is coupled to pepsin prior to immobilization onto BEH particles. The aldehyde group in triethoxysilyl butyraldehyde reacted with various amine groups in pepsin, including the N-terminus, to form a temporary Schiff base which was nearly immediately reduced by sodium cyanoborohydride (NaCNBH3). The hydroxyl groups on BEH surface were successfully linked with the triethoxysilyl side of the linker. Importantly, the attachment between pepsin and a linker did not disturb the proteolytic activity of pepsin, as shown below in digestion tests.
Successful immobilization was confirmed by the digestion performance of the BEH pepsin column. Cytochrome c could be digested online using either the BEH or POROS pepsin column with no pressurization and both column digestions resulted in peptides that covered 100% of the amino acid sequence of cytochrome c (with 34 and 29 reproducible peptic peptides for BEH and POROS, respectively, see Supporting Information, Fig. S1). The peptides were produced reproducibly and were found at least twice in triplicate digestions.
The BEH pepsin column was tested at a variety of digestion temperatures (Supporting Information, Fig. S2). Mass spectral identification of reproducibly produced peptides resulted in various amounts of sequence coverage (100%, 99%, and 63%) of cytochrome c at 25, 10, and 0 °C, respectively. As expected, decreased digestion efficiency (again monitoring only reproducible peptides found at least twice in triplicate digestions) was observed at lower digestion temperatures. The BEH pepsin column was used for over 200 digestions, stored for 1 year in 0.1% formic acid at 5 °C and then tested again by digesting an antibody (IgG). After this prolonged period, its digestion performance was comparable to freshly made BEH pepsin or a new POROS pepsin column (Supporting Information, Fig. S3). These data suggest not only that pepsin immobilization on BEH particles was successful, but also that stable immobilization was maintained over time without compromising enzyme activity.
To this point, we had only tested the BEH pepsin column at relatively low pressure (950 psi). To use the column at higher pressure, we introduced a flow restrictor. Figure 2 illustrates the online digestion flow-path for pressurization at continuous flow. The protein sample was delivered into the BEH pepsin column at 100 μL/min in trap mode. The residence time for digestion inside the BEH pepsin column was approximately 30-40 sec (considering flow rate and column volume) and the generated peptides were captured with a trapping column and desalted for 3 min. With a flow restrictor placed after a trap column (in trap mode), the digestion pressure was elevated from 950 to 9,800 psi and the digestion flow rate was maintained at 100 μL/min. The digestion time was not changed upon high-pressure digestion. After 3 min of desalting on the trap, the peptides were eluted to the analytical column for separation and mass analysis. The digestion and separation temperatures of 10 °C and at 0 °C, respectively, were independently controlled by the HDX manager.22 The setup as described allows for easy implementation of high-pressure online digestion in the Waters HDX system (or other comparable instrumentation) without modifying the existing valve configuration. Using these BEH particles at even higher pressurize is likely possible as the mechanical stability of BEH silica particles is well above 15,000 psi. In the present work, we report only on analyses for pressures up to 10,000 psi.
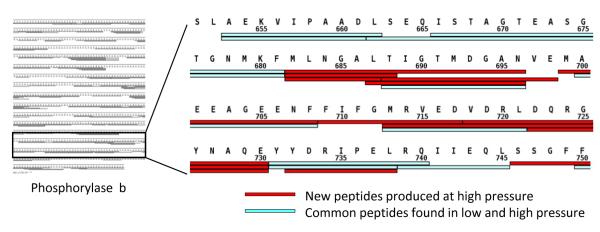
As predicted from previous digestion work at high pressure, improved digestion efficiency was achieved using a BEH pepsin column at high versus low pressure. Analysis of the reproducible peptides obtained after digestion with and without pressure (meaning at 9,800 psi versus 950 psi, respectively) showed that 83% and 76% sequence coverage of phosphorylase b was achieved with and without pressurized digestion, respectively. A small example of the peptide map from phosphorylase b is shown in Figure 3 (see Supporting Information Figure S4 for the full maps). Significant improvements in digestion efficiency were observed when the number of overlapping peptides were compared (Figure 3). Some peptides that were not found in low pressure digestions were newly produced in pressurized digestions (Figure 3, red), whereas other peptides were found in both low and high pressure digestions (Figure 3, cyan). A total of 112 additional peptides, which also were reproducible peptides, were found in the high-pressure digestion versus the low-pressure digestion of phosphorylase b. Table 1 lists the numbers of peptides found for both low- and high-pressure digestions of phosphorylase b. The majority of new peptides in high-pressure digestion were in the size range of 5 to 12 amino acids long. During HDX MS analyses, additional peptides can be essential for better defining the location of deuteration, all without involving longer digestion time or modification to the digestion buffer conditions (including denaturants, for example).
Figure 3.
Comparison of the map of peptic peptides from digestion of phosphorylase b under low- (950 psi) and high-pressure (9,800 psi) conditions. The left panel shows 83% coverage of phosphorylase b at 9,800 psi (see Supporting Information Figure S4 for the full map from the left panel). Part of the sequence map from residues 651-750 is shown in the right panel. The cyan bars indicate the peptic peptides found in both pressure conditions and the red bars indicate those new peptides found only with high-pressure digestion.
Table 1.
Number of peptides of various lengths found in low- and high-pressure digestion.
| Peptide Length (# of AA) |
Number of peptides | ||
|---|---|---|---|
| 950 psi (N=5) |
9800 psi (N=5) |
# of new peptides |
|
| 5 | 19 | 32 | 13 |
| 6 | 26 | 35 | 9 |
| 7 | 44 | 56 | 12 |
| 8 | 40 | 36 | - |
| 9 | 60 | 68 | 8 |
| 10 | 45 | 60 | 15 |
| 11 | 41 | 54 | 13 |
| 12 | 29 | 42 | 13 |
| 13 | 48 | 40 | - |
| 14 | 14 | 15 | 1 |
| 15 | 36 | 47 | 11 |
| 16 | 24 | 27 | 3 |
| 17 | 17 | 14 | - |
| 18 | 20 | 23 | 3 |
| 19 | 5 | 11 | 6 |
| 20 | 13 | 14 | 1 |
| 22 | 4 | 5 | 1 |
| 23 | 10 | 13 | 3 |
| Total | 112 | ||
It is paramount in any new HDX methodology to assess deuterium back-exchange. If deuterium losses are significant, regardless of the methodological improvements, the data will be useless. Significant deuterium loss during digestion can be a problem when measuring deuterium uptake of peptides in HDX experiemnts. As previously reported26, different types of pepsin columns can induce extensive deuterium loss incompatible with high-quality experiments; POROS is a good choice for HDX experiments in that it does not lead to increased back-exchange (see also Supporting Information, Figure S5). For fully deuterated angiotensin II (DRVYIHPF), Figure 4, we observed comparable levels of deuterium loss for a BEH pepsin column and a POROS pepsin column when the digestion chamber temperature was held at 0, 10, 25, or 37 °C (all analysis steps were identical after the digestion, see Experimental section). The deuteirum loss was only 2-3% different between BEH and POROS pepsin columns and the meaured deuteirum losses at 0, 10, and 25 °C were close to the theoretical values (a sum of deuterium losses from MS, LC, and digestion, see also 26 for description of the calculations). Another fully deuterated peptide, bradykinin, exhibited deuterium loss at 16% and 21% for BEH and POROS, repectively, at 0 °C digestion temperature (Supporting Information, Fig. S6). These deuterium recovery experiments strongly indicate that pepsin immobilized on BEH particles and online digestion using such particles at pressures of ~10,000 psi does not alter detuerium recovery. Online digestion at high pressure with BEH-immobilized pepsin is therefore highly compatible with HDX experiments and workflows.
Figure 4.
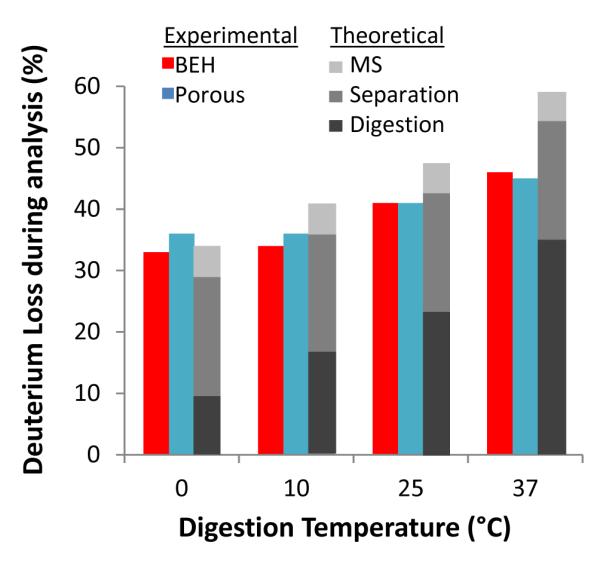
Comparison of deuterium loss from fully deuterated angiotensin II using BEH and POROS pepsin columns. The measured values of deuterium losses of BEH, POROS, and theoretical are shown in red, blue, and grey. All conditions past digestion were held constant (see Experimental section).
CONCLUSIONS
We have immobilized pepsin on BEH particles and prepared a prototype digestion column. The prototype column was evaluated to characterize the digestion performance of the new particles in a completely online format, with the specific intent of using continuous flow, high-pressure online digestion for HDX experiments. BEH particles are very mechanically strong silica particles, which were able to easily withstand prolonged high-pressure digestion at 10,000 psi. Enhanced digestion efficiency was reported at high pressure and more reproducibly produced peptides were found versus at lower pressures. Stable and robust digestions at high-pressure were exhibited and digestion activity was not lost over hundreds of digestions or after 1 year. While digestion at high-pressure has been described before for proteins, and for proteomic applications involving LC-MS, we believe this to be the first description of a continuous flow online digestion strategy at high pressure. As such, the approach can be easily adapted to existing instrumentation and then utilized in HDX MS workflows to provide more peptide coverage in experiments where fast, efficient, and reproducible online pepsin digestion is desired.
Supplementary Material
ACKNOWLEDGMENT
This work was supported in part by a grant from NIH, R01-GM086507 and by a research collaboration with the Waters Corporation. We thank Keith Fadgen, Daniel P. Walsh and Christopher Morgan for helpful discussions.
Footnotes
Supporting Information Available.
REFERENCES
- 1.Wang R, Chait BT. Curr Opin Biotechnol. 1994;5:77–84. doi: 10.1016/s0958-1669(05)80074-6. [DOI] [PubMed] [Google Scholar]
- 2.Slysz GW, Lewis DF, Schriemer DC. J Proteome Res. 2006;5:1959–1966. doi: 10.1021/pr060142d. [DOI] [PubMed] [Google Scholar]
- 3.Massolini G, Calleri E. J Sep Sci. 2005;28:7–21. doi: 10.1002/jssc.200401941. [DOI] [PubMed] [Google Scholar]
- 4(a).Cooper JW, Chen J, Li Y, Lee CS. Anal Chem. 2003;75:1067–1074. doi: 10.1021/ac025768b. [DOI] [PubMed] [Google Scholar]; (b) Temporini C, Calleri E, Campese D, Cabrera K, Felix G, Massolini G. J Sep Sci. 2007;30:3069–3076. doi: 10.1002/jssc.200700337. [DOI] [PubMed] [Google Scholar]; (c) Wang S, Regnier FE. J Chromatogr A. 2001;913:429–436. doi: 10.1016/s0021-9673(01)00604-5. [DOI] [PubMed] [Google Scholar]; (d) Zhao C, Jiang H, Smith DR, Bruckenstein S, Wood TD. Anal Biochem. 2006;359:167–175. doi: 10.1016/j.ab.2006.09.005. [DOI] [PubMed] [Google Scholar]; (e) Forsberg EM, Green JR, Brennan JD. Anal. Chem. 2011;83:5230–5236. doi: 10.1021/ac200534t. [DOI] [PubMed] [Google Scholar]
- 5(a).Yamaguchi H, Miyazaki M, Kawazumi H, Maeda H. Anal Biochem. 2010;407:12–18. doi: 10.1016/j.ab.2010.07.026. [DOI] [PubMed] [Google Scholar]; (b) Yuan H, Zhang L, Hou C, Zhu G, Tao D, Liang Z, Zhang Y. Anal Chem. 2009;81:8708–8714. doi: 10.1021/ac900310y. [DOI] [PubMed] [Google Scholar]
- 6.Krenkova J, Foret F. Electrophoresis. 2004;25:3550–3563. doi: 10.1002/elps.200406096. [DOI] [PubMed] [Google Scholar]
- 7.Cingoz A, Hugon-Chapuis F, Pichon V. J Chromatogr A. 2008;1209:95–103. doi: 10.1016/j.chroma.2008.08.120. [DOI] [PubMed] [Google Scholar]
- 8(a).Hsieh YL, Wang H, Elicone C, Mark J, Martin SA, Regnier F. Anal Chem. 1996;68:455–462. doi: 10.1021/ac950421c. [DOI] [PubMed] [Google Scholar]; (b) Riggs L, Sioma C, Regnier FE. J Chromatogr A. 2001;924:359–368. doi: 10.1016/s0021-9673(01)00900-1. [DOI] [PubMed] [Google Scholar]
- 9(a).Petro M, Svec F, Frechet JM. J Chromatogr A. 1996;752:59–66. doi: 10.1016/s0021-9673(96)00510-9. [DOI] [PubMed] [Google Scholar]; (b) Petro M, Svec F, Frechet JM. Biotechnol Bioeng. 1996;49:355–363. doi: 10.1002/(SICI)1097-0290(19960220)49:4<355::AID-BIT1>3.0.CO;2-O. [DOI] [PubMed] [Google Scholar]; (c) Petro M, Svec F, Gitsov I, Frechet JM. Anal Chem. 1996;68:315–321. doi: 10.1021/ac950726r. [DOI] [PubMed] [Google Scholar]
- 10(a).Jun SH, Chang MS, Kim BC, An HJ, Lopez-Ferrer D, Zhao R, Smith RD, Lee SW, Kim J. Anal Chem. 2010;82:7828–7834. doi: 10.1021/ac101633e. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Kim BC, Lopez-Ferrer D, Lee SM, Ahn HK, Nair S, Kim SH, Kim BS, Petritis K, Camp DG, Grate JW, Smith RD, Koo YM, Gu MB, Kim J. Proteomics. 2009;9:1893–1900. doi: 10.1002/pmic.200800591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kim J, Kim BC, Lopez-Ferrer D, Petritis K, Smith RD. Proteomics. 2010;10:687–699. doi: 10.1002/pmic.200900519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12(a).Zhang Z, Smith DL. Protein Science. 1993;2:522–531. doi: 10.1002/pro.5560020404. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Ehring H. Anal Biochem. 1999;267:252–259. doi: 10.1006/abio.1998.3000. [DOI] [PubMed] [Google Scholar]; (c) Wang L, Smith DL. Chapter 17. Curr Protoc Protein Sci. 2002 doi: 10.1002/0471140864.ps1706s28. Unit 17 16. [DOI] [PubMed] [Google Scholar]
- 13.Engen JR, Wales TE, Shi X. In: Encyclopedia of Analytical Chemistry. Meyers RA, editor. Wiley; 2011. pp. 2–17. [Google Scholar]
- 14.Wang L, Pan H, Smith DL. Mol Cell Proteomics. 2002;1:132–138. doi: 10.1074/mcp.m100009-mcp200. [DOI] [PubMed] [Google Scholar]
- 15(a).Cheftel JC. Effects of High Hydrostatic Pressure on Food Constituents: An Overveiw. John Libbey Eurotext, Ltd; London, UK: 1992. pp. 195–209. [Google Scholar]; (b) Munoz M, de Ancos B, Sanchez-Moreno C, Cano MP. J Food Prot. 2007;70:1587–1593. doi: 10.4315/0362-028x-70.7.1587. [DOI] [PubMed] [Google Scholar]; (c) Hernandez A, Cano MP. J Agric Food Chem. 1998;46:266–270. doi: 10.1021/jf970455g. [DOI] [PubMed] [Google Scholar]
- 16.Eisenmenger MJ, Reyes-De-Corcuera JI. Enzyme and Microbial Technology. 2009;45:331–347. [Google Scholar]
- 17.Dufour E, Herve G, Haertle T. Biopolymers. 1995;35:475–483. [Google Scholar]
- 18.Lopez-Ferrer D, Petritis K, Lourette NM, Clowers B, Hixson KK, Heibeck T, Prior DC, Pasa-Tolic L, Camp DG, 2nd, Belov ME, Smith RD. Anal Chem. 2008;80:8930–8936. doi: 10.1021/ac800927v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lopez-Ferrer D, Petritis K, Hixson KK, Heibeck TH, Moore RJ, Belov ME, Camp DG, 2nd, Smith RD. J Proteome Res. 2008;7:3276–3281. doi: 10.1021/pr7008077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lopez-Ferrer D, Petritis K, Robinson EW, Hixson KK, Tian Z, Lee JH, Lee SW, Tolic N, Weitz KK, Belov ME, Smith RD, Pasa-Tolic L. Mol Cell Proteomics. 2011;10 doi: 10.1074/mcp.M110.001479. M110 001479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jones LM, Zhang H, Vidavsky I, Gross ML. Anal Chem. 2010;82:1171–1174. doi: 10.1021/ac902477u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wales TE, Fadgen KE, Gerhardt GC, Engen JR. Anal Chem. 2008;80:6815–6820. doi: 10.1021/ac8008862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Silva JC, Gorenstein MV, Li GZ, Vissers JP, Geromanos SJ. Mol Cell Proteomics. 2006;5:144–156. doi: 10.1074/mcp.M500230-MCP200. [DOI] [PubMed] [Google Scholar]
- 24.Silva JC, Denny R, Dorschel C, Gorenstein MV, Li GZ, Richardson K, Wall D, Geromanos SJ. Molular Cell and Proteomics. 2006;5:589–607. doi: 10.1074/mcp.M500321-MCP200. [DOI] [PubMed] [Google Scholar]
- 25.Kavana D, Mana P. Intern. J. Mass Spectrom. 2011;302:53–58. [Google Scholar]
- 26.Wu Y, Kaveti S, Engen JR. Anal Chem. 2006;78:1719–1723. doi: 10.1021/ac0518497. [DOI] [PubMed] [Google Scholar]
- 27.Afeyan NB, Gordon NF, Mazsaroff I, Varady L, Fulton SP, Yang YB, Regnier FE. J Chromatogr. 1990;519:1–29. doi: 10.1016/0021-9673(90)85132-f. [DOI] [PubMed] [Google Scholar]
- 28.Phillips DJ, Capparella M, Neue UD, el Fallah Z. J Pharm Biomed Anal. 1997;15:1389–1395. doi: 10.1016/s0731-7085(96)02049-3. [DOI] [PubMed] [Google Scholar]
- 29.Swartz ME. Journal of Liquid Chromaotography and Related Technologies. 2005;28:11. [Google Scholar]
- 30.Wyndham KD, O’Gara JE, Walter TH, Glose KH, Lawrence NL, Alden BA, Izzo GS, Hudalla CJ, Iraneta PC. Anal Chem. 2003;75:6781–6788. doi: 10.1021/ac034767w. [DOI] [PubMed] [Google Scholar]
- 31.Mellors JS, Jorgenson JW. Anal Chem. 2004;76:5441–5450. doi: 10.1021/ac049643d. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.